Abstract
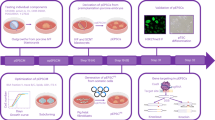
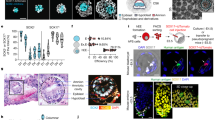
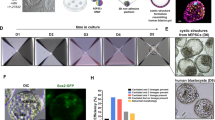
We previously reported the derivation of expanded potential stem cells (EPSCs) by modulating signaling pathways involved in preimplantation embryogenesis. These cells exhibit expanded developmental potential into embryonic and extraembryonic lineages, and we have shown that human EPSCs (hEPSCs) possess trophoblast differentiation potency for generating human trophoblast stem cells. Here we report protocols for deriving stable hEPSC lines directly from morula or early blastocyst stages of human preimplantation embryos (hEPSC-em) and by reprogramming human dermal fibroblasts (human induced EPSCs) using six exogenous factors, as an extension to our previous protocols on deriving porcine EPSCs from preimplantation embryos and by reprogramming somatic cells. These hEPSC lines proliferate robustly over long-term passaging and are amenable to both simple indels and precision genome editing. We provide guidance for characterizing these newly established hEPSCs, including cell-cycle analysis, pluripotency validation and karyotyping. The hEPSCs form teratomas with embryonic and extraembryonic cell lineages and readily differentiate into human trophoblast stem cells in vitro. At the molecular level, hEPSCs have unique features such as high expression of core histone genes and low H3K27me3 levels resembling eight-cell/morula stage embryos. These properties make hEPSCs a valuable tool not only for studying early human development but also for potential applications in regenerative medicine. The protocols presented in this manuscript can be readily performed by postgraduate students or postdoctoral fellows and completed within around 2 months.
Key points
-
This protocol describes detailed procedures for the establishment of human expanded potential stem cell lines from preimplantation embryos and by reprogramming somatic cells, including validation and genome editing.
-
Six-factor reprogramming of human dermal fibroblasts leads to the establishment of stable human induced expanded potential stem cell lines with 100% efficiency from picked colonies.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
Supporting data for this study can be found in our previous publications1,3 and are available from the corresponding authors upon request. All source data generated or analyzed during this study are included in this article. Source data are provided with this paper.
References
Gao, X. et al. Establishment of porcine and human expanded potential stem cells. Nat. Cell Biol. 21, 687–699 (2019).
Wilkinson, A. C. et al. Expanded potential stem cell media as a tool to study human developmental hematopoiesis in vitro. Exp. Hematol. 76, 1–12.e15 (2019).
Chen, A. C. H. et al. Expanded potential stem cells from human embryos have an open chromatin configuration with enhanced trophoblast differentiation ability. Adv. Sci. 10, e2204797 (2023).
Ruan, D. et al. Human early syncytiotrophoblasts are highly susceptible to SARS-CoV-2 infection. Cell Rep. Med. 3, 100849 (2022).
Xu, S., Wang, S., Tam, T., Liu, P. & Ruan, D. Derivation of trophoblast stem cells from human expanded potential stem cells. STAR Protoc. 4, 102354 (2023).
Ruan, D. et al. An optimized culture system for efficient derivation of porcine expanded potential stem cells from preimplantation embryos and by reprogramming somatic cells. Nat. Protoc. 19, 1710–1749 (2024).
Xia, W. et al. Resetting histone modifications during human parental-to-zygotic transition. Science 365, 353–360 (2019).
Kumar, B. et al. Polycomb repressive complex 2 shields naive human pluripotent cells from trophectoderm differentiation. Nat. Cell Biol. 24, 845–857 (2022).
Zijlmans, D. W. et al. Integrated multi-omics reveal polycomb repressive complex 2 restricts human trophoblast induction. Nat. Cell Biol. 24, 858–871 (2022).
Zhang, Q. et al. Adrenomedullin has a pivotal role in trophoblast differentiation: a promising nanotechnology-based therapeutic target for early-onset preeclampsia. Sci. Adv. 9, eadi4777 (2023).
Chen, Y. et al. SP6 controls human cytotrophoblast fate decisions and trophoblast stem cell establishment by targeting MSX2 regulatory elements. Dev. Cell 59, 1506–1522.e11 (2024).
Chao, Y. et al. Organoid-based single-cell spatiotemporal gene expression landscape of human embryonic development and hematopoiesis. Signal Transduct. Target Ther. 8, 230 (2023).
Cao, H. et al. PD-L1 regulates inflammatory programs of macrophages from human pluripotent stem cells. Life Sci. Alliance https://doi.org/10.26508/lsa.202302461 (2024).
Liu, X. et al. Recapitulating primary immunodeficiencies with expanded potential stem cells: proof of concept with STAT1 gain of function. J. Allergy Clin. Immunol. 153, 1125–1139 (2024).
Wei, X. et al. A human organoid drug screen identifies alpha2-adrenergic receptor signaling as a therapeutic target for cartilage regeneration. Cell Stem Cell 31, 1813–1830.e8 (2024).
Yang, Y. et al. Derivation of pluripotent stem cells with in vivo embryonic and extraembryonic potency. Cell 169, 243–257.e225 (2017).
Liu, B. et al. Chemically defined and xeno-free culture condition for human extended pluripotent stem cells. Nat. Commun. 12, 3017 (2021).
Xu, J. et al. Generation of pig induced pluripotent stem cells using an extended pluripotent stem cell culture system. Stem Cell Res. Ther. 10, 193 (2019).
Yu, Z. et al. Porcine pluripotent stem cells established from LCDM medium with characteristics differ from human and mouse extended pluripotent stem cells. Stem Cell 40, 751–762 (2022).
Zheng, R. et al. Derivation of feeder-free human extended pluripotent stem cells. Stem Cell Rep. 16, 2410–2414 (2021).
Guo, G. et al. Human naive epiblast cells possess unrestricted lineage potential. Cell Stem Cell 28, 1040–1056 e1046 (2021).
Yeung, W. S. et al. Frozen–thawed embryo transfer cycles. Hong. Kong Med. J. 15, 420–426 (2009).
McMahon, A. P. & Bradley, A. The Wnt-1 (int-1) proto-oncogene is required for development of a large region of the mouse brain. Cell 62, 1073–1085 (1990).
Wang, W. et al. Rapid and efficient reprogramming of somatic cells to induced pluripotent stem cells by retinoic acid receptor gamma and liver receptor homolog 1. Proc. Natl Acad. Sci. USA 108, 18283–18288 (2011).
Balaban, B. & Gardner, D. K. in Human Gametes and Preimplantation Embryos: Assessment and Diagnosis (eds Gardner, D. K., Sakkas, D., Seli, E. & Wells, D.) 31–43 (Springer, 2013).
Zeng, R. et al. Generation of four H1 hESC sublines carrying a hemizygous knock-out/mutant MECP2. Stem Cell Res. 40, 101533 (2019).
Yan, L. et al. Single-cell RNA-Seq profiling of human preimplantation embryos and embryonic stem cells. Nat. Struct. Mol. Biol. 20, 1131–1139 (2013).
Acknowledgements
This project is supported by the National Key Research and Development Program of China (grant nos. 2022YFA1105401, 2022YFA1105400, 2018YFA0902702 and 2022YFC2702503); Health@InnoHK, Innovation Technology Commission, HKSAR; Hong Kong Research Council (grant nos. GRF17116324, GRF17127219 and GRF17126421, 17112422, 17104523, 17109924. Germany/Hong Kong travel grant G-HKU704/21); ITF (MHKITFS) (MHP/079/20); National Natural Science Foundation of China/RGC Collaborative Research Scheme (CRS_HKU703); National Natural Science Foundation of China (grant nos. 32100639, 81570202, 32100639 and 32070869); The University of Hong Kong-Shenzhen Hospital Fund for Shenzhen Key Medical Discipline (grant no. SZXK2020089); Shenzhen Science and Technology Program (grant no. KQTD20190929172749226); Sanming Project of Medicine in Shenzen Municipality, SZSM202211014; High Level-Hospital Program, Health Commission of Guangdong Province, China (grant no. HKUSZH201902025). We thank R. Jauch for supplying the human dermal fibroblasts (HDF) cells. Instrument or analysis software was supported by Imaging and Flow Cytometry Core of HKU Centre for PanorOmic Sciences (CPOS).
Author information
Authors and Affiliations
Contributions
P.L., W.S.B.Y. and Y.L.L. conceived the project. A.C.H.C. and D.G.R. drafted the protocol. A.C.H.C., W.H., H.Z.R. and S.W.F. contributed to the generation of and performed the establishment of hEPSCs from preimplantation embryos. D.G.R. and X.Y.L. performed the establishment of hiEPSCs from HDFs. Y.L.L. and A.C.H.C. performed the generation of human CDX2 reporter cell lines. A.C.H.C., D.G.R. and S.X. contributed to the generation of hTSCs. A.C.H.C., W.H., H.Z.R., S.W.F., G.J.L. and S.X. contributed to the teratoma formation, karyotyping and the generation of human ACE2-knockout cell lines. D.G.R. and A.C.H.C. performed RT–qPCR analysis of gene expression levels, immunofluorescence staining and FACS assays. P.L., W.S.B.Y., Y.L.L. and T.T.K.K.T. contributed to the writing and the critical revision of the manuscript. A.C.H.C. performed scRNA-seq analysis. T.T.K.K.T., X.F.G. and W.S.B.Y. provided intellectual input. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
A patent application related to the data presented here is under preparation on behalf of the Center for Translational Stem Cell Biology. The other authors declare no competing interests.
Peer review
Peer review information
Nature Protocols thanks Norah Fogarty, Frederic Lluis, Tijs Vanhessche, and the other, anonymous, reviewer for their contribution to the peer review of this work.
Additional information
Key references
Gao, X. et al. Nat. Cell Biol. 21, 687–699 (2019): https://doi.org/10.1038/s41556-019-0333-2
Chen, A. C. H. et al. Adv. Sci. 10, e2204797 (2023): https://doi.org/10.1002/advs.202204797
Ruan, D. et al. Cell Rep. Med. 3, 100849 (2022): https://doi.org/10.1016/j.xcrm.2022.100849
Xu, S. et al. STAR Protoc. 4, 102354 (2023): https://doi.org/10.1016/j.xpro.2023.102354
Chen, Y. et al. Dev. Cell 59, 1506–1522.e11 (2024): https://doi.org/10.1016/j.devcel.2024.03.025
This protocol is an extension to: Nat. Protoc. 19, 1710–1749 (2024): https://doi.org/10.1038/s41596-024-00958-4
Supplementary information
Supplementary Information
Supplementary Table 1.
Source data
Source Data Fig. 3
Unprocessed gel—Fig. 3j,k.
Source Data Fig. 5
Source Data for Fig. 5g.
Source Data Fig. 8
Unprocessed gel—Fig. 8b,f.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ruan, D., Chen, A.C.H., Tam, T.T.K.K. et al. Establishment of human expanded potential stem cell lines via preimplantation embryo cultivation and somatic cell reprogramming. Nat Protoc 20, 2698–2734 (2025). https://doi.org/10.1038/s41596-025-01168-2
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41596-025-01168-2