Abstract
Severity of deceased donor kidney fibrosis impacts graft survival in deceased-donor kidney transplantation. Our aim was to identify potential miRNA biomarkers in urinary exosomes that mirror interstitial fibrosis and tubular atrophy (IFTA) severity. Among 109 urine samples from deceased donors, 34 displayed no IFTA in the zero-day biopsy (No IFTA group), while the remaining 75 deceased donor kidneys exhibited an IFTA score ≥ 1 (IFTA group). After analyzing previous reports and electronic databases, six miRNAs (miR-19, miR-21, miR-29c, miR-150, miR-200b, and miR-205) were selected as potential IFTA biomarker candidates. MiR-21, miR-29c, miR-150, and miR-205 levels were significantly higher, while miR-19 expression was significantly lower in the IFTA group. MiR-21 (AUC = 0.762; P < 0.001) and miR-29c (AUC = 0.795; P < 0.001) showed good predictive accuracy for IFTA. In the No IFTA group, the eGFR level at 1 week after transplantation was significantly higher compared to the IFTA group (41.34 mL/min/1.73m2 vs. 28.65 mL/min/1.73m2, P = 0.012). These findings signify the potential of urinary exosomal miRNAs as valuable biomarker candidates for evaluating the severity of IFTA in deceased donor kidneys before they undergo recovery.
Similar content being viewed by others
Introduction
Kidney transplantation (KT) is well-known as the most effective way to treat chronic kidney disease in terms of patient survival and cost1,2. The administration of immunosuppressants, such as tacrolimus and anti-thymocyte globulin, has improved long-term survival by reducing rejection rates3,4. The long-term outcomes of KT are also influenced by the chronic fibrosis status of the donor’s kidney5,6. The evaluation of kidney tissue through preimplantation biopsies has been identified as a standalone factor for predicting the success of grafts in expanded criteria donor (ECD) KT. Consequently, various research teams utilize renal histology to assess the viability of ECD kidneys7,8. However, the preimplantation kidney tissue necessitates prompt delivery to the pathology laboratory and expeditious examination by on-call pathologists within a few hours to mitigate cold ischemic time. Consequently, the universal adoption of this strategy poses considerable challenges.
MicroRNAs (miRNAs) are small noncoding RNAs consisting of about 18–23 nucleotides. In many studies, miRNAs are reported as regulators of gene expression at the post-transcriptional level9,10. In addition to these regulatory functions, miRNAs are known to be highly stable particles in plasma and urine10,11,12,13. Exosomes are small vesicles or membrane-bound particles that are secreted by many types of cells, including kidney cells14,15, and have been found to contain various biomolecules, including miRNAs, proteins, and lipids, and they play a role in intercellular communication. miRNA can also be contained within exosomes, which are then delivered between cells16. Exosomal miRNAs are of particular interest in research and clinical studies because they can play a role in cell signaling, and disease processes, and potentially serve as biomarkers for various conditions14,15,16. As such, many efforts have been made to develop miRNAs as diagnostic tools for various diseases, including cancers, metabolic diseases, and inflammatory diseases17.
Multiple studies reported that the expression levels of several miRNAs, including miR-142-5p or miR-142-3p, from kidney biopsy and peripheral blood mononuclear cell samples of kidney transplant recipients were closely associated with allograft rejection and interstitial fibrosis and tubular atrophy (IFTA)18,19,20,21. From a pilot study, we found that urinary transglutaminase 2 (TG2) is a potent biomarker for predicting IFTA in kidney allografts during the early post-transplant period in deceased donor kidney transplantation22.
The aim of this study was to identify a panel of new markers associated with IFTA in deceased donor kidney prior to KT. To this end, we isolated and analyzed the exosomal miRNA candidates related to kidney fibrosis and TG2 in urine samples from deceased donors to assess their correlation with kidney fibrosis.
Results
Baseline characteristics and zero-day biopsy
Among a total of 109 recipients, 34 (31%) were classified into the No IFTA group, while the remaining 75 were classified into the IFTA group (Fig. 1, Table 1). ECDs are characterized as donors who are either (1) aged 60 years or older, with or without comorbidities, or (2) aged 50–59 with at least two out of the following comorbid conditions (elevated serum creatinine levels [> 1.5 mg/dL], a history of hypertension, or death caused by cerebrovascular accident). There were 36 (33%) ECD donors and only one donor after cardiac death (DCD) in this cohort. There were no significant differences in the baseline characteristics of recipients and donors between the two groups, except for the lower level of terminal serum creatinine in the No IFTA group compared to the IFTA group (1.49 ± 1.16 mg/dL vs. 2.46 ± 2.15 mg/dL, P = 0.003). Of the induction regimen, basiliximab was more frequently used in both groups although there was no significant difference between the two groups. Almost all recipients had tacrolimus (98.2%), mycophenolic acid (96.3%), and steroid (100%) as a maintenance regimen.
Detailed Banff scores of the zero-day biopsy for interstitial fibrosis, tubular atrophy, and vascular fibrous intimal thickening are described in Table 2. The IFTA group had significantly higher scores for interstitial fibrosis (ci) (0.03 ± 0.17 vs. 0.48 ± 0.66, P < 0.001), tubular atrophy (ct) (0.03 ± 0.17 vs. 0.52 ± 0.53, P < 0.001), and vascular fibrous intimal thickening (cv) (0.00 ± 0.00, 0.19 ± 0.43, P < 0.001).
Selection of miRNA biomarker candidates for deceased donor kidney fibrosis
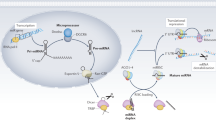
Based on previous reports, we selected six miRNAs that are associated with renal fibrosis and TG2. Subsequently, we verified the selected exosomal miRNAs using the ExoCarta database (http://www.exocarta.org/) (kidney fibrosis: miR-29c, miR-150, miR-200b, and miR-21; TG2: miR-19 and miR-205) (Fig. 2). In addition, we compared these potential biomarkers with the EVmiRNA database (http://bioinfo.life.hust.edu.cn/EVmiRNA) whether the candidates were urinary exosomal miRNA or kidney-specific exosomal miRNA (Fig. 3). In Fig. 3, two Venn diagrams serve to elucidate the intersection of potential biomarkers with established databases of extracellular vesicle microRNAs.
Intersection of identified proteins in published exosome datasets. Intersecting proteins in the compared public datasets are demonstrated by Venn diagrams (A and B). The left diagram (A) presents a binary comparison: the ExoCarta database is depicted by a blue circle, encompassing 1348 unique miRNAs, whereas the red circle, representing the current study, shows no unique miRNAs, with an intersection of six shared miRNAs between the two datasets. The right diagram (B) compares three distinct datasets—Urinary exosomal miRNA (blue), Kidney exosomal miRNA (red), and the current study (green). The urinary and kidney exosomal miRNA datasets possess 82 and 51 unique miRNAs respectively, while the current study identifies four unique miRNAs. Central to the diagram is a single miRNA shared across all three datasets. Notably, one miRNA, miR-29c, is exclusively shared between the urinary exosomal miRNAs and the current study, and another, miR-21, is shared between the kidney exosomal miRNAs and the current study, as indicated by the arrows. The pairwise overlap between urinary and kidney exosomal miRNAs reveals nine shared miRNAs, whereas no miRNAs are exclusively shared between kidney exosomal miRNAs and the current study, underscoring the specificity of the miRNA distribution within these biological contexts.
Exosome identification
Using transmission electron microscopy, the exosomes isolated from urine samples were negatively stained with 2% uranyl acetate (Fig. 4A). The diameter of each exosome was 100–300 nm, as determined by nanoparticle tracking analysis (Fig. 4B). In addition, the expression of TSG101, CD63, CD9, HSP70, and Actin, which are markers of exosomes, was also identified by Western blot analysis (Fig. 4C and Supplementary Fig. 1). There were no significant differences in concentration (no IFTA: 8.40e + 10 ± 1.18E + 11 vs. IFTA: 1.50E + 11 ± 3.46E + 11) and size (206.32 ± 34.63 nm vs. 208.50 ± 28.32 nm) of particles between the groups (Fig. 4D, E).
Urinary exosome characterization. (A) Transmission electron microscopy view of the exosomes isolated from urine samples that were negatively stained with 2% uranyl acetate. (B) The diameter of each exosome was 100–300 nm on nanoparticle tracking analysis. (C) Western blot analysis of the expression of exosomal markers TSG101, CD63, CD9, HSP70, and actin in urinary exosome proteins. There were no significant differences in the concentration (D) and size (E) of particles between the groups.
miRNA expression levels
The expression level of each miRNA was quantified using qRT-PCR, and the differences between the two groups were analyzed using an unpaired Mann–Whitney test (Fig. 5, Table 3). Among the six potential biomarkers, the relative expression levels of miR-21 (P < 0.001), miR-29c (P < 0.001), and miR-205 (P = 0.004) were significantly higher in the IFTA group. In contrast, the IFTA group showed significantly lower expression levels of miR-19 (P = 0.0046) and miR-150 compared to the No IFTA group. As MiR-150 was expressed in only 71.2% of the total samples, it had limited use as a biomarker.
Prognostic value of the miRNA panel
The ROC curves of the selected biomarkers showed statistically significant prognostic accuracy (Fig. 6). The ROC analysis of miR-21, miR-29c, and miR-19 showed significant sensitivity and specificity in predicting the grade of IFTA.
Next, we tested the prognostic value of combined miRNAs to enhance the accuracy of prognosis for IFTA. The sensitivities, specificities, and AUC values of the combined miRNA biomarkers are summarized in Table 4. Based on these prognostic parameters, model 3 demonstrated the highest prognostic accuracy compared to the other models.
Pathway enrichment analysis and human microRNA–disease network analysis
Figure 7 presents an integrative analysis employing the DIANA mirPath v.4 (https://diana-lab.e-ce.uth.gr/app/miRPathv4) computational tool for KEGG pathway enrichment and the STRING database for interaction network mapping of miRNA targets. In Fig. 7A, pathway enrichment analysis with a significance threshold of P < 0.001 unveils the top 10 pathways associated with the up-regulated miRNAs in the interstitial fibrosis and tubular atrophy (IFTA) group, with ‘Focal adhesion’ emerging as the pathway most significantly implicated, followed by pathways involved in oncogenesis and signaling such as ‘Pathways in cancer’ and ‘PI3K-Akt signaling pathway’. These pathways highlight a potential link between the molecular changes in IFTA and pivotal cellular processes. The focal adhesion pathway plays a crucial role in the development of fibrosis through its involvement in myofibroblast activation, TGF-β signaling, and cell–matrix interactions23,24,25. Figure 7B, constructed using STRING database, illustrates a complex interaction network of 58 gene targets, offering a visual representation of the proteomic landscape influenced by the dysregulated miRNAs. Figure 7C complements the preceding analyses by delineating the disease-specific associations of the 58 gene targets modulated by the up-regulated miRNAs in IFTA. This figure adopts a bar chart format to quantify the prevalence of each gene target across a spectrum of pathological states, drawing upon annotations from comprehensive biomedical databases and literature. The bar chart format allows for precise quantification of the number of gene targets associated with each specific condition. By aligning bars horizontally, we can systematically compare the extent to which different diseases are associated with the modulated gene targets. In addition, this format provides a straightforward visual representation that makes it easier to identify patterns and trends. Researchers can quickly discern which conditions have higher associations, facilitating a more intuitive understanding of the data. This multilayered analysis elucidates the broad biological ramifications of miRNA dysregulation, affirming their role as key molecular determinants in IFTA and possibly in a spectrum of other pathological contexts. It is hypothesized that the three validated miRNAs will be involved in the progression of IFTA by regulating common target factors identified through pathway analysis.
Target gene pathway (A), gene network analysis for focal adhesion pathway (B) and Human microRNA–disease network analysis (C). In (A), pathway enrichment analysis with a significance threshold of P < 0.001 unveils the top 10 pathways associated with the up-regulated miRNAs in the interstitial fibrosis and tubular atrophy (IFTA) group, with 'Focal adhesion' emerging as the pathway most significantly implicated, followed by pathways involved in oncogenesis and signaling such as ‘Pathways in cancer’ and ‘PI3K-Akt signaling pathway’. In (B), constructed using STRING database, illustrates a complex interaction network of 58 gene targets, offering a visual representation of the proteomic landscape influenced by the dysregulated miRNAs. Nodes represent individual gene targets, while edges denote the interactions, underscoring the extensive cross-talk and potential synergistic effects on cellular functions. In (C) complements the preceding analyses by delineating the disease-specific associations of the 58 gene targets modulated by the up-regulated miRNAs in IFTA. Each bar represents a discrete condition—ranging from ‘ovarian neoplasm’ to ‘cirrhosis’ to ‘bipolar disorder’—and is proportioned to reflect the count of implicated gene targets. The anticipated color-coding scheme would facilitate distinction among categories such as neoplastic, metabolic, and neuropsychiatric disorders, allowing for an immediate visual appraisal of the data. The configuration of bars would thus illustrate not only the concentrated involvement of these miRNAs in renal fibrotic transformation but also their potential systemic influence, reinforcing the concept of miRNAs as critical regulators with extensive biomedical relevance.
Comparison of clinical outcomes between the groups
We then compared clinical outcomes such as delayed graft function, eGFR, BPAR of kidney allograft, kidney allograft survival, and patient survival. There was no significant difference in the incidence of delayed graft function between the two groups (17.6% vs 25.3%, P = 0.377). The eGFR level of the No IFTA group was significantly higher than that of the IFTA group at 1 week post-transplant (41.34 mL/min/1.73m2 vs. 28.65 mL/min/1.73m2, P = 0.012). According to the linear mixed model, the No IFTA group showed a significantly earlier improving pattern of eGFR over time compared to the IFTA group (Time*Group P = 0.031; Supplementary Fig. 2A). During the 3-year follow-up period, there were no significant differences observed in patient survival (Supplementary Fig. 2B), kidney allograft survival (Supplementary Fig. 2C), and the incidence of BPAR (Supplementary Fig. 2D) between the two groups.
Discussion
In this study, we identified several promising biomarkers in urine specimens from deceased KT donors that correlated with the fibrotic status of deceased donor kidneys. In the IFTA group, miR-21, miR-29c, and miR-205 were expressed at higher levels compared to the No IFTA group, while miR-19 exhibited lower expression levels. Moreover, miR-21 and miR-29c demonstrated significant prognostic value in the ROC analysis.
Several studies have attempted to find non-invasive methods to predict the progression of kidney fibrosis and allograft rejection26,27,28. Schaub et al.29 analyzed the urine protein profiles of kidney transplant recipients. Some researchers have demonstrated specific biomarkers related to kidney allograft fibrosis; for example, Cheng et al.30 suggested that connective tissue growth factor could serve as both a biomarker and a therapeutic target for kidney allograft fibrosis. Still, these biomarkers have not replaced allograft biopsy as the primary method for diagnosing kidney allograft pathology.
Recently, miRNAs have emerged as promising candidates for biomarkers to monitor allograft injury or rejection following KT. These single-stranded RNAs are transcribed by RNA polymerase II enzymes and processed by RNases. The role of miRNAs is to regulate gene expression in intercellular communication31. These molecules can act in distant organs from their origins because they bind to specific proteins such as Ago2 or are transported in exosomes32,33. In this way, miRNAs can be maintained stably in human body fluids. Because of their stability and accessibility, miRNAs are already considered excellent biomarkers for diagnosis and therapeutic targets in the field of oncology34. Moreover, Wang et al.35 reported that urinary levels of miR-29b and miR-29c were higher in patients with IgA nephropathy, and these levels were found to be associated with the progression of renal fibrosis in IgA nephropathy. Lv et al.36 also suggested that there is a significant dysregulation of exosomal miR-29c and miR-21, which correlates with renal fibrosis.
The miRNAs selected as potential biomarkers in this study were previously reported as possible biomarkers for renal fibrosis in other studies. It is reported that urinary miR-21 plays an important role in renal injury and is closely associated with kidney fibrosis37,38,39,40,41,42. miR-29c is also noted as a molecule that inhibits renal interstitial fibrosis through the activation of HIF-α and the PI3K-PKB pathway43,44, and miR-205 and miR-19 influence renal injury through the regulation of PTEN45,46.
Different from previous studies, we aimed to assess the potential of urinary exosomal miRNA as non-invasive biomarkers for determining the IFTA grade of a deceased donor kidney prior to KT. We discovered that the disparity in renal function following KT was associated with the grade of IFTA observed in the zero-day biopsy. Taken together, it may be reasonable to explain that urinary exosomal miRNAs play essential roles as biomarkers in reflecting the grade of IFTA of deceased donor kidneys. To our knowledge, this study is the first to assess the relationship between urinary exosomal miRNA and the IFTA status of a deceased donor kidney prior to kidney transplantation. The timeframe for obtaining results from miRNA analysis indeed varies based on several factors, including the specific methods employed, the availability of equipment and reagents, and the overall efficiency of the laboratory workflow. However, from the isolation of urinary exosomes to conducting quantitative reverse transcription polymerase chain reaction (qRT-PCR), the process generally takes between 7 and 16 h. Given this manageable timeframe, it is both practical and feasible to utilize urinary exosomal miRNAs as biomarkers in daily clinical practice.
The combination of multiple miRNAs demonstrates a higher AUC than that of a single marker (miR-29c), yet lower sensitivity, specificity, PPV, and NPV. This discrepancy raises important considerations in the interpretation of our results. The higher AUC value achieved by the combined miRNA panel underscores the potential synergistic effect of leveraging multiple markers for predictive purposes, enhancing the overall discriminatory power of the model. However, the observed reduction in sensitivity, specificity, PPV, and NPV when transitioning from a single miRNA to a combined panel highlights the trade-off between maximizing AUC and maintaining diagnostic accuracy across all parameters. We recognize the need to further investigate the underlying factors contributing to this phenomenon, including the interplay between different miRNAs, their individual diagnostic strengths, and potential interactions within the combined panel. Additionally, we acknowledge the importance of conducting additional analyses, such as establishing cut-off values in a development set and validating performance in an independent cohort, to refine and validate the diagnostic efficacy of the miRNA panel in the near future.
There are several limitations to this study that should be noted. First, the mean Banff ‘ci’ and ‘ct’ scores were less than 1 in the IFTA group as well. Initially, we attempted to classify the cohort based on the severity of IFTA and ‘cv’ and ‘cg’ scores. However, we encountered limitations: the combined ‘ci’ and ‘ct’ scores exceeded 2 in only two zero-day biopsies. Additionally, the ‘cg’ score was 0 in nearly all zero-day biopsies, with only two exceptions showing a score of 1. In terms of vascular fibrous intimal thickening, the ‘cg’ score was 1 or higher (1 in ten biopsies and 2 in one biopsy) in eleven biopsies within the IFTA group. These observations indicate that further division of the IFTA group by different histological lesions is constrained within our cohort. Therefore, we expect that our findings be validated in an independent cohort classified by these different histological lesions. Second, The follow-up period in this study was only 3 years. Therefore, further observation is necessary to determine the impact of urinary exosomal miRNAs from deceased donors on long-term clinical outcomes including kidney allograft survival and renal function. Third, comprehensive data on 24-h proteinuria or additional urinary biomarkers of tubular injury were unattainable due to variations in the protocols employed for managing each deceased donor at differing centers. Fourth, The small sizes of EVs pose technical challenges in detection and characterization. Additionally, their heterogeneity in biogenesis, size distribution, and molecular cargos complicates their study. Surface proteins such as tetraspanins (CD63 and CD9) are important for EV characterization, but they may form dimers or clusters, adding another layer of complexity. These factors limit the precision and consistency of our methodologies on EV characterization. Lastly, this study did not aim to reveal the mechanism by which these miRNAs influence kidney fibrosis. However, the mechanism by which IFTA is induced could be predicted through target gene network analysis of the verified miRNA. We expect to discover the process by which biomarkers regulate kidney fibrosis in further studies and, eventually, develop new agents to prevent kidney allograft fibrosis. Although it is reasonable to select six miRNA biomarkers using a bioinformatic approach, employing a miRNA array is recommended in the future study. The miRNA array can provide a comprehensive overview of miRNA expression and quantitative expression data. Furthermore, it combines both unbiased discovery and validation in a single step enabling robust statistical analysis and comparison.
In conclusion, our study showed that urinary exosomal miRNAs are promising biomarker candidates for assessing the severity of IFTA in kidneys prior to recovery from a deceased donor.
Methods
Patients, classification, and endpoint
This multicenter, prospective observational study involved four medical centers in Korea, including Asan Medical Center (AMC), Korea University Medicine Anam, Ulsan University Hospital, and Paik Hospital Ilsan. After obtaining written informed consent from all participants, the medical records were assessed. All methods and experimental protocols were approved by the institutional review board of Asan Medical Center (approval number: 2019-0337). All clinical activities in this study were conducted in compliance with the Declaration of Helsinki and the ethical principles stated in the Declaration of Istanbul on Organ Trafficking and Transplant Tourism. Urine specimens were collected from donors at the time of solid organ recovery and prior to transplantation. Zero-day biopsies of the grafts were performed on the back table. Among 141 deceased kidney donors between May 2019 and June 2021 at these centers, 32 cases were excluded due to insufficient zero-day biopsy data (n = 28) or lost urine samples (n = 4). Consequently, a total of 109 cases were included in this study (Fig. 1).
Based on the histologic findings using the Banff 2018 classification, the enrolled cases were classified into two groups. A recipient was classified into the “No IFTA group” if both the ‘ci’ and ‘ct’ scores were 0 in the zero-day biopsy, and into the “IFTA group” if the sum of the ‘ci’ and ‘ct’ scores was 1 or higher. The purpose of this study was to identify specific exosomal miRNAs from donor urine samples that reflect the severity of IFTA. The primary endpoint was the correlation between the levels of selected urine biomarkers and the status of IFTA on the zero-day biopsy. Additionally, we analyzed clinical outcomes such as the estimated glomerular filtration rate (eGFR), biopsy-proven acute rejection (BPAR), allograft survival, and patient survival for both groups. All the rejection episodes were diagnosed using a for-cause biopsy for clinical indications such as progressive proteinuria, elevated serum creatinine, and decreased urine output.
Urine processing
During the recovery procedures, urine samples (≥ 30 mL) were obtained from deceased donors in the operating room, and a protease inhibitor mix (consisting of 4-(2-aminoethyl) benzenesulfonyl fluoride hydrochloride (AEBSF-HCl, Sigma-Aldrich), leupeptin-hemisulfate (Sigma-Aldrich), and NaN3 (Sigma-Aldrich)) was promptly added. The samples were centrifuged at 4000 rpm for 15 min at 4 ℃ to remove urinary sediments. After centrifugation, the supernatants were collected and stored at – 80 ℃ until the extraction of exosomes.
Exosome extraction
Upon thawing 15 mL of frozen urine samples, they were vortexed for one minute and centrifuged at 17,000 g for 15 min at room temperature. The collected supernatant was then subjected to ultracentrifugation at 200,000 g for 70 min at room temperature using a Beckman Coulter Optima L-80xp ultracentrifuge equipped with an SW40Ti rotor (Beckman Coulter; Brea, CA, USA). The supernatant was discarded, and the resulting pellet was dissolved in 11 mL of DPBS for washing. Then, another round of ultracentrifugation was performed at 200,000 × g for 70 min at room temperature. The supernatant was removed, and the exosome pellet was used for miRNA isolation or quantification of exosome particles. Transmission electron microscopy was used to confirm the presence of exosomes. The isolated urinary exosomes were then examined and characterized through nanoparticle tracking analysis using NanoSight NS300 (Malvern Instruments Ltd., UK) and Western blot analysis47. The blots were probed with primary antibodies including TSG101, HSP70, CD9, CD63 (Exosome panel kit, Ab275018, Abcam, Cambridge, UK), and Actin (A3854, Sigma-Aldrich).
RNA isolation
Total RNA was extracted using the miRNeasy Mini Kit from Qiagen, according to the manufacturer's instructions. RNA quality was assessed using the Agilent 2100 Bioanalyzer with the RNA 6000 Pico Chip (Agilent Technologies, Amstelveen, The Netherlands). RNA quantification was performed using the NanoDrop 2000 Spectrophotometer system (Thermo Fisher Scientific, Waltham, MA, USA).
Quantitative real-time PCR
The expression of miRNAs in urinary exosomes was validated by qRT-PCR. To quantify microRNAs, a TaqMan RT-PCR assay was used in this experiment. The process involved isolating total RNA, followed by reverse transcription of RNA using the Applied Biosystems™ TaqMan™ Advanced miRNA cDNA Synthesis Kit. The TaqMan RT-PCR assay was performed in duplicate using TaqMan™ Advanced miRNA Assays and TaqMan™ Fast Advanced Master Mix. To ensure the accuracy of the results, the expression of hsa-miR-16-5p was used as an endogenous control for normalization48,49. The relative expression levels of miRNAs were calculated using the comparative 2−△△Ct method. The primers used for RT-PCR are provided in Supplementary Table 1.
Human miRNA–disease association dataset
The human miRNA–disease association dataset was downloaded from the HMDD database (version 4.0) (http://www.cuilab.cn/hmdd/). The HMDD database is a collection of experimentally verified evidence for associations between human miRNAs and diseases.
Statistical analysis
Categorical variables were assessed using the chi-squared test, while continuous variables were examined with either the Student’s t-test or the Wilcoxon rank-sum test. The unpaired Mann–Whitney test was used to assess differences in miRNA expression levels between the two groups. We compared the patterns of eGFR improvement over time using a linear mixed model. Patient and graft survival were estimated using Kaplan–Meier survival estimates and assessed with the log-rank test.
Statistical analyses were conducted using IBM SPSS Statistics for Windows, version 28.0 (https://www.ibm.com/products/spss-statistics) (IBM Corp., Armonk, NY, USA), and GraphPad Prism, version 9.4.1 (https://www.graphpad.com) (GraphPad Software Inc., Boston, MA, USA). A significance level of P < 0.05 was considered statistically significant. Receiver operating characteristic (ROC) curves were calculated using MedCalc Statistical Software, version 20.106 (https://www.medcalc.org/download.php) (MedCalc Software Ltd., Ostend, Belgium).
Data availability
The datasets used and/or analyzed in the present study can be obtained upon reasonable request from the corresponding author, who can be contacted at sshin@amc.seoul.kr.
References
Wolfe, R. A. et al. Comparison of mortality in all patients on dialysis, patients on dialysis awaiting transplantation, and recipients of a first cadaveric transplant. N. Engl. J. Med. 341, 1725–1730 (1999).
Merion, R. M. et al. Deceased-donor characteristics and the survival benefit of kidney transplantation. Jama 294, 2726–2733 (2005).
Halloran, P. F. Immunosuppressive drugs for kidney transplantation. N. Engl. J. Med. 351, 2715–2729 (2004).
Waichi, W., Venetz, J.-P., Tolkoff-Rubin, N. & Pascual, M. Inmunosuppressive strategies in kidney transplantation: Which role for the calcineurin inhibitors. [Estrategias Inmunosupresoras en el trasplante renal: El papel de los inhibidores de la calcineurina]. Transplantation 80, 289–296 (2005).
Bae, S. et al. Changes in discard rate after the introduction of the Kidney Donor Profile Index (KDPI). Am. J. Transplant. 16, 2202–2207 (2016).
Gaber, L. W. et al. Glomerulosclerosis as a determinant of posttransplant function of older donor renal allografts. Transplantation 60, 334–338 (1995).
Ibernon, M. et al. Donor structural and functional parameters are independent predictors of renal function at 3 months. Transplant. Proc. 39, 2095–2098. https://doi.org/10.1016/j.transproceed.2007.06.026 (2007).
Lopes, J. A. et al. Evaluation of pre-implantation kidney biopsies: Comparison of Banff criteria to a morphometric approach. Kidney Int. 67, 1595–1600. https://doi.org/10.1111/j.1523-1755.2005.00241.x (2005).
Bartel, D. P. MicroRNAs: Target recognition and regulatory functions. Cell 136, 215–233 (2009).
Bartel, D. P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 116, 281–297 (2004).
Krol, J., Loedige, I. & Filipowicz, W. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 11, 597–610 (2010).
Turchinovich, A., Weiz, L. & Burwinkel, B. Extracellular miRNAs: The mystery of their origin and function. Trends Biochem. Sci. 37, 460–465 (2012).
Mall, C., Rocke, D. M., Durbin-Johnson, B. & Weiss, R. H. Stability of miRNA in human urine supports its biomarker potential. Biomark. Med. 7, 623–631 (2013).
Karpman, D., Ståhl, A. L. & Arvidsson, I. Extracellular vesicles in renal disease. Nat. Rev. Nephrol. 13, 545–562. https://doi.org/10.1038/nrneph.2017.98 (2017).
Kwon, S. H. Extracellular vesicles in renal physiology and clinical applications for renal disease. Korean J. Intern. Med. 34, 470–479. https://doi.org/10.3904/kjim.2019.108 (2019).
Wang, Y. & Zhang, M. Urinary exosomes: A promising biomarker for disease diagnosis. Lab. Med. 54, 115–125. https://doi.org/10.1093/labmed/lmac087 (2023).
Condrat, C. E. et al. miRNAs as biomarkers in disease: Latest findings regarding their role in diagnosis and prognosis. Cells 9, 276 (2020).
Aomatsu, A. et al. MicroRNA expression profiling in acute kidney injury. Transl. Res. 244, 1–31 (2022).
Van de Vrie, M., Deegens, J., Eikmans, M., van der Vlag, J. & Hilbrands, L. Urinary microRNA as biomarker in renal transplantation. Am. J. Transplant. 17, 1160–1166 (2017).
Amrouche, L. et al. MicroRNA-146a in human and experimental ischemic AKI: CXCL8-dependent mechanism of action. J. Am. Soc. Nephrol. 28, 479–493 (2017).
Soltaninejad, E. et al. Altered expression of microRNAs following chronic allograft dysfunction with interstitial fibrosis and tubular atrophy. Iran. J. Allergy Asthma Immunol. 2015, 615–623 (2015).
Kim, J. Y. et al. Urinary transglutaminase 2 as a potent biomarker to predict interstitial fibrosis and tubular atrophy of kidney allograft during early posttransplant period in deceased donor kidney transplantation. Ann. Surg. Treatment Res. 97, 27–35 (2019).
Yu, W. K. et al. Nintedanib inhibits endothelial mesenchymal transition in bleomycin-induced pulmonary fibrosis via focal adhesion kinase activity reduction. Int. J. Mol. Sci. 2022, 23. https://doi.org/10.3390/ijms23158193 (2022).
Zhao, X. K. et al. Focal adhesion kinase regulates hepatic stellate cell activation and liver fibrosis. Sci. Rep. 7, 4032. https://doi.org/10.1038/s41598-017-04317-0 (2017).
Leask, A. Focal adhesion kinase: A key mediator of transforming growth factor beta signaling in fibroblasts. Adv. Wound Care (New Rochelle) 2, 247–249. https://doi.org/10.1089/wound.2012.0363 (2013).
Böhmig, G. & Regele, H. Diagnosis and treatment of antibody-mediated kidney allograft rejection. Transplant. Int. 16, 773–787. https://doi.org/10.1007/s00147-003-0658-3 (2003).
Loupy, A. & Lefaucheur, C. Antibody-mediated rejection of solid-organ allografts. N. Engl. J. Med. 379, 1150–1160. https://doi.org/10.1056/NEJMra1802677 (2018).
Seron, D. et al. Proposed definitions of T cell-mediated rejection and tubulointerstitial inflammation as clinical trial endpoints in kidney transplantation. Transplant. Int. 35, 10135. https://doi.org/10.3389/ti.2022.10135 (2022).
Schaub, S. et al. Proteomic-based detection of urine proteins associated with acute renal allograft rejection. J. Am. Soc. Nephrol. 15, 219–227 (2004).
Cheng, O. et al. Connective tissue growth factor is a biomarker and mediator of kidney allograft fibrosis. Am. J. Transplant. 6, 2292–2306 (2006).
Fu, G., Brkić, J., Hayder, H. & Peng, C. MicroRNAs in human placental development and pregnancy complications. Int. J. Mol. Sci. 14, 5519–5544 (2013).
Théry, C. Exosomes: Secreted vesicles and intercellular communications. F1000 Biol. Rep. 2011, 3 (2011).
Théry, C., Zitvogel, L. & Amigorena, S. Exosomes: Composition, biogenesis and function. Nat. Rev. Immunol. 2, 569–579 (2002).
Ho, P. T., Clark, I. M. & Le, L. T. MicroRNA-based diagnosis and therapy. Int. J. Mol. Sci. 23, 7167 (2022).
Wang, G. et al. Urinary miR-21, miR-29, and miR-93: Novel biomarkers of fibrosis. Am. J. Nephrol. 36, 412–418 (2012).
Lv, C.-Y. et al. A PEG-based method for the isolation of urinary exosomes and its application in renal fibrosis diagnostics using cargo miR-29c and miR-21 analysis. Int. Urol. Nephrol. 50, 973–982 (2018).
Kang, Z. et al. Remote ischemic preconditioning upregulates microRNA-21 to protect the kidney in children with congenital heart disease undergoing cardiopulmonary bypass. Pediatr. Nephrol. 33, 911–919 (2018).
Larrue, R. et al. The versatile role of miR-21 in renal homeostasis and diseases. Cells 11, 3525 (2022).
Saikumar, J. et al. Expression, circulation, and excretion profile of microRNA-21,-155, and-18a following acute kidney injury. Toxicol. Sci. 129, 256–267 (2012).
Du, J. et al. MicroRNA-21 and risk of severe acute kidney injury and poor outcomes after adult cardiac surgery. PloS One 8, e63390 (2013).
Chen, C. et al. Urinary miR-21 as a potential biomarker of hypertensive kidney injury and fibrosis. Sci. Rep. 7, 17737 (2017).
Zhao, S. et al. Exosomal miR-21 from tubular cells contributes to renal fibrosis by activating fibroblasts via targeting PTEN in obstructed kidneys. Theranostics 11, 8660 (2021).
Feng, W. et al. miR-29c inhibits renal interstitial fibrotic proliferative properties through PI3K-AKT pathway. Appl. Bionics Biomech. 2022, 1–8 (2022).
Fang, Y. et al. miR-29c is downregulated in renal interstitial fibrosis in humans and rats and restored by HIF-α activation. Am. J. Physiol.-Renal Physiol. 304, F1274–F1282 (2013).
Zhang, Y., Zhang, G.-X., Che, L.-S., Shi, S.-H. & Lin, W.-Y. miR-19 promotes development of renal fibrosis by targeting PTEN-mediated epithelial-mesenchymal transition. Int. J. Clin. Exp. Pathol. 13, 642 (2020).
Zhang, Y. et al. MiR-205 influences renal injury in sepsis rats through HMGB1-PTEN signaling pathway. Eur. Rev. Med. Pharmacol. Sci. 23, 10950–10956 (2019).
Street, J. M., Koritzinsky, E. H., Glispie, D. M. & Yuen, P. S. Urine exosome isolation and characterization. Drug Saf. Eval. Methods Protocols 2017, 413–423 (2017).
Lange, T. et al. Identification of miR-16 as an endogenous reference gene for the normalization of urinary exosomal miRNA expression data from CKD patients. PLoS One 12, e0183435. https://doi.org/10.1371/journal.pone.0183435 (2017).
Wang, X. Y. et al. Evaluation of the performance of serum miRNAs as normalizers in microRNA studies focused on cardiovascular disease. J. Thorac. Dis. 10, 2599–2607. https://doi.org/10.21037/jtd.2018.04.128 (2018).
Funding
This work was supported by the Korea Science and Engineering Foundation (KOSEF) grant funded by the Korea government (NRF-2021R1G1A1004361) and the Asan Institute for Life Sciences, Asan Medical Center, Seoul, Korea (2020IP0063-1).
Author information
Authors and Affiliations
Contributions
S.S., J.G., S.P., and H.J. conceived the study design and analysis plan. M.K., H.E.K., H.J., and J.K. collected data and performed experiments. M.K., H.E.K., J.L., J.J., Y.K., H.K., and Y.K. performed data analysis and generated all figures and tables. S.S., J.G., S.P., and H.J. drafted and revised the manuscript. S.S., J.G., S.P., and H.J. approved the final version of the manuscript. S.S. supervised the work.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Kim, M.J., Kwon, H.E., Jang, HW. et al. Multicenter, prospective, observational study for urinary exosomal biomarkers of kidney allograft fibrosis. Sci Rep 14, 20319 (2024). https://doi.org/10.1038/s41598-024-71279-5
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-024-71279-5