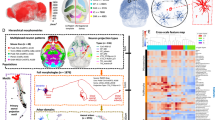
Abstract
Neuron morphology and sub-neuronal patterns offer vital insights into cell typing and the structural organization of brain networks. The community-collaborative BRAIN Initiative Cell Census Network (BICCN) project has yielded a vast amount of whole-brain imaging data. However, reconstructing multi-scale neuron morphometry at a whole-brain scale requires not only the integration of diverse hardware devices, tools, and algorithms but also a dedicated production workflow. To address these challenges, we developed a cloud-based, collaborative platform capable of handling peta-scale imaging data. Using this platform, we generated the largest multi-scale morphometry dataset from hundreds of sparsely labeled mouse brains. The morphometry dataset comprises 182,497 annotated cell bodies, 15,441 locally traced morphologies, and 1,876 fully reconstructed morphologies. We also identified sub-neuronal arborizations for both axons and dendrites, along with the primary axonal tracts connecting them. In addition, we identified 2.63 million putative boutons. All morphometric data were registered to the Allen Common Coordinate Framework (CCF) atlas. The morphometry dataset has proven to be an invaluable resource for whole-brain cross-scale morphological studies in mouse.
Similar content being viewed by others
Background & Summary
Digital reconstruction of complete neuron morphology1 is pivotal in identifying cell types2,3, elucidating the relation between morphology and function4, and understanding the organization of the brain network5,6. Despite recent advances in labeling7,8, whole-brain imaging9,10, and annotation11,12,13,14 have greatly facilitated the reconstruction of complete neuron morphology, the generation of large-scale morphological datasets in mammalian brain remains to be a formidable challenge15.
On one hand, reconstructing mammalian neurons with long-range axonal projection necessitates the processing of tera- to peta-scale voxels on a submicron-imaged brain16,17. On the other hand, the current sparse labeling18,19 is confined to hundreds of neurons for each brain, thus producing a large-scale morphological dataset requires hundreds of brains. Dealing with these challenges entails the handling of peta-scale storage and computing resources, in addition to efficient visualization and processing of heterogeneous morphometry data4,20,21.
Numerous automated algorithms22,23,24,25,26 have been developed to trace neuron morphologies for different species since the 1970s. These algorithms have recently been comprehensively benchmarked within the BigNeuron project27,28. Despite these advancements, there remains a dearth of algorithms capable of correctly tracing the tangled neurites of neurons and generating a complete reconstruction from whole-brain data4,21,29. One feasible approach for data collection would involve a phased divide-and-conquer strategy. One of such approach would be to combine automated algorithms with manual proofreading, supported by high-throughput tools, to obtain high-quality data in a timely manner.
Several datasets4,21,29 reconstructing long-projection neurons in mammalian have been published in the last five years. These datasets, consisting of thousands of neurons generated using semi-automated reconstruction pipelines, are valuable but insufficient for comprehensive characterization of the whole-brain connectome. Given the vast morphological diversity of neurons, there is a critical need for multi-scale morphometry, encompassing sub-neuronal arborizations, structural motifs, and synaptic sites30,31.
The community-collaborative BRAIN Initiative Cell Census Network (BICCN) project5,32 has released thousands of mouse brain images across multiple modalities. As a part of the program’s synergetic efforts, we utilized nearly two hundreds light microscopy-imaged mouse brains and obtained a bunch of multi-scale morphometry from them. To do that, we developed a cloud-based platform, Collaborative Augmented Reconstruction (CAR)33, which features cloud-oriented, multi-clients, virtual reality powered, and real-time collaboration. The platform provides real-time peta-scale data processing capacity and multiple clients including virtual reality headsets, workstations, mobiles, and game consoles. By leveraging this platform and collecting a large-scale whole-brain image database, we have demonstrated that neuron morphometry can be efficiently produced and analyzed across multiple scales31,34,35,36,37.
In this study, we utilized a mobile application named CAR-Mobile and a desktop application called CAR-WS within CAR to semi-automatically identify 182,497 cell bodies (SEU-S182K) (Figs. 1a and 2a). Based on SEU-S182K, we automatically reconstructed 15,441 local dendritic morphologies by combining results from two widely-used auto-tracing algorithms25,26. This process followed a collaborative annotation protocol involving multiple domain experts20, resulting in 1,876 full morphologies of neurons (SEU-A1876). Additionally, we employed a Gaussian model to identify presynaptic structures, yielding a substantial dataset of 2.63 million putative boutons (Fig. 1a).
Schematic overview of the multi-morphometry production workflow. (a) An example of producing multi-scale morphometric data from imaging data. Initially, annotators employed CAR-Mobile to mark the locations of cell bodies identified through automatic detection. Subsequently, the auto-tracing algorithms APP2 and neuTube were utilized to reconstruct neuronal morphologies from image volumes centered on these cell bodies. Each image volume had a fixed size of 512 voxels in the X and Y dimensions and 256 voxels in the Z dimension, corresponding to approximately 256 μm in each spatial direction. The subset comprising 1,876 complete morphologies (SEU-A1876) was finalized using the CAR platform, following a collaborative reconstruction protocol. Finally, putative axonal boutons were identified based on these full morphologies and the corresponding images. (b) and (d) Morphometric data of the fMOST brain (HUST ID: 18454) mapped on the original raw brain (c) and on the CCFv3 atlas (d). (c) Examples of sub-neuronal structures, including a dendritic arbor (left), axonal arbors (middle), and a primary axonal tract (right) that connects those arborizations.
To facilitate comparative analysis, all the morphometric data were registered to the Allen Common Coordinate Framework version 3 (CCFv3) atlas38 (Fig. 1b,d). The identified somas span major brain structures, including the Isocortex, CTXsp, striatum (STR), hippocampal formation (HPF), and cerebellum (CB) (Fig. 2a). The traced local morphometry was predominantly located preferentially in Isocortex and STR (Fig. 2b), while the traced full morphometry was mainly found in the Isocortex and thalamus (TH), with others in STR and HPF (Fig. 2c).
We then generated various sub-neuronal structures, including arbors and motifs. We initially segregated a neuron into dendritic and axonal arbors (Fig. 1c). To further elucidate and simplify axonal organization, we identified primary axonal tracts connecting various arborizations, facilitating the visualization of single-cell connectivity on a panoramic scale. Subsequently, we utilized a graph-based classification tool, AutoArbor4,31, to automatically partition the neuronal axons into densely-packed arbors, identifying a total of 3,776 distinct axonal arbors. These arbors offer valuable insights into the arborization patterns of individual neurons.
The primary difference between our multi-morphometry dataset and existing morphological datasets, such as those published by Janelia21 and ION29,39,40, lie in two aspects. First, our dataset includes neuronal morphometry covering six different scales that are absent from other datasets. Second, the primary brain regions covered by our data differ significantly. While local morphometry in our dataset spans a broader range of brain regions, the full morphometry data are primarily derived from the cortex, striatum, and thalamus.
Methods
The following subsections describe mouse brain image collection, multi-morphometry data identification, as well as the utilized platform. Although the majority of the methods employed have been previously described in our studies31, we provide a more detailed description here.
Whole-brain imaging dataset
In this study, we collected a whole-brain image dataset31 comprising 181 images of mouse brains at submicron resolution from the BICCN community. The image data can be downloaded from Brain Image Library41 (BIL, www.brainimagelibrary.org). A whole-brain image can occupy approximately 20 terabytes of storage, comprising over 10,000 coronal planes with a resolution of 0.2–0.35 µm. To manage the tera-voxel scale imaging data, we employed TeraFly11, a powerful multidimensional visualization and annotation tool for big-data. As part of this process, some of the downloaded images may need to be converted into a hierarchical format using the tool teraconverter provided by the Vaa3D42,43.
CAR platform
Considering the complex nature of neuron morphology and extensive interconnections among neuronal compartments, an effective annotation platform should enable user-friendly data interaction while catering to the demands of collaborative processing at scale. We established a cloud-based collaborative augmented reconstruction platform33 (CAR), allowing for collaborative annotation of neuron morphology using a variety of client devices, such as desktop workstations (CAR-WS), VR headsets (CAR-VR), mobile app (CAR-Mobile) and a crowd-sourcing game console. By incorporating several AI-based automation tools, CAR takes advantage in addressing challenges associated with large-scale neuron reconstruction tasks. In this study, CAR was primarily used for manual annotations and semi-automatic data generation, such as the collaborative production of soma locations and full morphometry. Yet, some customized automatic algorithms such as neuron tracing with neuTube, automatically identification of putative axonal boutons, graph-based detection of axonal arbors, and axonal tract extraction, are not integrated in the current version.
Soma localization
Soma localization serves as the initial step for subsequent morphometric production. We employed two strategies to generate those kinds of three-dimensional coordinates from the whole-brain imaging database.
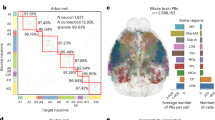
The first strategy entailed a semi-automatic annotation process using our CAR platform, which comprised two primary steps (Fig. 3). We initially filtered out imaging blocks from the whole-brain image that exhibited low intensity values. Specifically, we defined potential soma-containing image blocks with a minimum intensity threshold of 250 for unsigned 16-bit images. The remaining blocks were then normalized using Z-score normalization and converted to the unsigned 8-bit range. Subsequently, these blocks were binarized using the 99th percentile as a threshold, and the resulting images were transformed utilizing the gray-scale distance transform (GSDT) algorithm. Candidates were identified as voxels with intensities ranging from 5 to 30 in the transformed image. These candidate voxels underwent additional processing using a Non-Maximal Suppression (NMS)-like approach to eliminate redundancy. In the second step, we cropped image blocks at the second-highest resolution, centering them on the potential soma locations with dimensions of 128 pixels in each direction. These cropped image blocks were then uploaded to the CAR server, enabling remote users to access and annotate them through CAR-Mobile (Fig. 3a,b).
Soma generation workflow. (a) The diagram illustrates the semi-automatic soma detection process. The whole-brain image, formatted in TeraFly, was divided into image volumes of approximately 256 voxels in each dimension. In the initial step, a multi-threaded process was employed to filter out image blocks containing potential somas and estimate their locations. Subsequently, a Non-Maximal Suppression (NMS)-like approach was applied to eliminate duplicate soma labels for the same cells. Following this, manual annotation was performed using CAR-Mobile to proofread and refine the annotations. Finally, Mean-Shift was utilized to reduce deviation in the annotated results. (b) Examples of the images after several key processing steps. (c) A showcase of soma morphometry produced using Vaa3D-TeraFly is presented. The left panel displays all somas reconstructed from a specific brain image (HUST ID: 18454), while the right panel provides a zoom-in view for a more detailed examination. The somas were rendered using red markers.
The second strategy involved a fully manual approach using Vaa3D-TeraFly platform. Users utilized the instant zoom-in/out function to navigate to the soma-containing imaging blocks and pinpoint markers at the soma locations (Fig. 3c).
Following the generation of raw soma data, a refinement step was applied using the mean-shift algorithm to ensure that the soma coordinates were accurately centered on the cell body. Before implementing this algorithm, voxels with intensities below the sum of the mean and standard deviation of the soma block were removed via zero-clipping. For the mean-shift optimization of the soma location, a window radius of 15 voxels was used. In cases where two somas were within 15 voxels of each other and the intensity at their center point was lower than the average intensity of the two somas, duplicate entries were eliminated. As a result, we identified a total of 182,497 somas from whole-brain images, including 136,833 somas produced using the CAR platform and 45,664 somas identified using the Vaa3D-TeraFly platform.
Local morphometry production
Local morphometry of an individual neuron refers to the morphology within a confined range centered around the cell body. In our study, we utilized the second-highest resolution image of TeraFly-formatted whole-brain data to expedite the tracing process, defining this range as 512 voxels in the X and Y directions and 256 voxels in the Z direction. This range corresponds approximately to a 125 μm extension in both the X and Y directions from the cell body and a 250 μm extension in the Z direction. In certain brain images, multiple cells may be labeled within the local range, resulting in intertwined morphologies that complicate the automatic reconstruction of individual neuronal structures. To mitigate interference from neighboring neurons during local reconstruction, we excluded instances where more than five cells were present within a 128 μm range around the soma.
We combined two automated neuron tracing algorithms, namely APP225 and neuTube26, to trace the morphologies of individual neurons from local image blocks. APP2 and neuTube are automated tracing methods that employ different reconstruction strategies22. APP2 reconstructs a neuron by considering information from the entire image, while neuTube initiates from a seed point and gradually extends along the likely direction of fibers. Therefore, APP2 and neuTube have complementary roles in many scenarios such as weak signals or gaps. However, it may produce erroneous connections in areas with dense signals. On the other hand, the neuTube method effectively addresses the local errors in connection that may arise in APP2. The algorithms were executed using default parameters, with the exception of the threshold applied to segment the background in APP2. We adjusted this threshold to a value of μ + 0.5σ, where μ represents the mean and σ represents the standard deviation of the image block. The reconstructions obtained from APP2 underwent a segment-pruning pipeline, detailed below, to rectify potential loops, errant branches, and intersections with other neurons. Each pruning stage served as an independent filter that processed the raw neuronal tree, resulting in a final structure representing the intersection of all filtered reconstructions. Subsequently, the reconstructions from neuTube were employed to refine the pruned APP2 reconstructions by removing nodes that lacked corresponding nodes within a 5-voxel range in neuTube (Fig. 4).
A comprehensive description of the segment-pruning pipeline is outlined as follows:
-
1.
Abnormal Branch Pruning: Branches that present an angle of less than 80 degrees to their parent or exhibit a radius increase greater than 1.5 times that of the parent branch were eliminated.
-
2.
Crossover Branch Pruning: This step involves purging branches from presumptive crossover structures. We first identified all potential crossover structures consisting of densely packed branching nodes with more than two child branches. Next, we examined all connections between the current branch and its child branches for each identified crossover structure, removing branches with small angles (less than 80 degrees). Finally, we assessed branches with moderate turning angles (80–100 degrees) to determine if another branch formed a sufficiently large angle with them (greater than 150 degrees); if such a branch was found, the moderately turning branch was removed.
-
3.
Soma Pruning: This step involves removing branches originating from potential somas. We identified a potential soma as a candidate node with a radius larger than 1 voxel. Neurites that were too close to the traced soma (less than 50 voxels) were excluded. For each potential soma, we estimated the integral of deviation angles along the fiber path connecting the potential soma and the traced soma to determine the optimal cutting position. The deviation angle is defined as the angle between a local branch and the radial line connecting the soma to the nearest end of the same branch, similar to the G-Cut method. We calculated the integral of deviation angles on both sides—from the current branch to the traced soma and from the current branch to the potential soma—while considering branch length. Branches with a lower integral of deviation angles leading to the potential soma were subsequently removed.
-
4.
Winding Pruning: Any branch forming a tortuous path to the soma was eliminated if the ratio of path distance to Euclidean distance exceeded 3.
-
5.
Final Deletion: Reconstructions containing fewer than 20 nodes were excluded from the analysis.
Full morphometry production
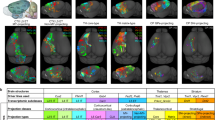
We employed a multi-level annotation protocol4,20, executed within the CAR platform, to annotate the morphologies of neurons based on traced morphology (Fig. 5). Initially, the annotation was divided into two levels (L1 and L2) based on the analysis purposes of morphological data. Level 1 (L1) consists of complete dendritic arbors and primary axonal skeletons without dedicated arborizations. Level 2 (L2) reconstructs the complete axonal arborizations capable of quantifying the neuron projections (Fig. 5b).
Full morphology annotation workflow. (a) Collaborative multi-level protocol for single neuron annotation. The manual annotation process consists of two levels: L1 and L2, each responsible for different aspects of morphological reconstruction. Initially, annotators collaboratively annotate a neuron within the CAR platform, starting from an automatic reconstruction (indicated by blue dots). Once the data meets the requirements of L1, it is submitted as L1A. A senior annotator, also known as an inspector then verifies the accuracy, marking missing segments with green lines and over-traced segments with black lines. This produces L1B data, which is collaboratively reviewed by the annotators to resolve any issues before submission as L1C. The inspector performs a final review, producing the finalized L1 data. A similar process is followed for L2. Finally, a post-processing pipeline is applied to standardize and enhance the quality of the annotations before their final release. During phases A (L1A and L2A) and C (L1C and L2C) of each level (cyan arrows), at least two annotators collaborate on reconstructing the neuron to meet the criteria of each level. (b) Skeletons generated in level 1 (L1) were labeled in black, while those in level 2 (L2) were labeled in red. (c) The collaborative tracing process in phase A. Traced skeletons from different annotators are differently colored (e.g., yellow and cyan). (d) and (e) Zoomed-in views of identified missing segments.
At each level, we established several rounds of generation-validation (GV) steps. In round 1, annotators reconstructed data to meet the requirements of level 1 and submitted them for quality validation (Fig. 5a,c). Subsequently, inspectors assessed the accuracy and completeness of the submitted data, fixed correct structures, and pushed to the next round (Fig. 5d,e). This iterative process continued until it met the quality control criteria. Typically, we employ two rounds of GV steps with the collaborative involvement of at least two annotators and one inspector. Therefore, the complete neuron morphology data in our research undergoes participation from a minimum of six individuals (Fig. 5a).
The immersive data interaction tools provided by the CAR platform, along with its collaborative annotation mode, help improve the accuracy and completeness of neuron skeleton reconstruction. Additionally, the embedded AI tools assist annotators by identifying potential quality issues in the reconstructed data, such as errors at branching points and breaks in tree structures. Further quality control procedures were performed to ensure compliance with community formatting standards and to facilitate subsequent analyses. These procedures included single-tree structure checking, loop and trifurcation detection, short terminal branch pruning, node resampling, and skeleton refinement44,45. With these extensive quality control, the reconstruction accuracy is always over 90%33. The relevant processing tools are described in the Usage Notes section.
Putative axonal bouton identification
In previous studies, axonal boutons have been characterized as high-intensity swellings along axonal shafts in light microscopy data20,46,47. In our study, we approached bouton detection as a peak detection process within one-dimensional signal processing, utilizing the radius and intensity profiles of axonal branches to identify potential bouton locations (Fig. 6a). We began by estimating the intensity and radius profiles along the shafts of the reconstructed axonal arbors. To address discrepancies between the axial resolution of whole-brain images (typically 1 µm) and the planar resolution (0.2–0.35 µm), we applied an image upsampling method to achieve isotropic resolution in all three dimensions during radius calculations. Additionally, we employed an image enhancement pipeline, as described in our previous work48, to improve the accuracy of radius extraction.
Axonal bouton identification workflow. (a) The workflow started with extracting the axonal arbor from the full morphology of a neuron. Subsequently, potential synaptic bouton sites were identified independently for each branch. (b) The profiled axonal arbor and its branches were rendered using Vaa3D, allowing for a clear distinction between branches containing boutons and those without (b1 and b2). Additionally, each branch of the axonal arbor was assigned a different color to facilitate a more intuitive understanding of the axonal branching structure. (c) Two types of axonal branches were presented in both raw data and enhanced image data space. The first type, referred to as null bouton branches, had a consistent diameter throughout. The second type, referred to as bouton branches, exhibited variations in brightness and diameter, forming a waved pattern. Additionally, the feature distributions of intensity and radius were compared between these two types of branches to further demonstrate those patterns.
The profiled axonal arbor exhibited inconsistent radius distributions between branches with boutons and those without (Fig. 6b), in which branches containing boutons showing significant fluctuations in radius. Given that bouton locations are typically found in areas where axonal arbors exhibit a higher branching density, we assumed that the lengths of neuronal branches follow a normal distribution and excluded branches exceeding the mean plus three standard deviations. Furthermore, branches longer than 20 μm were segmented into smaller segments not exceeding this length.
By analyzing the radius and intensity distributions, we identified peak points across the two profiles. Candidate boutons were defined by two criteria: they must exhibit a radius at least 1.5 times larger than adjacent points and exceed 1 μm in size, with voxel values greater than 120 in 8-bit images. Finally, we eliminated bouton candidates that were topologically connected and within 5 voxels of each other, retaining only the bouton with the largest radius and highest brightness (Fig. 6c).
Arbor detection
Here a neuronal arbor is defined as a densely packed subtree structure, rather than the traditional division of dendritic or axonal trees. For dendrites, we retained the intact apical and basal dendrites as dendritic arbor structures. Meanwhile, we used spectral clustering to subdivide axons into arbors. This was realized as follows. We considered each neuronal reconstruction as an undirected graph, where vertices represent nodes in the original tree and weights of edges between pairs of vertices are represented by two key metrics, connectivity and distance. This weight was represented as a negative exponent of inter-node distances, mainly determined by connectivity. Our implementation ensured that neurons were segmented into tightly packed arbors while preserving the original connectivity relationships. To better compare neurons of the same brain regions, we calculated the number of dominant arbors in each neuron after the clustering process using the major vote method.
Primary axonal tract motif detection
The furthest-reaching axonal arbor of each neuron provides insights into their projecting targets, while the collective assembly of neurons belonging to the same types or classes delineates their projection patterns. To simplify the analysis of distal axonal arborization, we focused on extracting the longest axonal tract while eliminating all terminal short branches, which we refer to as the primary axonal tract. More specifically, our approach involved the initial identification of the terminal tip with the largest path distance from the soma, followed by an iterative process of eliminating short branches with a distal-to-local protocol. A branch was considered short if its length was less than that of the second-longest axonal branch.
Image and morphometry registration
Brain image registration was conducted using the cross-modal registration tool mBrainAligner49. All images were registered to the CCFv3 template (25 μm version). The registration followed a similar pipeline stated in the mBrainAligner instructions on GitHub, as described below:
-
1.
Down-sample the image (registration channel) to approximately isotropic 25 μm resolution with skimage.transform.resize function (scikit-image version 0.17.2) using linear interpolation method.
-
2.
Perform linear min-max intensity normalization using the 1st and 99th percentiles as minimum and maximum values, followed by conversion of the image from 16-bit unsigned integer (uint16) to 8-bit unsigned integer.
-
3.
Manually remove superfluous tissues slice by slice using ITK-SNAP (version 3.8.0) if necessary.
-
4.
Manually remove artifacts and stripe noises.
-
5.
Coarse alignment to CCFv3 atlas using the automatic global registration module. For those brain images with poor alignment accuracy, an alternative was to manually label 10 to 14 corresponding landmarks on both subject and target images as input to the affine transformation.
-
6.
Apply the automatic local registration to the output of global alignment to achieve a more accurate mapping through a nonlinear transformation.
The forward deformation field computed during the registration process was applied to generate anatomical segmentation of down-sampled images based on the CCFv3 atlas. All multi-morphometry, including full morphologies, local morphologies, arbors, and putative axonal boutons were inversely transformed into CCFv3 space using the inverse deformation matrices.
Data Records
The reconstructed morphometries are summarized in Table 1. Soma morphometry was stored in an Excel spreadsheet, while other morphometric data were archived in SWC format. The SWC format50 is widely used to represent neuron morphology through a sequence of nodes that model the position, connectivity, radius, and type of neurites. This format comprises seven attributes, separated by spaces, which are listed from left to right as follows: ‘index’, ‘type’, ‘x’, ‘y’, ‘z’, ‘radius’, and ‘parent’. The ‘index’ and ‘parent’ attributes define the logical relationships between nodes, while the ‘x’, ‘y’, ‘z’, and ‘radius’ attributes document the coordinates and signal radius of each node. The ‘type’ attribute designates the node type, with values assigned as follows: 1 for soma, 2 for axon, 3 for dendrite, 4 for apical dendrite, and 5 for axonal bouton sites. Notable, the ‘type’ attribute in the SWC data didn’t have implications for local morphometry data; however, in the context of axonal arbor data, we employed the ‘type’ attribute to differentiate among various arbors.
To accommodate various applications, we presented two versions of morphometry data: one included the coordinates generated from the raw brain (labeled as “Raw brain” in Table 1), and the other included the coordinates mapped to the Allen Common Coordinate Framework version 3 (labeled as “CCFv3” in Table 1). For axonal arbor, axonal tract and dendritic arbor, only coordinates in CCFv3 were provided.
The dataset is available for downloading on Zenodo51 (https://doi.org/10.5281/zenodo.13944322). The file structure in Zenodo is illustrated in Fig. 7. For soma morphometry, we provided meta information regarding the brain region where the neuron was located, the reconstruction platform employed, and the availability of corresponding local morphometry. Users can obtain the matching local morphometry data according to the soma ID. For the full morphometry data, we summarized the morphology name, the whole brain ID, the soma coordinates, the brain region and cortical location of the soma, and the projection type in the “Full_morphometry.xlsx” Excel spreadsheet.
Organization of the shared morphometries on Zenodo. The shared data consist of two Excel spreadsheets and nine folders, with the folders uploaded to Zenodo in ZIP format. All data are categorized into two major groups: full morphometry and local morphometry. Metadata for the full morphometry data are recorded in the “Full_morphometry.xlsx” file, while metadata for the local morphometry data are stored in “Soma_morphometry.xlsx.”. The full morphometry data include four types of morphometry: full morphology, axonal bouton, axonal arbor, and axonal tract. For each type, the “RAW” flag indicates that the data are of coordinate in the raw brain, while the “CCFv3” flag means that the coordinates are aligned with the CCFv3. Regarding file naming conventions, the soma ID for local morphometry data is documented in “Soma_morphometry.xlsx,” while the “Name” for full morphometry data is recorded in “Full_morphometry.xlsx”.
Technical Validation
While conducting neuronal diversity and stereotypy analysis using the multi-scale morphometry dataset31, we performed a comprehensive assessment of the dataset’s quality. These included comparing local morphometric features with fully reconstructed local morphometry within image blocks of the same size, and evaluating the robustness of neuron morphometry registration into the CCFv3 atlas. In this study, we carried out quantitative comparisons to further validate our dataset.
The quality of the digitalized neuronal morphometry retains the quality of whole-brain images acquired from the fMOST imaging system9. We have employed TeraFly11 to organize and merge the original 2-dimensional brain slice data into a 3-dimensional format. During this process, the values of the image data remain unaltered, thereby preserving the original quality of the images.
Soma locations serve as the starting point for neuron identification and tracing of neuron morphology, with quality ensured by a semi-automatic production pipeline involving at least two annotators. To further validate our dataset, we conducted quantitative comparisons. We used the Janelia21 dataset and SEU-A1876 to demonstrate the reliability of our data. To avoid possible inaccuracy raised by neurons from different brain regions, we compared only neurons from somatomotor areas (MO) in both datasets. We observed similar distributions in major morphological characteristics, such as total length, maximum Euclidean distance, number of bifurcations, and average remote bifurcation angle (Fig. 8).
Comparison among two datasets (SEU-A1876, and Janelia) using the distribution of four key morphological features of somatomotor areas (MO) neurons as an example: total length, maximum Euclidean distance, number of bifurcations, and average remote bifurcation angle. The analysis included 127 neurons from the SEU-A1876 dataset and 324 neurons from the Janelia dataset.
The efficacy of our bouton data production method has been both published and verified. Our method demonstrated 95% precision and 89% recall when compared with manually annotated bouton data20. Additionally, previous studies have shown that neurons of the same type tend to exhibit similar bouton distribution patterns31,34,36. The axonal tract and axonal arbor data are derived from the complete morphological data and can be adjusted using various parameters of the production method, resulting in different versions of data extraction.
Usage Notes
As part of the BICCN initiative, we are dedicated to generating neuronal data at the whole-brain level. With the focus on collecting and analyzing such data, BICCN has already made significant contributions by publishing thousands of invaluable whole-brain datasets5,32,41. These datasets are expected to benefit researchers across multiple domains of neuroscience. As members within BICCN, we firmly adhere to the FAIR principle52 in creating the resources. Additionally, we emphasize the importance of enabling accessibility to intermediate data, as it not only provides insights into the data but also facilitates a more efficient production process.
The produced dataset serves multiple purposes. The annotated somas (SEU-S182K) provide a solid starting point for anatomical delineation (e.g., cortical layer identification) and morphological analyses. The detected neurites and derived region-region correlations may aid in uncovering the intrinsic modularity of the mouse brain. The large-scale local morphologies offer insights into the spatial morphological divergence both across the whole mouse brain and within brain regions. The single-neuron full morphometry data for long-range projection neurons in mammalian is invaluable in cell typing, projection characterization, as well as sub-cellular analyses such as synaptic site identification. Furthermore, the qualified full morphologies will benefit the development of reconstruction algorithms and provide gold standards for training and evaluation of these algorithms.
The putative boutons can be utilized in characterizing wiring rules, calculation of functional connectivity strength, and simulation of neurons. Similarly, both axonal arbor data and axonal tract data simplify the highly arborized full morphology at the sub-cellular level, facilitating visualization and quantification of neuron projections. Moreover, mapping morphometry to the Common Coordinate Framework (CCF) enables the comparison of distribution differences and connections among various brains and brain regions.
In addition to directly utilizing our multi-morphometry dataset to investigate the diversity and stereotypy of neuronal morphologies, our dataset, combined with the corresponding imaging dataset, can serves as “gold standard data” for neuron image recognition tasks. These tasks include segmentation of neuron skeletons, bouton detection, and testing automatic tracing methods. The brain images used in our study are well-hosted on the Brain Image Library (BIL) for community access. Furthermore, we have documented the name of each neuronal morphology’s associated brain image and its corresponding download link in the work of characterizing neuron morphology at multiple scales31.
We have developed a repertoire of tools for processing and visualizing large-scale images (TeraFly, TeraVR, and CAR) as the Vaa3D plugins or derivatives, as well as reconstructing, processing, and analyzing neuron morphology. Users can develop their own tools on top of these tools to better suit their needs. We have also compiled a list of additional plugins that assist in processing and analyzing morphology data.
-
Image thresholding: “Simple_Adaptive_Thresholding”.
-
Automatic neuron tracing: GD53, APP24, APP225 and neuTube26.
-
Neuron morphology feature extraction: “global_neuron_feature”.
-
Neuron morphology processing: “resample_swc”, “sort_neuron_swc”, “neuron_radius”, “inter_node_pruning” and “refine_swc”44.
-
Neuron morphology exploration: NeuroXiv54.
Code availability
Source codes to produce and process the dataset are publicly accessible (Table 2).
References
Parekh, R. & Ascoli, G. A. Neuronal Morphology goes Digital: A Research Hub for Cellular and System Neuroscience. Neuron 77, 1017–1038 (2013).
Zeng, H. What is a cell type and how to define it? Cell 185, 2739–2755 (2022).
Zeng, H. & Sanes, J. R. Neuronal cell-type classification: challenges, opportunities and the path forward. Nat Rev Neurosci 18, 530–546 (2017).
Peng, H. et al. Morphological diversity of single neurons in molecularly defined cell types. Nature 598, 174–181 (2021).
Callaway, E. M. et al. A multimodal cell census and atlas of the mammalian primary motor cortex. Nature 598, 86–102 (2021).
Zingg, B. et al. Neural Networks of the Mouse Neocortex. Cell 156, 1096–1111 (2014).
Nern, A., Pfeiffer, B. D. & Rubin, G. M. Optimized tools for multicolor stochastic labeling reveal diverse stereotyped cell arrangements in the fly visual system. Proceedings of the National Academy of Sciences 112, E2967–E2976 (2015).
Cai, D., Cohen, K. B., Luo, T., Lichtman, J. W. & Sanes, J. R. Improved tools for the Brainbow toolbox. Nat Methods 10, 540–547 (2013).
Gong, H. et al. High-throughput dual-colour precision imaging for brain-wide connectome with cytoarchitectonic landmarks at the cellular level. Nature Communications 7, 12142 (2016).
Economo, M. N. et al. A platform for brain-wide imaging and reconstruction of individual neurons. eLife 5, e10566 (2016).
Bria, A., Iannello, G., Onofri, L. & Peng, H. TeraFly: real-time three-dimensional visualization and annotation of terabytes of multidimensional volumetric images. Nature Methods 13, 192–194 (2016).
Pietzsch, T., Saalfeld, S., Preibisch, S. & Tomancak, P. BigDataViewer: visualization and processing for large image data sets. Nature Methods 12, 481–483 (2015).
Wang, Y. et al. TeraVR empowers precise reconstruction of complete 3-D neuronal morphology in the whole brain. Nature Communications 10, 3474 (2019).
Peng, H. et al. Automatic tracing of ultra-volumes of neuronal images. Nature Methods 14, 332–333 (2017).
Meijering, E. Neuron tracing in perspective. Cytometry Part A 77A, 693–704 (2010).
Gong, H. et al. Continuously tracing brain-wide long-distance axonal projections in mice at a one-micron voxel resolution. Neuroimage 74, 87–98 (2013).
Ropireddy, D., Scorcioni, R., Lasher, B., Buzsáki, G. & Ascoli, G. A. Axonal morphometry of hippocampal pyramidal neurons semi-automatically reconstructed after in vivo labeling in different CA3 locations. Brain Struct Funct 216, 1–15 (2011).
Aransay, A., Rodríguez-López, C., García-Amado, M., Clascá, F. & Prensa, L. Long-range projection neurons of the mouse ventral tegmental area: a single-cell axon tracing analysis. Front. Neuroanat. 9 (2015).
De Paola, V. et al. Cell Type-Specific Structural Plasticity of Axonal Branches and Boutons in the Adult Neocortex. Neuron 49, 861–875 (2006).
Jiang, S. et al. Petabyte-Scale Multi-Morphometry of Single Neurons for Whole Brains. Neuroinform 20, 525–536 (2022).
Winnubst, J. et al. Reconstruction of 1,000 Projection Neurons Reveals New Cell Types and Organization of Long-Range Connectivity in the Mouse Brain. Cell 179, 268–281.e13 (2019).
Liu, Y., Wang, G., Ascoli, G. A., Zhou, J. & Liu, L. Neuron tracing from light microscopy images: automation, deep learning and bench testing. Bioinformatics 38, 5329–5339 (2022).
Acciai, L., Soda, P. & Iannello, G. Automated Neuron Tracing Methods: An Updated Account. Neuroinformatics 14, 353–367 (2016).
Peng, H., Long, F. & Myers, G. Automatic 3D neuron tracing using all-path pruning. Bioinformatics 27, i239 (2011).
Xiao, H. & Peng, H. APP2: automatic tracing of 3D neuron morphology based on hierarchical pruning of a gray-weighted image distance-tree. Bioinformatics 29, 1448–1454 (2013).
Feng, L., Zhao, T. & Kim, J. neuTube 1.0: A New Design for Efficient Neuron Reconstruction Software Based on the SWC Format. eNeuro 2 (2015).
Manubens-Gil, L. et al. BigNeuron: a resource to benchmark and predict performance of algorithms for automated tracing of neurons in light microscopy datasets. Nat Methods 1–12 https://doi.org/10.1038/s41592-023-01848-5 (2023).
Peng, H. et al. BigNeuron: Large-scale 3D Neuron Reconstruction from Optical Microscopy Images. Neuron 87, 252–256 (2015).
Gao, L. et al. Single-neuron projectome of mouse prefrontal cortex. Nat Neurosci 25, 515–529 (2022).
Iascone, D. M. et al. Whole-Neuron Synaptic Mapping Reveals Spatially Precise Excitatory/Inhibitory Balance Limiting Dendritic and Somatic Spiking. Neuron 106, 566–578.e8 (2020).
Liu, Y. et al. Neuronal diversity and stereotypy at multiple scales through whole brain morphometry. Nat Commun 15, 10269 (2024).
Hawrylycz, M. et al. A guide to the BRAIN Initiative Cell Census Network data ecosystem. PLOS Biology 21, e3002133 (2023).
Zhang, L. et al. Collaborative augmented reconstruction of 3D neuron morphology in mouse and human brains. Nat Methods 1–11 https://doi.org/10.1038/s41592-024-02401-8 (2024).
Qian, P., Manubens-Gil, L., Jiang, S. & Peng, H. Non-homogenous axonal bouton distribution in whole-brain single-cell neuronal networks. Cell Reports 43 (2024).
Xiong, F. et al. DSM: Deep sequential model for complete neuronal morphology representation and feature extraction. Patterns 5, 100896 (2024).
Xiong, F., Liu, L. & Peng, H. Reconstruct a Connectome of Single Neurons in Mouse Brains by Cross-Validating Multi-Scale Multi-Modality Data. 2024.10.01.616182 Preprint at https://doi.org/10.1101/2024.10.01.616182 (2024).
Liu, Y., Zhao, S., Yun, Z., Xiong, F. & Peng, H. Constructing a Mouse Brain Atlas of Dendritic Microenvironments Helps Discover Hidden Associations Between Anatomical Layout, Projection Targets and Transcriptomic Profiles of Neurons. 2024.09.22.614330 Preprint at https://doi.org/10.1101/2024.09.22.614330 (2024).
Wang, Q. et al. The Allen Mouse Brain Common Coordinate Framework: A 3D Reference Atlas. Cell 181, 936–953.e20 (2020).
Gao, L. et al. Single-neuron analysis of dendrites and axons reveals the network organization in mouse prefrontal cortex. Nat Neurosci 26, 1111–1126 (2023).
Qiu, S. et al. Whole-brain spatial organization of hippocampal single-neuron projectomes. Science 383, eadj9198 (2024).
Kenney, M. et al. The Brain Image Library: A Community-Contributed Microscopy Resource for Neuroscientists. Sci Data 11, 1212 (2024).
Peng, H., Bria, A., Zhou, Z., Iannello, G. & Long, F. Extensible visualization and analysis for multidimensional images using Vaa3D. Nature Protocols 9, 193–208 (2014).
Peng, H., Ruan, Z., Long, F., Simpson, J. H. & Myers, E. W. V3D enables real-time 3D visualization and quantitative analysis of large-scale biological image data sets. Nature Biotechnology 28, 348–353 (2010).
Li, Y., Jiang, S., Ding, L. & Liu, L. NRRS: a re-tracing strategy to refine neuron reconstruction. Bioinformatics Advances 3, vbad054 (2023).
Peng, H., Long, F., Zhao, T. & Myers, E. Proof-editing is the Bottleneck Of 3D Neuron Reconstruction: The Problem and Solutions. Neuroinform 9, 103–105 (2011).
Cheng, S. et al. DeepBouton: Automated Identification of Single-Neuron Axonal Boutons at the Brain-Wide Scale. Front. Neuroinform. 13 (2019).
Gala, R. et al. Computer assisted detection of axonal bouton structural plasticity in in vivo time-lapse images. eLife 6, e29315 (2017).
Guo, S., Zhao, X., Jiang, S., Ding, L. & Peng, H. Image enhancement to leverage the 3D morphological reconstruction of single-cell neurons. Bioinformatics 38, 503–512 (2022).
Qu, L. et al. Cross-modal coherent registration of whole mouse brains. Nat Methods 19, 111–118 (2022).
Mehta, K. et al. Online conversion of reconstructed neural morphologies into standardized SWC format. Nat Commun 14, 7429 (2023).
Jiang, S. et al. A Multi-Scale Neuron Morphometry Dataset from Peta-voxel Mouse Whole-Brain Images. Zenodo https://doi.org/10.5281/zenodo.13944322 (2024).
Wilkinson, M. D. et al. The FAIR Guiding Principles for scientific data management and stewardship. Sci Data 3, 160018 (2016).
Peng, H., Ruan, Z., Atasoy, D. & Sternson, S. Automatic reconstruction of 3D neuron structures using a graph-augmented deformable model. Bioinformatics 26, i38–i46 (2010).
Jiang, S. et al. Beyond Static Brain Atlases: AI-Powered Open Databasing and Dynamic Mining of Brain-Wide Neuron Morphometry. 2024.09.22.614319 Preprint at https://doi.org/10.1101/2024.09.22.614319 (2024).
Acknowledgements
We thank organizers and collaborators within the BICCN initiative in data collection and coordination, including Yong Yao, Hongkui Zeng, Giorgio A. Ascoli, Qingming Luo, Hongwei Dong, Partha Mitra, Michael Hawrylycz, Hui Gong, Pavel Osten, Zhuhao Wu, Josh Huang, Silvia De Rubeis, Wei Wang, Marta Garcia-Forn, Brain Image Library (BIL, www.brainimagelibrary.org) for hosting of the BICCN database, Lei Qu and Yuanyuan Li for assisting in brain registration, Lijuan Liu, Linus Manubens-Gil, Penghao Qian, Qiaobo Gong, Xin Chen, Gaoyu Wang, Zuo-Han Zhao, Yuning Hang, Yuanyuan Song, Lulu Yin for assistance of the morphometry generation, Jiangshan Liang, Luchen Deng, Shize Chen, Fei Xing, Yihang Zhu, Lei Huang and Kaixiang Li for help in platform implementation, Xuan Zhao, Ye Zhong, Jingzhou Yuan, and other 7 anonymous users for help in proofreading somas and Yiwei Li for the assistance of skeleton refinement. H.P. is a SANS Senior investigator and a New Cornerstone Investigator. This work is also supported by a Natural Science Foundation of Jiangsu Province under Grant No. BK20243064, “the Fundamental Research Funds for the Central Universities” (No. 2242023K5005), and a MOST (China) Brain Research Project, “Mammalian Whole Brain Mesoscopic Stereotaxic 3D Atlas” (2022ZD0205200 and 2022ZD0205204).
Author information
Authors and Affiliations
Contributions
H.P. conceptualized this study. H.P. and Y.Liu managed this study. S.J. collected and preprocessed the brain images, developed tools for data management, co-developed the skeleton refinement method, and detected varicosities. Y.L., L.Z. and Y.Liu led the soma detection. Y.Liu led the reconstruction of local dendritic morphologies. S.Z. was responsible for the production of neuronal arbors, and Y.Liu developed the method for producing primary axonal tracts. Z.Y. conducted the image registration and morphometry mapping to the CCFv3. S.J. drafted the majority of the figures and tables, and S.Z. prepared the Technical Validation section and Fig. 8. S.J. and Y.Liu wrote the manuscript with contributions from all authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Jiang, S., Zhao, S., Li, Y. et al. A Multi-Scale Neuron Morphometry Dataset from Peta-voxel Mouse Whole-Brain Images. Sci Data 12, 683 (2025). https://doi.org/10.1038/s41597-025-04379-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41597-025-04379-0