Abstract
Observational studies have found associations between urinary sodium (UNa) with obesity, body shape and composition; but the findings may be biased by residual confounding. The objective of this two-sample Mendelian randomization (MR) study was to analyze their causal associations in both sex-combined and sex-specific models. Genome-wide association studies of UNa, body mass index (BMI), BMI-adjusted waist-to-hip ratio (WHR), body fat (BF) percentage and estimated glomerular filtration rate (eGFR) were identified. We initially extracted fifty SNPs associated with UNa at significance level of 5 × 10–8, but further removed those SNPs with potential horizontal pleiotropy. Univariable and multivariable MR with adjustment for eGFR were performed. Inverse-variance weighted MR was performed as the primary analysis, with MR-Egger methods as sensitivity analysis. The potential bidirectional association between BMI and UNa was investigated. All exposure and outcomes were continuous, and the effect measure was regression coefficients (beta) and their 95% confidence intervals (95% CI). The total sample size was up to 322 154. UNa was causally associated with increased BMI in both men [eGFR-adjusted beta 0.443 (0.163–0.724)] and women [0.594 (0.333–0.855)]. UNa caused BF percentage increase in men [0.622 (0.268–0.976)] and women [0.334 (0.007–0.662)]. UNa significantly elevated BMI-adjusted WHR in men [0.321 (0.094–0.548)], but not in women [0.170 (− 0.052 to 0.391)]. Additionally, we found that BMI causally increased UNa [0.043 (0.023–0.063)]. UNa increased BMI and BF percentage. Salt intake affects male body shape by increasing BMI-adjusted WHR, but showed no effects on female body shape. The bidirectional association between BMI and UNa suggested that salt reduction measures and weight reduction measures should be implemented simultaneously to break the vicious cycle and gain more health benefits.
Similar content being viewed by others
Introduction
Obesity has been a global public health concern with increasingly substantial disease burdens1. Obesity is believed to result from a complex relationship among psychosocial, biological and behavioral factors, such as genetic susceptibility, excessive calorie intake and physical inactivity2. Other factors also contribute independently to development of obesity2,3. Obesity, commonly measured with body mass index (BMI, body weight in kilogram/squared height in meter), is a well-recognized risk factor for a range of health conditions and diseases, including cardiovascular diseases4, diabetes5 and cancers6. BMI is easy to measure and calculate; nevertheless, body shape and body composition are stronger predictors for health outcomes than BMI7,8.
Dietary salt intake and urinary sodium (UNa) are known to be associated with a range of health outcomes, such as hypertension, cardiovascular diseases and death9,10,11. Urinary sodium is a surrogate measure for salt intake12. Observational studies13,14,15,16,17 have suggested their effects on obesity. For example, a systematic review of observational studies13 found that both dietary salt intake and UNa were positively associated with BMI. A recent cross-sectional study14 showed that this positive association was directionally concordant across Asian and Western populations, with varying effect sizes. Furthermore, UNa was also demonstrated to be correlated with body shape and body composition measures, such as waist circumference (WC)15,16, waist-to-hip ratio (WHR, waist circumference/hip circumference)16, body fat (BF) mass and lean mass17.
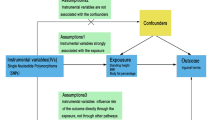
However, so far, the majority of the evidence has been generated from observational studies, mainly cross-sectional studies, in which residual confounding and reverse causation are likely. Evidence from randomized controlled trials has been limited. Although there have been many randomized trials investigating the effects of salt reduction on health outcomes18,19, very few of them reported obesity-related outcomes. Furthermore, they were flawed with small sample size, short follow-up and restricted generalizability20. Mendelian randomization (MR), by using single nucleotide polymorphisms (SNPs) as instrumental variables to investigate the causal link between two phenotypes, is helpful in causal inference, because it removes reverse association and residual confounding21. The objective of this study is to examine the causal association between UNa with body mass, shape and composition using two-sample MR approach.
Methods
We used summary-level statistics from relevant GWA studies, where ethical approval and patient consent had been obtained. The exposure of interest was UNa. The outcomes of interest included BMI, body shape and body composition. For body shape, we used BMI-adjusted WHR as the primary outcome, while WC, hip circumference (HC), WHR, BMI-adjusted WC, BMI-adjusted HC were secondary outcomes. For body composition, we used BF percentage as the primary outcome, while whole-body lean mass (WLM) and appendicular lean mass (ALM) as secondary outcomes. Since sex has been suggested as a potential effect modifier in obesity-related outcomes22,23, we further investigated sex-specific effects in men and women separately.
SNP selection
A GWA study including 446,237 European-ancestry individuals from UK Biobank24 identified 50 lead SNPs associated with UNa that reached significance level of 5 × 10–8 (Supplementary Table S1). Spot urine sample collection and storage in UK Biobank have been described elsewhere11. UNa concentration was measured by the ion-selective electrode method. The effect between genetic variants and log-transformed UNa concentration was adjusted for age, sex, array information and ancestral principal components. The SNPs are located in different gene regions and distributed independently (not in linkage disequilibrium), and explain 6.4% of the total variance of the trait24. If an SNP could not be matched in the GWA studies of an outcome, a proxy in linkage disequilibrium with the SNP (R2 > 0.80) would be identified; if no proper proxy was identified, the unmatched SNP would have to be removed from MR analysis. Relevant traits associated with the SNPs were also searched in the PhenoScanner v2 database at a significance level of 5 × 10–8 for potential pleiotropy.
Outcome data
We extracted summary-level data of the outcomes from four GWA studies of BMI25, body shape26 and body composition27,28. The BMI GWA study25 included 322,154 European-ancestry individuals (152,882 men and 171,963 women) from 125 cohorts. The effect was adjusted for age, squared age, ancestral principal components and study-specific covariates. The body shape GWA study26 included totally 211,088 European-ancestry individuals (93,480 men and 116,742 women), and provided data for BMI-adjusted and unadjusted WC, HC and WHR, additionally adjusted for age, squared age and ancestry principal components. The BF percentage GWA study28 included 89,297 European-ancestry individuals (44,429 men and 45,525 women), and was adjusted for age, squared age and ancestral principal components. The GWA study for WLM and ALM27 included 38,292 and 28,330 European-ancestry individuals, respectively, and was adjusted for age, sex, squared age, height, body fat percentage and ancestral principal components. Body composition (BF, WLM and ALM) was measured either with bioimpedance analysis or dual-energy X-ray absorptiometry26,27. Sex-combined and sex-specific outcome data were available from the GWA studies of BMI, body shape and BF percentage. The GWA study for WLM and ALM provided sex-combined data only.
Covariate
Renal function is associated with both UNa29 and obesity30,31, which is a potential confounder in the associations of interest. We identified a GWA study32 of estimated glomerular filtration rate (eGFR), which included 567,460 European-ancestry individuals. Linear regression of log-transformed eGFR was adjusted for age, sex and ancestry principal components.
Statistical analysis
Previous studies24 have suggested that some of the UNa-associated SNPs are related to adiposity phenotypes, which could introduce horizontal pleiotropy and violate one of the underlying assumptions in MR analysis. To minimize potential horizontal pleiotropy, we further removed SNPs that: (1) were associated with the outcomes of interest at significance level of 5 × 10–8, or (2) were identified as outliers by the MR Pleiotropy RESidual Sum and Outlier (MR-PRESSO) outlier test33. Therefore, the included SNPs for each outcome actually differed but remained a subset of the 50 SNPs (or their proxies) identified by the UNa GWA study24. Supplementary file 1 shows the characteristics of the SNPs included in the MR analysis for each outcome. F statistic was used to measure the strength of instrumental variables, and an F statistic > 10 suggests strong instruments34.
All GWA summary-level data were harmonized with the method of Hartwig et al.35, so that the effect estimates of SNPs on all phenotypes were shown for the same alleles. Univariable MR analyses were performed with inverse-variance weighted method as primary analysis, while weighted median method and MR-Egger regression method as sensitivity analysis. Multivariable MR analyses adjusted for eGFR with inverse-variance weighted method were performed as primary analysis, while MR-Egger method as secondary analysis. The intercept test for MR-Egger regression was used to examine residual horizontal pleiotropy, with a P value < 0.05 suggesting pleiotropy.
To investigate sex-specific effects, we extracted male-specific and female-specific outcome data from the GWA studies25,26,28, and performed MR analyses separately. Sex-specific effects on WLM and ALM were not performed, because the data were not available. To examine the effect difference between men and women, we performed a heterogeneity test similar to that in meta-analysis, using Cochran’s Q test. A P value for Cochrane’s Q test < 0.10 indicates statistical significance.
To investigate potential bidirectional association between UNa and BMI, we also performed MR analysis with BMI as the exposure and UNa as the outcome. Ninety-seven BMI-associated SNPs were extracted from the BMI GWA study25, but 84 SNPs were finally included after removing those unmatched with UNa GWA data24 and/or identified as outliers by the MR-PRESSO outlier test. Similarly, univariable and multivariable MR analyses were performed.
A second way to deal with potential confounding effect of eGFR, alternative to multivariable MR, was to remove the SNPs associated with eGFR. We found that rs1260326 (located in GCKR gene) was associated with eGFR at a significance level of 5 × 10–8 in Phenoscanner v2 database; therefore, a sensitivity analysis by removing rs1260326 in univariable MR was performed as sensitivity analysis. Additional sensitivity analyses were performed by including outlier SNPs detected by the MR-PRESSO outlier tests. Since all outcomes were continuous, the effect measure was regression coefficient beta with its 95% confidence interval (CI), which should be interpreted as the change in units of standard deviation (SD) in the outcome when the log-UNa level increases 1 SD. Statistical significance was indicated by P value < 0.05. We did not apply Bonferroni correction, although we employed three primary outcomes, because their effects were measured from three independent samples of individuals, instead of one single sample. All statistical analysis was conducted with “MendelianRandomization” (version 0.4.1. https://CRAN.R-project.org/package=MendelianRandomization) and “MRPRESSO” (version 1.0. https://rdrr.io/github/rondolab/MR-PRESSO/) packages in R environment (R Core Team, 2003. https://www.R-project.org ).
We planned to perform power calculations based on the method of Brion et al.36, which required the beta estimates from our study and another beta estimates from ordinary least squares estimation (i.e., observational analysis). However, the UNa GWA study analyzed UNa concentration after log transformation (in log-mmol/L), while previous observational studies analyzed UNa concentration in a natural unit (mmol/L) or converted it to 24-h UNa (mmol/day or g/day) or daily salt intake (g/day), thus making it impossible to make direct comparison of the two beta estimates in the absence of individual participant data.
Results
The proportions of exposure variance explained by the instrument variables ranged from 4.036 to 5.565% (mean 4.780%), and the according F statistics ranged from 536.146 to 613.340 (mean 587.078), suggesting strong instrumental variables (Table 1). Supplementary Information S1 showed detailed information on the SNPs included in the MR analysis for each outcome.
Univariable MR
In sex-combined model, univariate MR analyses demonstrated positive causal associations between UNa with BMI (Fig. 1), BMI-adjusted WHR (Fig. 2) and BF percentage (Fig. 3) in both sex-combined and sex-specific models (Table 1). MR-Egger intercept tests revealed no horizontal pleiotropy for these outcomes. BMI was positively related to UNa [beta (95% CI): 0.042 (0.022–0.062)], which suggested a bidirectional association between UNa and BMI.
Scatter plots for Mendelian randomization analysis of urinary sodium and BMI. (A) sex-combined, (B) male-specific, (C) female-specific. Horizontal axis: SNPs’ association with urinary sodium. Vertical axis: SNPs’ association with BMI. The fitted line showed the results of the inverse-variance weighted method in univariable MR.
Scatter plots for Mendelian randomization analysis of urinary sodium and BMI-adjusted waist-to-hip ratio. (A) Sex-combined, (B) male-specific, (C) female-specific. Horizontal axis: SNPs’ association with urinary sodium. Vertical axis: SNPs’ association with BMI-adjusted waist-to-hip ratio. The fitted line showed the results of the inverse-variance weighted method in univariable MR.
Scatter plots for Mendelian randomization analysis of urinary sodium and body fat percentage. (A) Sex-combined, (B) male-specific, (C) female-specific. Horizontal axis: SNPs’ association with urinary sodium. Vertical axis: SNPs’ association with body fat percentage. The fitted line showed the results of the inverse-variance weighted method in univariable MR.
Multivariable MR
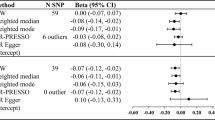
UNa showed independent positive association with BMI [0.406 (0.202–0.610)], BMI-adjusted WHR [0.222 (0.029–0.416)] and BF percentage [0.471 (0.198–0.744)], after adjusted for eGFR. No association was observed with BMI-adjusted HC, BMI-adjusted WC, WLM or ALM. MR-Egger intercept tests revealed no pleiotropy (Table 2). The bidirectional association between BMI and UNa remained after adjusted for eGFR. The eGFR-adjusted effect of BMI on UNa was 0.043 (0.023–0.063) with no pleiotropy (P for MR-Egger intercept test = 0.149).
For men, UNa was positively associated with BMI [eGFR-adjusted beta 0.443 (0.163–0.724)], BMI-adjusted WHR [0.321 (0.094–0.548)], and BF percentage [0.622 (0.268–0.976)], but negatively with BMI-adjusted HC [− 0.368 (− 0.673 to − 0.063)]. No pleiotropy was observed for these outcomes, although pleiotropy was observed in secondary outcomes, such as HC and WHR (Table 2).
For women, UNa was associated with BMI [eGFR-adjusted beta 0.594 (0.333–0.855)] and BF percentage [0.334 (0.007–0.662)], but not BMI-adjusted WHR [0.170 (− 0.052 to 0.391)]. No pleiotropy was observed (Table 2).
Overall, UNa was associated with increased BMI and BF percentage in both men and women. Higher UNa affected male body shape by increasing BMI-adjusted WHR and decreasing BMI-adjusted HC. UNa seemed to have little effects on female body shape after adjusted for BMI and eGFR. Formal heterogeneity test showed significant difference between men and women on BMI-adjusted HC (Table 2).
In sensitivity analysis including the SNPs that were identified as outliers by MR-PRESSO outlier tests, both the univariable and multivariable MR showed results consistent with primary results (Supplement Tables S2, S3). Removing the eGFR-associated SNP rs1260326 also generated concordant results (Supplementary Table S4).
The scatter plots showed a potential outlier SNP rs4803378, located in gene CTC-490E21.10 and associated with urine creatinine according to PhenoScanner v2 database, the post hoc sensitivity analyses by removing this SNP showed similar results to the primary results (Supplementary Table S5).
Discussion
UNa was causally associated with increased BMI and BF percentage. Higher UNa changed body shape towards central obesity in men, by reducing BMI-adjusted HC and increasing BMI-adjusted WHR, but probably not in women. There was a bidirectional positive association between UNa and BMI.
UNa has been widely used as a surrogate for dietary salt intake9,10,37. UNa is mainly determined by dietary sodium intake and kidney function, and 24-h UNa is favored than spot UNa. Some validated equations were used to estimate 24-h urinary sodium from spot urinary sodium, which additionally considers urinary creatinine or kidney function, such as Kawasaki equation, INTERSALT equation38,39,40. Spot urine sample was collected in UK Biobank24. Although we did not estimate 24-h UNa due to lack of individual data in this study, which is one of the limitations, we adjusted for kidney function (eGFR) in multivariable analysis, which enhanced the reliability of our estimation of the effect size of dietary salt intake on the outcomes. Nevertheless, future studies using individual 24-h UNa or dietary sodium intake are warranted.
These results were consistent with previous findings. Moosavian et al.13 found in a meta-analysis that both dietary salt intake and UNa were positively associated with BMI. This study computed the effect measure as the BMI mean difference between highest and lowest categories of exposure, which introduced heterogeneity due to various cutoff values used in included studies and made it difficult to directly compare the effect size with other studies. Zhou et al.14 found that for each 1 gram per day (g/day) increase in salt intake, BMI increased by 0.42 and 0.52 kg/m2, overweight/obesity risk increased by 29% and 24% in UK and US population, respectively, after adjustment for total energy intake and physical activity.
UNa showed an effect on body composition by increasing BF percentage, which is consistent with previous observational findings. An increase of 1 g/d salt intake was associated with an increase of 0.91 kg BF mass in the UK population21, and 0.79 kg BF mass and 0.44% BF percentage in US population17. Oh et al.41 observed this positive association but only in people under 65 years old. Zhu et al.42 found salt intake was associated with increased BF mass, BF percentage, and subcutaneous abdominal adipose tissue. Interestingly, a randomized trial20 found that low-salt diet intervention significantly decreased body weight, BMI and extracellular water, but reductions in BF mass and lean mass were insignificant. However, this trial was limited by small sample size (84 obese people) and short follow-up period (2 months), which might make it underpowered to detect significant reductions. Salt intake has an instant effect on water retention, but its effect on fat accumulation may take longer.
We failed to find the effect of UNa on lean mass, neither WLM nor ALM. This remains uncertain. Previously, Ma et al.15 showed that 1 g/d increase in salt intake was associated with significant WLM increase by 0.32 kg, but 1 g/2000 kcal increase in salt intake density insignificantly reduced WLM by 0.007 kg. Zhang et al.17 found that salt intake significantly increased WLM by 0.53 kg, but the effect became insignificant when using salt intake density (g/kcal) to adjust for energy intake. It seems that there are two possible effects of salt intake on body composition: (1) increase both fat and lean mass, but fat mass increases at a higher rate; (2) increase fat mass solely. The first explanation was supported by the findings that salt intake was associated with more BF mass than lean mass in both UK (0.91 kg versus 0.32 kg for fat and lean mass increase, respectively)15 and US populations (0.79 kg versus 0.53 kg)17. The second explanation was supported by the the null effect of salt intake on lean mass after adjustment for energy intake15,17. Our findings also support the second explanation. Nevertheless, more research is warranted on this topic.
There are several potential explanations for how salt intake affects body weight and composition. It has been well recognized that excessive salt intake increases thirst and fluid consumption, thus increasing extracellular water while keeping urine volume almost unchanged43. Higher dietary salt intake also increases sweetened-beverage intake44, food appetite, food consumption and calorie intake13,45, which all contribute to weight gain. More importantly, high sodium intake has independent biological effects on fat accumulation. Lanaspa et al.46 demonstrated that high salt intake activated the aldose reductase-fructokinase pathway in liver and hypothalamus, causing endogenous fructose production and the development of leptin resistance and hyperphagia, which eventually cause obesity. Fonseca-Alaniz et al.47 observed that the high salt-induced adiposity in rats was characterized by high plasma leptin concentrations and adipocyte hypertrophy, which might be mediated by lipogenic capacity of white adipose mass. Lee et al.48 revealed that high salt increased the expression of adipogenic and lipogenic genes (such as PPAR-γ, SREBP1c, ACC, C/EBPα, and FAS), but decreased lipolysis gene expression (such as AMPK). However, its effect on lean mass requires more replications and mechanism research.
We found that UNa had an impact on body shape. Body shape, rather than simple BMI, is more predictive of obesity-related mortality and health outcomes49,50. Overall, UNa increased both BMI-adjusted and unadjusted WHR, leading to central obesity, which is in line with previous findings13,16,17,42. However, we observed an effect modification caused by sex. UNa increased WHR in both sexes, but a significant increase in BMI-adjusted WHR was seen in men only. Nam et al.16 observed a similar sex difference. This may suggest that UNa affects body shape independent of BMI in men, but its effect on female body shape is, at least partly, mediated by BMI.
This sexual dimorphism of body fat accumulation and distribution may be related to sex hormones51,52, sex chromosome53 and sex-specific autosomal genetic heritability22,23,54,55. Estrogens favor fat accumulation in subcutaneous depot in women. Estrogens and their receptor regulations also enhance the expandability of adipose cells in subcutaneous depot and inhibits it in visceral depots51. Elevated free androgens have also been shown to be related to increased abdominal visceral fat accumulation and increased WC56. The Four Core Genotypes mouse model studies, have shown that X chromosome dosage is a risk factor for fat accumulation and obesity, but the presence of Y chromosome is not54. Other studies have also shown that that adipocytes from different subcutaneous depots (abdominal versus gluteal) are developmentally distinct in a sex-dependent manner57,58. The GWA study of BMI-adjusted WHR showed a higher heritability in women than in men (2.4% versus 0.8%)26. This indicates that male body shape is more likely to be influenced by environmental factors54, such as salt intake. However, mechanistic studies are needed to elucidate this issue.
In this MR study, we observed that high BMI was causally linked to higher UNa. High UNa could be resulted from two possible mechanisms: obesity-related glomerulopathy and high salt intake. Obesity is known to increase risk of glomerulopathy, characterized with altered renal hemodynamics, increased eGFR, filtration fraction, tubular sodium reabsorption, glomerular hyperfiltration, hormonal and neurohormonal activation59; however, the overall effect of glomerulopathy on UNa remains unclear. On the other hand, the association between BMI and UNa remained consistent after adjusting for renal function (eGFR) in multivariable MR, therefore, we believe that salt intake has independent mediation effect on the association between BMI and UNa, that is, BMI increases UNa by increasing salt intake, independent of renal function. Li et al.60 found that overweight/obese people had lower salt sensitivity and higher salt preference, and consumed 2.3 g/day more dietary salt than normal-weight people.
The bidirectional association between obesity and salt intake indicates the formation of a vicious cycle, in which people with higher BMI are prone to consume more salt, which then further increases BMI. Considering that high salt intake and obesity, both prevalent worldwide, are shared risk factors for many chronic conditions and diseases, and that they also have their own adverse health consequences and disease burdens61, urgent interventions are required to break this cycle in high-risk individuals.
Salt reduction and weight reduction programs have been being widely implemented in various settings, but separately. Salt reduction programs do not consider weight change as an objective, while weight reduction programs pay little emphasis on salt reduction measures. We are not advocating weight control by only restricting salt. Instead, our findings provide evidence to combine salt reduction and weight reduction measures, to facilitate people to acquire a healthier lifestyle and achieve higher health benefits. Although the actual effect size of salt reduction on weight reduction has not been precisely estimated yet, which warrants future well-designed randomized trials, salt reduction per se costs less and brings additional health benefits18,41,62.
There are some limitations. First, we did not include all identified UNa-associated SNPs due to unmatched data and pleiotropy issues; however, our instrument variables were strong (high F statistics) and the results are consistent in primary analyses, sensitivity analyses, and with previous studies. Second, more accurate UNa measure is via 24-h urine sample collection, but the UK Biobank used spot urine sample. 24-h urine collection is more expensive, time-consuming, labor-intensive and difficult for an epidemiological study as large as UK Biobank with half a million participants. We adjusted for kidney function in multivariable MR, which is considered in previous validated equations converting spot UNa to 24-h UNa38,39. Nevertheless, future studies with 24-h UNa measurement or dietary salt intake are warranted. Third, there might still be residual pleiotropy, although we have removed SNPs that were significantly associated with outcomes and that were identified as outliers by the MR-PRESSO outlier test. MR-Egger intercept test showed pleiotropy for a few secondary outcomes (e.g., HC, WC), but not for the primary outcomes. Fourth, the effect size estimated in this study was not directly comparable to other studies in which the effect was measured at natural units of outcome by 1 g/day salt intake increase. Although several equations have been proposed to estimate salt intake in g/day from 24 h urine sodium secretion in g/d or spot urine sodium concentration in mmol/L63, the UNa GWA study log-transformed the original spot UNa concentration, which makes back-transformation of beta estimate to natural unit impossible. Due to the same reason, formal power calculations were not performed as we planned, which was another limitation, but our study has employed strong instrumental variables and so far the largest GWA studies available. Lastly, we only included European-ancestry population, thus generalizing the results to other ethnicities requires cautions.
Conclusion
UNa increased BMI and BF percentage, and changes male body shape towards central obesity by increasing BMI-adjusted WHR. Given the bidirectional association between salt intake and obesity, salt reduction measures and weight reduction measures should be implemented together to achieve more health benefits.
Data availability
All data and analytic code are available upon request.
References
Tremmel, M., Gerdtham, U. G., Nilsson, P. M. & Saha, S. Economic burden of obesity: A systematic literature review. Int. J. Environ. Res. Public. Health 14, 435 (2017).
Apovian, C. M. Obesity: Definition, comorbidities, causes, and burden. Am. J. Manag. Care 22, s176–s185 (2016).
Lee, E. Y. & Yoon, K. H. Epidemic obesity in children and adolescents: Risk factors and prevention. Front. Med. 12, 658–666 (2018).
Ortega, F. B., Lavie, C. J. & Blair, S. N. Obesity and cardiovascular disease. Circ. Res. 118, 1752–1770 (2016).
Malone, J. I. & Hansen, B. C. Does obesity cause type 2 diabetes mellitus (T2DM)? Or is it the opposite?. Pediatr. Diabetes 20, 5–9 (2019).
Avgerinos, K. I., Spyrou, N., Mantzoros, C. S. & Dalamaga, M. Obesity and cancer risk: Emerging biological mechanisms and perspectives. Metabolism 92, 121–135 (2019).
Chen, G.-C. et al. Association between regional body fat and cardiovascular disease risk among postmenopausal women with normal body mass index. Eur. Heart J. 40, 2849–2855 (2019).
Huxley, R., Mendis, S., Zheleznyakov, E., Reddy, S. & Chan, J. Body mass index, waist circumference and waist:hip ratio as predictors of cardiovascular risk—a review of the literature. Eur. J. Clin. Nutr. 64, 16–22 (2010).
Mente, A. et al. Urinary sodium excretion, blood pressure, cardiovascular disease, and mortality: A community-level prospective epidemiological cohort study. Lancet Lond. Engl. 392, 496–506 (2018).
O’Donnell, M. et al. Urinary sodium and potassium excretion, mortality, and cardiovascular events. N. Engl. J. Med 371, 612–623 (2014).
Welsh, C. E. et al. Urinary sodium excretion, blood pressure, and risk of future cardiovascular disease and mortality in subjects without prior cardiovascular disease. Hypertension 73, 1202–1209 (2019).
Meyer, H. E., Johansson, L., Eggen, A. E., Johansen, H. & Holvik, K. Sodium and potassium intake assessed by spot and 24-h urine in the population-based Tromsø Study 2015–2016. Nutrients 11, 1619 (2019).
Moosavian, S. P., Haghighatdoost, F., Surkan, P. J. & Azadbakht, L. Salt and obesity: A systematic review and meta-analysis of observational studies. Int. J. Food Sci. Nutr. 68, 265–277 (2017).
Zhou, L. et al. Salt intake and prevalence of overweight/obesity in Japan, China, the United Kingdom, and the United States: The INTERMAP Study. Am. J. Clin. Nutr. 110, 34–40 (2019).
Ma, Y., He, F. J. & MacGregor, G. A. High salt intake: Independent risk factor for obesity?. Hypertension 66, 843–849 (2015).
Nam, G. E. et al. Association between 24-h urinary sodium excretion and obesity in Korean adults: A multicenter study. Nutrition 41, 113–119 (2017).
Zhang, X., Wang, J., Li, J., Yu, Y. & Song, Y. A positive association between dietary sodium intake and obesity and central obesity: Results from the National Health and Nutrition Examination Survey 1999–2006. Nutr. Res. 55, 33–44 (2018).
He, F. J., Li, J. & MacGregor, G. A. Effect of longer term modest salt reduction on blood pressure: Cochrane systematic review and meta-analysis of randomised trials. BMJ 346, f1325–f1325 (2013).
Hyseni, L. et al. Systematic review of dietary salt reduction policies: Evidence for an effectiveness hierarchy?. PLoS One 12, e0177535 (2017).
Kang, H. J., Jun, D. W., Lee, S. M., Jang, E. C. & Cho, Y. K. Low salt and low calorie diet does not reduce more body fat than same calorie diet: A randomized controlled study. Oncotarget 9, 8521–8530 (2018).
Emdin, C. A., Khera, A. V. & Kathiresan, S. Mendelian randomization. JAMA 318, 1925–1926 (2017).
Rask-Andersen, M., Karlsson, T., Ek, W. E. & Johansson, A. Genome-wide association study of body fat distribution identifies adiposity loci and sex-specific genetic effects. Nat. Commun. 10, 339 (2019).
Winkler, T. W. et al. The influence of age and sex on genetic associations with adult body size and shape: A large-scale genome-wide interaction study. PLoS Genet 11, e1005378 (2015).
Pazoki, R. et al. GWAS for urinary sodium and potassium excretion highlights pathways shared with cardiovascular traits. Nat. Commun. 10, 3653 (2019).
Locke, A. E. et al. Genetic studies of body mass index yield new insights for obesity biology. Nature 518, 197–206 (2015).
Shungin, D. et al. New genetic loci link adipose and insulin biology to body fat distribution. Nature 518, 187–196 (2015).
Zillikens, M. C. et al. Large meta-analysis of genome-wide association studies identifies five loci for lean body mass. Nat. Commun. 8, 80 (2017).
Lu, Y. et al. New loci for body fat percentage reveal link between adiposity and cardiometabolic disease risk. Nat. Commun. 7, 10495 (2016).
Nomura, K., Asayama, K., Jacobs, L., Thijs, L. & Staessen, J. A. Renal function in relation to sodium intake: A quantitative review of the literature. Kidney Int. 92, 67–78 (2017).
Jamshidi, P. et al. Investigating associated factors with glomerular filtration rate: Structural equation modeling. BMC Nephrol. 21, 30 (2020).
Whaley-Connell, A. & Sowers, J. R. Obesity and kidney disease: From population to basic science and the search for new therapeutic targets. Kidney Int. 92, 313–323 (2017).
Wuttke, M. et al. A catalog of genetic loci associated with kidney function from analyses of a million individuals. Nat. Genet. 51, 957–972 (2019).
Verbanck, M., Chen, C. Y., Neale, B. & Do, R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat. Genet. 50, 693–698 (2018).
Pierce, B. L., Ahsan, H. & Vanderweele, T. J. Power and instrument strength requirements for Mendelian randomization studies using multiple genetic variants. Int. J. Epidemiol. 40, 740–752 (2011).
Hartwig, F. P., Davies, N. M., Hemani, G. & Davey Smith, G. Two-sample Mendelian randomization: Avoiding the downsides of a powerful, widely applicable but potentially fallible technique. Int. J. Epidemiol. 45, 1717–1726 (2016).
Brion, M.-J.A., Shakhbazov, K. & Visscher, P. M. Calculating statistical power in Mendelian randomization studies. Int. J. Epidemiol. 42, 1497–1501 (2013).
Mente, A. et al. Associations of urinary sodium excretion with cardiovascular events in individuals with and without hypertension: A pooled analysis of data from four studies. Lancet Lond. Engl. 388, 465–475 (2016).
Kawasaki, T., Itoh, K., Uezono, K. & Sasaki, H. A simple method for estimating 24 h urinary sodium and potassium excretion from second morning voiding urine specimen in adults. Clin. Exp. Pharmacol. Physiol. 20, 7–14 (1993).
Mente, A. et al. Validation and comparison of three formulae to estimate sodium and potassium excretion from a single morning fasting urine compared to 24-h measures in 11 countries. J. Hypertens. 32, 1005–1014 (2014) ((discussion 1015)).
Brown, I. J. et al. Estimating 24-hour urinary sodium excretion from casual urinary sodium concentrations in Western populations: The INTERSALT study. Am. J. Epidemiol. 177, 1180–1192 (2013).
Oh, S. W., Koo, H. S., Han, K. H., Han, S. Y. & Chin, H. J. Associations of sodium intake with obesity, metabolic disorder, and albuminuria according to age. PLoS One 12, e0188770 (2017).
Zhu, H. et al. Dietary sodium, adiposity, and inflammation in healthy adolescents. Pediatrics 133, e635-642 (2014).
Bankir, L., Perucca, J., Norsk, P., Bouby, N. & Damgaard, M. Relationship between sodium intake and water intake: The false and the true. Ann. Nutr. Metab. 70(Suppl 1), 51–61 (2017).
Grimes, C. A., Riddell, L. J., Campbell, K. J. & Nowson, C. A. Dietary salt intake, sugar-sweetened beverage consumption, and obesity risk. Pediatrics 131, 14–21 (2013).
Bolhuis, D. P., Costanzo, A., Newman, L. P. & Keast, R. S. Salt promotes passive overconsumption of dietary fat in humans. J. Nutr. 146, 838–845 (2016).
Lanaspa, M. A. et al. High salt intake causes leptin resistance and obesity in mice by stimulating endogenous fructose production and metabolism. Proc. Natl. Acad. Sci. USA 115, 3138–3143 (2018).
Fonseca-Alaniz, M. H. et al. High dietary sodium intake increases white adipose tissue mass and plasma leptin in rats. Obes. Silver Spring 15, 2200–2208 (2007).
Lee, M., Sorn, S. R., Lee, Y. & Kang, I. Salt induces adipogenesis/lipogenesis and inflammatory adipocytokines secretion in adipocytes. Int. J. Mol. Sci. 20, 20 (2019).
Fu, J., Hofker, M. & Wijmenga, C. Apple or pear: Size and shape matter. Cell Metab. 21, 507–508 (2015).
Golzarri-Arroyo, L., Mestre, L. M. & Allison, D. B. What’s new in understanding the risk associated with body size and shape?: Pears, apples, and olives on toothpicks. JAMA Netw. Open 2, e197336 (2019).
Palmer, B. F. & Clegg, D. J. The sexual dimorphism of obesity. Mol. Cell Endocrinol. 402, 113–119 (2015).
Valencak, T. G., Osterrieder, A. & Schulz, T. J. Sex matters: The effects of biological sex on adipose tissue biology and energy metabolism. Redox Biol. 12, 806–813 (2017).
Zore, T., Palafox, M. & Reue, K. Sex differences in obesity, lipid metabolism, and inflammation-A role for the sex chromosomes?. Mol. Metab. 15, 35–44 (2018).
Link, J. C. & Reue, K. Genetic basis for sex differences in obesity and lipid metabolism. Annu. Rev. Nutr. 37, 225–245 (2017).
Pulit, S. L., Karaderi, T. & Lindgren, C. M. Sexual dimorphisms in genetic loci linked to body fat distribution. Biosci. Rep. 37, BSR20160184 (2017).
Blouin, K., Boivin, A. & Tchernof, A. Androgens and body fat distribution. J. Steroid Biochem. Mol. Biol. 108, 272–280 (2008).
Fried, S. K., Lee, M.-J. & Karastergiou, K. Shaping fat distribution: New insights into the molecular determinants of depot- and sex-dependent adipose biology. Obes. Silver Spring Md 23, 1345–1352 (2015).
Karastergiou, K. et al. Distinct developmental signatures of human abdominal and gluteal subcutaneous adipose tissue depots. J. Clin. Endocrinol. Metab. 98, 362–371 (2013).
D’Agati, V. D. et al. Obesity-related glomerulopathy: Clinical and pathologic characteristics and pathogenesis. Nat. Rev. Nephrol. 12, 453–471 (2016).
Li, Q. et al. Enhancement of neural salty preference in obesity. Cell. Physiol. Biochem. 43, 1987–2000 (2017).
Cheung, B. M. Y. & Li, C. Diabetes and hypertension: Is there a common metabolic pathway?. Curr. Atheroscler. Rep. 14, 160–166 (2012).
Hoffmann, I. S. & Cubeddu, L. X. Salt and the metabolic syndrome. Nutr. Metab. Cardiovasc. Dis. 19, 123–128 (2009).
Huang, L. et al. Mean population salt intake estimated from 24-h urine samples and spot urine samples: A systematic review and meta-analysis. Int. J. Epidemiol. 45, 239–250 (2016).
Author information
Authors and Affiliations
Contributions
Q.F. and B.M.Y.C. designed research. Q.F., Y.L. and R.J.F.L. collected data. Q.F. performed statistical analysis. Q.F. wrote paper. S.Y. and Q.Y. provided methodological guidance. All authors were involved in result interpretation and critical revision of manuscript. Q.F. had primary responsibility for final content.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Feng, Q., Yuan, S., Yang, Q. et al. Causal associations between urinary sodium with body mass, shape and composition: a Mendelian randomization study. Sci Rep 10, 17475 (2020). https://doi.org/10.1038/s41598-020-74657-x
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-020-74657-x
This article is cited by
-
High salt intake and its dual impact on obesity and cardiovascular mortality in individuals over 50: a paradoxical relationship
BMC Public Health (2025)
-
Dietary salt intake and kidney function in rural Senegalese populations: a cross-sectional study
Journal of Health, Population and Nutrition (2024)
-
The Association Between Insulin Use and Asthma: An Epidemiological Observational Analysis and Mendelian Randomization Study
Lung (2023)
-
The association between salt intake and blood pressure is mediated by body mass index but modified by hypertension: The ELSA-Brasil study
Journal of Human Hypertension (2022)