Abstract
Accurate segmentation of COVID-19 lesions from medical images is essential for achieving precise diagnosis and developing effective treatment strategies. Unfortunately, this task presents significant challenges, owing to the complex and diverse characteristics of opaque areas, subtle differences between infected and healthy tissue, and the presence of noise in CT images. To address these difficulties, this paper designs a new deep-learning architecture (named MD-Net) based on multi-scale input layers and dense decoder aggregation network for COVID-19 lesion segmentation. In our framework, the U-shaped structure serves as the cornerstone to facilitate complex hierarchical representations essential for accurate segmentation. Then, by introducing the multi-scale input layers (MIL), the network can effectively analyze both fine-grained details and contextual information in the original image. Furthermore, we introduce an SE-Conv module in the encoder network, which can enhance the ability to identify relevant information while simultaneously suppressing the transmission of extraneous or non-lesion information. Additionally, we design a dense decoder aggregation (DDA) module to integrate feature distributions and important COVID-19 lesion information from adjacent encoder layers. Finally, we conducted a comprehensive quantitative analysis and comparison between two publicly available datasets, namely Vid-QU-EX and QaTa-COV19-v2, to assess the robustness and versatility of MD-Net in segmenting COVID-19 lesions. The experimental results show that the proposed MD-Net has superior performance compared to its competitors, and it exhibits higher scores on the Dice value, Matthews correlation coefficient (Mcc), and Jaccard index. In addition, we also conducted ablation studies on the Vid-QU-EX dataset to evaluate the contributions of each key component within the proposed architecture.
Similar content being viewed by others
Introduction
The coronavirus disease 2019 (COVID-19) has emerged as an unparalleled global health crisis that poses a serious threat to human life and well-being. Since its initial appearance, the virus has spread rapidly across all continents and caused significant damage to countries around the world. This highly contagious disease poses a major challenge to public health systems, with symptoms including fever, cough, fatigue and respiratory distress, often accompanied by gastrointestinal disorders such as nasal congestion, rhinorrhea and diarrhea. If we can detect infected people in a timely manner, it will help curb the spread of novel coronavirus pneumonia, which requires an extremely sensitive and effective screening method that can identify both symptomatic cases and asymptomatic infected people and their close contacts. Currently, reverse transcription polymerase chain reaction (RT-PCR) is the gold standard for diagnosing COVID-19. Despite its widespread use, RT-PCR still has its limitations, including low sensitivity, high false-negative rates, and inability to detect the virus comprehensively. Moreover, the long turnaround time of RT-PCR results exacerbates the challenge of epidemic control, leaving health care workers vulnerable to infection during sampling and testing. Conversely, computed tomography (CT) technology offers a promising alternative for COVID-19 detection, leveraging its superior spatial resolution and detection efficiency. By analyzing characteristic lung images, such as subtle ground-glass opacities, interstitial changes, and bilateral lung infiltrates, CT scans provide valuable insights into disease progression and severity. However, the interpretation of CT images by radiologists presents multiple challenges, including a heavy workload, vulnerability to human error, and resource constraints exacerbated by a shortage of experienced professionals during the pandemic. In response to these problems, it is particularly important to train the detection model based on deep learning technology and develop an effective auxiliary diagnostic system to assist professionals in the automatic analysis of CT images.
Currently, deep-learning technology has been at a relatively advanced level in the field of image segmentation. For example, convolutional neural networks (CNNs) feed the original image into the network and uses convolutional operations to carefully extract complex features embedded in the image. Subsequently, the network proceeds to assign class labels to each pixel block to produce the final split output. This property gives CNNs the ability to achieve segmentation with amazing precision and accuracy, but it also imposes a huge computational burden. Unlike traditional CNNs, full convolutional network (FCN)1 replaces fully connected layers with full convolutional layers, allowing them to seamlessly process input images of any size. However, the upsampling of FCN uses a large multiple, which may unintentionally damage the ability of the network to effectively integrate the context feature information, and reduce the accuracy of image segmentation. Building on the foundation laid by FCN, Ronneberger et al.2 proposed the pioneering U-Net using the symmetrical codec-decode structure. At its core, U-Net employs a coding structure comprising convolutional layers and pooling layers to extract features from input images. In the decoding stage, the encoded features undergo a process of recovery facilitated by up-sampling operations, effectively reconstructing the image’s salient characteristics. Crucially, U-Net incorporates skip connections that facilitates the fusion of feature information from multiple levels within the network architecture. This efficiency not only streamlines the training process but also mitigates resource constraints, making U-Net an appealing choice for medical image segmentation tasks.
Although U-Net has demonstrated strong performance in medical image segmentation, it faces challenges in meeting the increasingly demanding requirements of modern medical applications. One of the key limitations lies in the restricted receptive field of convolutional layers, which hampers their ability to capture global context information essential for accurate segmentation. Additionally, the integration of features at different scales remains a complex task, especially when dealing with medical images that often exhibit varying anatomical structures and lesion sizes. To tackle these challenges, various advanced technologies are integrated with deep learning framework, such as self-supervised learning, contrastive learning, and transfer learning3,4,5,6,7. Among them, You et al.8 presented a new framework called CASTformer, which develops a class-aware transformation block that identifies and learns discriminant regions based on the semantic structure of an object. In addition, the authors introduced adversarial training strategies that enable transform-based discriminators to capture a rich mix of high-level semantic content and low-level anatomical details. Jin et al.9 introduced a novel two-stage network segmentation model that leverages a pseudo mask-guided feature aggregation technique. Instead of introducing extra components to handle uncertainty, the approach incorporated an uncertainty regularization strategy, which streamlines the process and reduces computational complexity. You et al.10,11 further introduced a contrastive voxel-wise representation learning approach aimed at improving the network’s capacity to effectively capture both low-level and high-level features. By leveraging detailed background information and rich anatomical structures, this method enhances the network’s ability to discriminate between different features across diverse scenarios. Additionally, it offers greater robustness against representation collapse, ensuring consistent performance and more reliable feature learning. Subsequently, You et al.12 introduced a comprehensive implicit neural rendering framework designed to enhance the process of medical image segmentation. This framework aims to improve the accuracy of segmentation by continuously aligning initial, coarse predictions with fuzzy representations derived from coordinate-based point data.
Additionally, various U-Net variants13,14,15,16,17 have been proposed and successfully applied to the segmentation of COVID-19 lesions. Among them, Chen et al.18 introduced a groundbreaking cascading architecture, which utilized the synergy of boundary monitoring, multi-scale attribute convolution, and dual attention mechanisms to achieve high-precision and efficient segmentation of COVID-19 lung infections. Building upon the foundational U-Net, Zhou et al.19 introduced a transformer module to address the challenge of capturing global context while maintaining the U-Net’s efficacy in handling local features. Drawing inspiration from the biological vision, Zhao et al.20 devised an innovative approach centered around spatial and channel-based attention networks. By designing specialized block for spatial intelligence and channel-based attention, it can extract pertinent features from areas afflicted by opacity at both the pixel and channel levels. To refine the complex segmentation process in medical imaging, Devi et al.21 introduced a pioneering framework specifically designed to segment COVID-19 lung infections. The approach is innovative in the strategic integration of these multi-special blocks with convolutional blocks at the encoder and bridge phases of the architecture, so that context clues and COVID-19 infection-specific characteristics can be fully exploited. To address the challenge posed by the ambiguity in both the shape and positioning of COVID-19 lesion areas, Liu et al.22 introduced a novel approach grounded in multi-scale representation learning, which provides a comprehensive solution to the problems encountered in identifying and characterizing COVID-19 lesion areas. Saha et al.23 utilized the power of deep learning to develop a deep neural network architecture for predicting COVID-19 that integrates deep supervision principles and strategically combines attention mechanisms among encoder, skip connection, and decoder components for dynamically regulating information flow and allocating focus to significant areas in the image. This attention mechanism not only enhances the network’s ability to recognize relevant features, but also effectively balances the integration of high and lower level functional components, thereby optimizing overall performance and segmentation accuracy. Fan et al.24 introduced a groundbreaking deep learning framework, known as Inf-Net. First, it uses parallel partial decoders to aggregate high-level features, which helps in generating comprehensive global feature maps. Next, two distinct attention mechanisms focuse on refining the core regions of infection, while the explicit edge attention sharpens the boundary delineation. This approach significantly reduces the need for large amounts of labeled data by allowing the network to primarily rely on unlabeled images while still maintaining high segmentation accuracy.
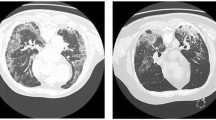
Despite the continuous development of various innovative algorithms, researchers encounter significant challenges in effectively segmenting COVID-19 infected areas within lung CT images. Figure 1 illustrates chest CT scans from three different categories: normal, COVID-19 infection, and other diseases infections (such as non-COVID-19 infections, pneumonia and pulmonary edema). These non-COVID-19 infections and abnormal lung manifestations are often visually similar to COVID-19 infections, complicating the task of accurately segmenting affected areas in medical images. Therefore, the main challenges in segmenting such lesions arise from the following factors: (1) The infected areas are scattered rather than concentrated, and they are distributed in different areas of the lungs, which creates a big barrier to accurate detection. (2) There are many disjoint boundaries within the infected area, which complicates the task of segmentation. However, it is often difficult for traditional algorithms to produce clear and distinct boundaries, which leads to fuzzy segmentation results. (3) The complexity of the lung CT image background brings more complications because it is susceptible to various disturbances, such as non-COVID-19 inflamed areas, which adds another layer of complexity to the segmentation task. Therefore, how to solve these multifaceted problems is crucial to improve the accuracy and reliability of COVID-19 lung infection segmentation in CT imaging.
In response to address the above challenges, we introduced a new deep-learning architecture called MD-Net specifically designed to segment COVID-19 lesions. The basic goal of our framework is to integrate multi-scale input layers with a dense decoder aggregation network to increase the segmentation accuracy of lesion region by taking the advantages of both approaches. Our contributions are as follows:
-
(1)
The multi-scale input layers are integrated into the architecture to allow a thorough examination of fine-grained details and contextual information in the input image. This allows MD-Net to capture a wider range of features, enhancing its ability to distinguish between COVID-19 lesions and surrounding tissue.
-
(2)
The SE-Conv module is introduced into the encoder network to facilitate the identification of relevant lesion information while attenuating the transmission of irrelevant data. By selectively focusing salient features and suppressing noise, the module contributes to the overall robustness and specificity of the segmentation process.
-
(3)
A dense decoder aggregation module is presented, which effectively integrates the feature distribution and critical damage information of adjacent encoders. This module helps to integrate information extracted from different scales and spatial locations, making segmentation results more consistent and accurate.
Methods
In this section, we embark on a thorough exploration of the proposed MD-Net, delving into its intricacies to offer a comprehensive understanding. First, we delineate the overarching architecture of the network, and then the key modules are described to illustrate their capabilities and contributions. Finally, we will introduce the loss function.
Overall architecture
Among the various architectures used in medical image segmentation tasks, U-Net model has become the most widely used and influential model, which uses convolutional blocks as the main components of feature extraction and spatial resolution recovery in both its encoder and decoder paths. However, due to the fixed kernel size in convolution, it is difficult to simulate remote dependencies and global contexts in images, especially in COVID-19 images. In addition, skipped connections in U-Net and its variants often involve directly adding or connecting corresponding feature maps from different levels and may not take full advantage of the complementary information that exists at various scales and depths of the network because they lack a global view of feature cross-fusion. Based on the above principles, we propose a new deep-learning architecture based on multi-scale input layers and dense decoder aggregation network for COVID-19 lesion segmentation, as shown in Fig. 2. The MD-Net consists of a 5-layer encoder on the left and a 4-layer decoder on the right, each component to fulfill a specific role in the overall framework of the network. As the initial stage of information processing, encoders are tasked with extracting and abstracting significant features from input images through a series of hierarchical convolution and pooling operations. Instead, the decoder works in tandem with the encoder, aiming to reconstruct spatial information and semantic context from the extracted features. Compared to the traditional U-Net, an important innovation of MD-Net is the integration of a dense decoder aggregation (DDA) module in the jump connection. The DDA module performs meticulous analysis and fusion of feature maps from multiple encoder layers, enabling the network to capture complex spatial relationships and semantic nuances that are critical for accurate lesion detection and analysis. In addition, MD-Net further distinguishes itself by adopting the SE-Conv module, which differs from traditional convolution layers. The SE-Conv module gives MD-Net the ability to adaptively emphasize the information channel while attenuating the effects of redundant or noisy signals, thereby enhancing the discrimination and robustness of the network. To enhance the fine-grained detail and contextual information inherent in the original image, MD-Net introduces a multi-scale input layers as a key component of its architecture. Whereas traditional single-scale input configurations may miss key details or fail to capture contextual nuances, the multi-scale input layers enable MD-Net to process original images at multiple resolutions simultaneously. By adapting to different scales of input, the network can fully understand the spatial hierarchy and semantic relationships embedded in the input data. With these improvements, MD-Net strives to achieve a delicate balance between feature abstraction and information reconstruction, making it stand out in a variety of image segmentation tasks.
Multi-scale input layers
Multi-scale input is a complex technique for feeding images of different scales into a neural network, which is good at identifying nuances and subtleties in the input images. In the field of semantic segmentation, a large number of studies have proved the effectiveness of multi-scale input25,26 to improve segmentation quality. This technique not only refines the description of an object, but also ensures a fuller understanding of the object’s spatial background and structural complexity. In this paper, multi-scale images are input into each layer of MD-Net network to make up for the lost feature information in the process of feature extraction. Initially, the input image undergoes a series of averaging pooling operations, down-sampling it to 1/2, 1/4 and 1/8 of its original size. Then SE-Conv operation is used to make the input image and the feature matrix of each layer have the same number of channels. Finally, the two feature matrices are fused to make up for the image information and suppress the redundancy of the input information.
SE-Conv module
The classical encoder–decoder architecture often faces limitations in its acceptable domain, which leads to two significant shortcomings. One is that it tends to produce locally constrained features. The second is that the broader context has been ignored. Taking inspiration from the work of Szegedy et al.27, we introduce an innovative SE-Conv module to replace the traditional double-convolution structure, as shown in Fig. 3. Firstly, the SE-Conv module employs a sequence of 3 \(\times\) 3 convolution, 3 \(\times\) 3 depthwise convolution, and 1 \(\times\) 1 pointwise convolution operations to generate feature maps spanning various spatial domains. Among them, the 3 \(\times\) 3 convolution is effective at capturing spatial dependencies and small patterns in the input data, and it preserves spatial resolution to a certain extent. The depthwise convolution applies the 3 \(\times\) 3 filter to each input channel separately, which reduces computational complexity while still allowing the model to capture local dependencies within each channel. The pointwise convolution uses a 1 \(\times\) 1 kernel to combine the output from depthwise convolution across channels, thus integrating information across channels without affecting the spatial resolution. Notably, the outputs of each convolution operation are concatenated along the channel dimension, yielding multi-scale feature maps that encapsulate both local details and global context. Moreover, we integrated the squeeze-and-excitation (SE) block into the SE-Conv module to selectively inhibit irrelevant features and amplify feature expression, as shown in Fig. 4. This strategic enhancement contributes significantly to the model’s discriminative power and efficacy in capturing intricate data patterns. Finally, to mitigate the issue of gradient vanishing stemming from network depth, we introduce residual structures within the SE-Conv module. These residual connections facilitate smoother gradient flow, thereby mitigating the adverse effects of network depth on training stability. The formula below serves as its representation of the above process.
Overall, the proposed SE-Conv module offers a comprehensive solution to the limitations of traditional encoder–decoder architectures. By employing a sequence of convolutional operations, incorporating the SE block, and introducing residual connections, SE-Conv module not only ensures robust feature extraction but also alleviates the inherent challenges posed by deep network architectures.
Dense decoder aggregation module
Numerous experiments have shown that the shallow layer of the network is good at capturing complex local details using its high-resolution and small receptive domain feature maps, while the deeper layer uses its broader receptive domain to extract semantic context and understand global patterns in input images. To make the most of the above features, we introduced an innovative approach called a dense decoder aggregation (DDA) module, as shown in Fig. 5. Unlike traditional skip connection, DDA modules employ a more complex mechanism for feature integration and optimization. In our method, the high-level features and low-level features are separately fed into the SE module, which dynamically recalibrates their importance based on channel-wise relationships. By intelligently fusing these complementary features, we ensure that crucial information is retained throughout the encoding process. Following the fusion stage, the feature maps undergo further refinement through 3 \(\times\) 3 convolution with expansion rates of 2 and 4, and subsequently joined along the channel dimensions, facilitating the integration of multi-scale information. To further refine the obtained results, a final convolution operation with a 3 \(\times\) 3 kernel is applied. The calculation of the above process is described below.
In summary, our approach leverages the SE module and a series of carefully orchestrated convolutional operations to mitigate information loss while enhancing feature representation and producing accurate output results.
Loss function
Selecting an appropriate loss function is crucial for optimizing machine learning models, as it directly influences the model’s ability to minimize the disparity between predicted and actual values. In our approach, we employed the Dice loss28,29 function to facilitate pixel-level binary classification, aiming to accurately identify and segment COVID-19 lesions within medical imaging data. The Dice loss is defined as follows:
where N stands for the total number of pixels in the segmentation mask or image, \({{\hat{y}}_{i}}\) symbolizes the true probability value assigned to pixel i , and \({{y}_{i}}\) denotes the predicted probability value assigned to the same pixel i by the segmentation model under evaluation.
Experimental results
Description of datasets
In assessing the efficacy of our proposed method, we conducted a rigorous evaluation using two widely recognized COVID-19 lesions segmentation datasets: Vid-QU-EX30 and QaTa-COV19-v231. These datasets, renowned for their comprehensive coverage and diverse array of COVID-19 lesions images, serve as invaluable benchmarks for gauging the performance and generalizability of our approach. Table 1 provides a detailed overview of the specifications associated with each dataset, including the number of images, resolution, and any pertinent metadata crucial for contextualizing the experimental results. In addition, to more intuitively understand the content and variability of these datasets. Figure 6 provides researchers with tangible examples of the COVID-19 lesions images used in our evaluation.
Vid-QU-EX
The researchers of Qatar University have compiled the Vid-QU-EX dataset specifically to address the urgent need for a comprehensive dataset in the context of COVID-19. It consists of 1864 training images, 466 validation images and 583 test images, and each image within the dataset is imbued with a wealth of information. Researchers seeking to delve deeper into the dataset’s intricacies and specifications can avail themselves of detailed information accessible via the provided link: https://www.kaggle.com/datasets/anasmohammedtahir/covidqu.
QaTa-COV19-v2
A collaborative effort between the esteemed researchers at Qatar University and Tampere University has yielded the QaTa-COV19-v2 dataset, which consists of 5359 training images, 1786 validation images and 2113 test images. This dataset represents a paradigm shift with its COVID-19 chest X-ray images to encompass the wide range of manifestations and changes observed in clinical practice. Unlike previous iterations, this dataset introduces a breakthrough feature, including a true segmentation mask for the COVID-19 pneumonia segmentation task. These masks serve as valuable annotations, providing pixel-scale descriptions of COVID-19 pneumonia lesions in chest X-ray images. Detailed information is available at: https://www.kaggle.com/datasets/aysendegerli/qatacov19-dataset/data.
Implementation details
Our research work is built on the powerful PyTorch platform, a dynamic framework renowned for its versatility and scalability in deep learning tasks. To execute our experiments with precision and efficiency, we harnessed the computational prowess of an NVIDIA Quadro RTX 6000 graphics card, equipped with a 24 GPU memory capacity that underpins the computational demands of our methodologies. Prior to commencing the training process, we meticulously prepared our dataset through a series of rigorous pre-processing procedures, and the images were cropped into small pieces of size 256 \(\times\) 256 that were served as the foundational inputs to our method. During the training phase, we employed the Adam optimizer, a state-of-the-art optimization algorithm revered for its efficacy in converging towards optimal solutions. With hardware limitations in mind, the initial learning rate is set to 10-3, the batch-size to 32 and the epochs to 250. Figure 7 illustrates the fluctuation of loss and accuracy values throughout the iterative training and verification process. The training loss consistently decreases over time, indicating that the model is progressively learning from the data. It starts around 0.5 and declines steadily, reaching very low values (approximately 0.05) by the 250th epoch. The training accuracy rises quickly in the initial epochs, reaching about 95% accuracy after only 50 epochs. It continues to improve slightly as training progresses, reaching close to 99% towards the end of training. This indicates that the model is learning to classify the training data correctly with high precision. However, the validation loss and validation accuracy show a more fluctuating behavior, especially in the earlier epochs. Despite these fluctuations, the validation accuracy still stabilizes at around 95%, which shows relatively strong performance.
Evaluation metrics
To ensure a rigorous and unbiased assessment of our model’s performance, we employed a comprehensive suite of three key performance evaluation metrics: the Dice value32,33, Matthews correlation coefficient34,35, and Jaccard index36,37. These metrics serve as indispensable yardsticks for quantitatively gauging the efficacy and accuracy of our model’s predictions across various tasks and datasets, which are defined as:
where TP and TN indicate instances correctly identified as positive and negative, FP and FN refer to instances incorrectly classified as positive and negative.
Ablation studies
Table 2 provides a comprehensive overview of the meticulous ablation experiment carried out on the Vid-QU-EX dataset. The evaluation criteria encompass Dice value, Matthews correlation coefficient, and Jaccard index, indicative of segmentation accuracy. The baseline model, represented by the U-Net architecture, serves as the foundation for comparison against a series of augmented models. The baseline model, represented by the U-Net architecture, serves as the foundation for comparison against a series of augmented models. Notably, the addition of SE-Conv, designed to recalibrate channel-wise feature responses, yields a noticeable improvement across all metrics. Similarly, MIL aims to help further improve performance by resolving label noise and ambiguity through instance-level monitoring. In addition, DDA facilitates adaptive enhancement strategies tailored to data set features. However, the most compelling findings emerge from the synergistic integration of these methodologies. The combination of Baseline+MIL+DDA+SE-Conv recorded the highest score in segmentation accuracy: Dice value was 0.8425, Mcc was 0.8176, and Jaccard index was 0.7292. This nuanced analysis not only validates the effectiveness of individual strategies, but also underscores the importance of their cohesive integration in advancing the latest semantic segmentation.
Furthermore, we present the visualization outcomes stemming from our meticulous module ablation experiment. As depicted in Fig. 8, the first row is the images of the test set, and the second row is the corresponding ground-truth label images. The third to last rows are the predicted segmentation visual renderings after the introduction of MIL, SE-Conv, DDA, MIL+DDA, MIL+SE-Conv, SE-Conv+DDA, MIL+DDA+SE-Conv, respectively. Through detailed comparative analysis of these segmentation visualizations, it is clear that the segmentation effect of the MD-Net network model is significantly better than that of the basic U-Net backbone network and the combination of MIL, SE-Conv and DDA networks. In addition, MD-Net shows commendable adaptability in handling complex and challenging scenes characterized by low contrast and blurred boundaries. This adaptability is due to MD-Net’s inherent ability to seamlessly blend deep and shallow features extracted from feature maps in its decoding structure, thus helping to obtain more precise target region boundaries.
Visualization of ablation results on the Vid-QU-EX dataset. (a,b) original images and corresponding gold labels on the Vid-QU-EX dataset. (c–j) are the results of Baseline, Baseline+MIL, Baseline+SE-Conv, Baseline+DDA, Baseline+MIL+DDA, Baseline+MIL+SE-Conv, Baseline+SE-Conv+DDA, Baseline+MIL+DDA+SE-Conv.
Comparisons with the state-of-the-art methods
To validate the effectiveness of our proposed method in accurately segmenting infected areas within CT images, we conducted a comprehensive evaluation using various models on the Vid-QU-EX dataset. The comparison networks included U-Net, Attention-U-Net, DCANet, M-Net, DCSAU-Net, MCDAU-Net, META-Unet, MSRAformer, Swin-Transformer, MCAFNet, MDUNet, and DualA-Net, all of which were conducted under the same experimental environment. Following 250 iterations of training with meticulously processed COVID-19 datasets, we conducted rigorous testing and compared the segmentation results of different networks based on meticulously recorded numerical evaluation indicators. As shown in Table 3, U-Net initially displayed commendable performance across all metrics, boasting Dice score of 0.8265, Mcc of 0.7992, and Jaccard index of 0.7051. When analyzing the performance metrics, it is evident that MSRAformer, Swin-Transformer, and DualA-Net consistently underperform in comparison to the traditional U-Net across several key evaluation measures. Attention-U-Net, DCANet, M-Net, DCSAU-Net, MCDAU-Net, META-Unet, MCAFNet, and MDUNet outperform U-Net across all metrics, with improvements in Dice, MCC, and Jaccard index. However, the MD-Net achieves the best results in the three evaluation indicators of Dice score, Mcc and Jaccard index, which indicated that the segmentation results of the MD-Net had a high similarity with the real labeled lesion areas. Moreover, the boundary similarity between the segmentation results and the real labeled areas was also high. Notably, our MD-Net demonstrates robust capabilities in accurately identifying COVID-19 lesion areas, and even has decent segmentation performance for accurately delineating smaller areas.
In order to compare each model more clearly, we made a visual analysis of the segmentation results, as shown in Fig. 9. In the COVID-19 lesion segmentation task, the U-Net network has obvious over-segmentation problems, resulting in rough edges, uneven contours, and insufficient placement of details. Taking inspiration from the effectiveness of the attention mechanism, Attention-U-Net managed to achieve performance comparable to U-Net. However, despite this improvement, U-Net and Attention-U-Net still fall short in providing satisfactory segmentation results. Transformer models such as MSRAformer and Swin-Transformer excel at capturing global context information through remote dependencies in the image. However, in the COVID-19 focus segmentation task, their inability to focus on local features with sufficient precision resulted in inaccurate or incomplete segmentation. DCANet, MCDAU-Net, META-Unet and DualA-Net have difficulty in effectively preserving edge detail textures, resulting in blurred images and instances of missing or error-detecting areas. Due to multi-scale and attentional mechanisms, MCAFNet and MDUNet are able to produce visually clearer and more accurate segmentation maps, but they can struggle when small, irregular areas of infection are often involved. In contrast, M-Net, a variant of U-Net, has emerged as a promising solution by integrating multi-scale input layers and side output layers, yielding commendable results. In addition, by the introduction of primary feature conservation mechanism, DCSAU-Net cleverly utilizes both low-level and high-level semantic information, showing excellent segmentation performance. However, the MD-Net method is able to segment even the smallest infections scattered throughout the COVID-19 lesion region, which highlights the superior accuracy of our method.
Visualization of different models on the Vid-QU-EX dataset. The first and second rows: original images and corresponding gold labels on the Vid-QU-EX dataset. The third to last rows are the predicted results of U-Net, Attention-U-Net, DCANet, M-Net, DCSAU-Net, MCDAU-Net, META-Unet, MSRAformer, Swin-Transformer, MCAFNet, MDUNet, DualA-Net and MD-Net.
Second, we performed the evaluation on the QaTa-COV19-v2 dataset, and the results were shown in Table 4. Notably, our model achieved impressive scores on key evaluation metrics, with Dice scores of 0.8395, Mcc of 0.8232, and Jaccard Index of 0.7311. These measures serve as robust indicators of the model’s ability to accurately portray diseased areas, even in areas with low contrast. Compared to U-Net, MD-Net demonstrated notable improvements across all metrics, with increases of 1.02% in Dice score, 1.33% in Mcc, and 1.67% in Jaccard index. Furthermore, when compared to the sub-optimal DCANet method, MD-Net exhibited marginal yet noteworthy improvements. The Dice score, Mcc, and Jaccard index increased by 0.08%, 0.03%, and 0.08%, respectively. However, although the improvement in accuracy is small, it has superior advantages in terms of parameters and efficiency. Thus, based on a comprehensive evaluation considering both performance and computational complexity, MD-Net emerges as the optimal choice. Its ability to achieve high segmentation accuracy while maintaining reasonable computational demands positions it as a promising solution for the precise detection and segmentation of COVID-19 lesions in medical imaging applications.
Furthermore, we complement our quantitative analysis with a visual examination of the segmentation outcomes generated by our model, as illustrated in Fig. 10. After closely examining the visual results, it was clear that MD-Net does an excellent job of capturing local detail with amazing accuracy. Overall, our model had the best results in COVID-19 lesions, confirming that our model had better generalization. Through a combination of quantitative and qualitative evaluations, we affirm the advantages of MD-Net as a universal and reliable tool for COVID-19 lesion segmentation in medical imaging.
Visualization of different models on the QaTa-COV19-v2 dataset. The first and second rows: original images and corresponding gold labels on the Vid-QU-EX dataset. The third to last rows are the predicted results of U-Net, Attention-U-Net, DCANet, M-Net, DCSAU-Net, MCDAU-Net, META-Unet, MSRAformer, Swin-Transformer, MCAFNet, MDUNet, DualA-Net and MD-Net.
Efficiency analysis
To ensure a fair and thorough comparison, we performed an extensive efficiency analysis across thirteen state-of-the-art models, utilizing both the number of parameters (Params) and frames per second (FPS) as key evaluation criteria, as indicated in Tables 3 and 4. U-Net stands out as a model that optimally balances computational resources, with relatively low parameters, minimal model size, and high FPS, positioning it as one of the most computationally efficient networks in our study. Similarly, DCSAU-Net and DualA-Net are efficient models that use fewer parameters and require shorter training times, which enhances their suitability for real-time applications. Despite their advanced architecture, models like MSRAformer and Swin-Transformer demand significantly higher computational resources, both in terms of parameters and extended training times. However, this increased complexity does not necessarily translate into superior segmentation performance. In contrast, MD-Net offers a compelling alternative by striking an ideal balance between precision and efficiency. With a compact network size of just 8.5747 MB and an impressive frame rate of 73 to 75 milliseconds per frame, MD-Net proves to be a highly practical solution that provides advanced capabilities for diagnosing COVID-19 lesions without requiring a significant amount of computing power.
Conclusion
This paper introduces MD-Net, a novel deep learning architecture specifically designed to segment COVID-19 lesions from medical images. By addressing the challenges posed by opaque regions, subtle organizational differences, and image noise, MD-Net utilizes a U-shaped structure to enhance multi-scale input layers, SE-Conv module, and dense decoder aggregation network. Through comprehensive quantitative analysis of Vid-QU-EX and QaTa-COV19-v2 datasets and comparison with existing methods, MD-Net showed higher performance on Dice value, Matthews correlation coefficient and Jaccard index. The experimental results not only show the robustness and versatility of MD-Net, but also highlight its effectiveness in capturing fine-grained detail and contextual information, which is critical for accurately segmenting lesions. In addition, ablation studies conducted on the Vid-QU-EX dataset provided insights into the effectiveness of key components integrated into MD-Net, further validating its advantages over competing approaches. MD-Net represents a significant advance in the field of segmentation of COVID-19 lesions and provides a powerful tool for accurate diagnosis and the development of effective treatment strategies.
Data availability
The authors have used publicly available data in this manuscript. The dataset link is mentioned in the paper.
References
Long, J., Shelhamer, E. & Darrell, T. Fully convolutional networks for semantic segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 39, 640–651 (2015).
Ronneberger, O., Fischer, P. & Brox, T. U-net: Convolutional networks for biomedical image segmentation. In International Conference on Medical Image Computing and Computer-Assisted Intervention 234–241 (2015).
You, C., Dai, W., Min, Y., Staib, L. & Duncan, J. S. Bootstrapping semi-supervised medical image segmentation with anatomical-aware contrastive distillation. In International Conference on Information Processing in Medical Imaging 641–653 (Springer, 2023).
You, C. et al. Mine your own anatomy: Revisiting medical image segmentation with extremely limited labels. In IEEE Transactions on Pattern Analysis and Machine Intelligence (2024).
You, C. et al. Rethinking semi-supervised medical image segmentation: A variance-reduction perspective. Adv. Neural Inf. Process. Syst. 36, 1 (2024).
You, C. et al. Action++: Improving semi-supervised medical image segmentation with adaptive anatomical contrast. In International Conference on Medical Image Computing and Computer-Assisted Intervention 194–205 (Springer, 2023).
You, C., Yang, J., Chapiro, J. & Duncan, J. S. Unsupervised wasserstein distance guided domain adaptation for 3d multi-domain liver segmentation. In Interpretable and Annotation-Efficient Learning for Medical Image Computing: Third International Workshop, iMIMIC 2020, Second International Workshop, MIL3ID 2020, and 5th International Workshop, LABELS 2020, Held in Conjunction with MICCAI 2020, Lima, Peru, October 4–8, 2020, Proceedings 3 155–163 (Springer, 2020).
You, C. et al. Class-aware adversarial transformers for medical image segmentation. Adv. Neural. Inf. Process. Syst. 35, 29582–29596 (2022).
Jin, Q. et al. Inter-and intra-uncertainty based feature aggregation model for semi-supervised histopathology image segmentation. Expert Syst. Appl. 238, 122093 (2024).
You, C., Zhao, R., Staib, L. H. & Duncan, J. S. Momentum contrastive voxel-wise representation learning for semi-supervised volumetric medical image segmentation. In International Conference on Medical Image Computing and Computer-Assisted Intervention, 639–652 (Springer, 2022).
You, C., Zhou, Y., Zhao, R., Staib, L. & Duncan, J. S. Simcvd: Simple contrastive voxel-wise representation distillation for semi-supervised medical image segmentation. IEEE Trans. Med. Imaging 41, 2228–2237 (2022).
You, C., Dai, W., Min, Y., Staib, L. & Duncan, J. S. Implicit anatomical rendering for medical image segmentation with stochastic experts. In International Conference on Medical Image Computing and Computer-Assisted Intervention 561–571 (Springer, 2023).
Chen, Y. et al. Joint margin adaption and multiscale feature fusion for covid-19 ct images segmentation. Biomed. Signal Process. Control 91, 105912 (2024).
Li, Y. et al. Cdrime-mtis: An enhanced rime optimization-driven multi-threshold segmentation for covid-19 X-ray images. Comput. Biol. Med. 169, 107838 (2024).
Li, D., Fu, Z. & Xu, J. Stacked-autoencoder-based model for covid-19 diagnosis on ct images. Appl. Intell. 51, 2805–2817 (2021).
Ding, X. et al. A novel approach to the technique of lung region segmentation based on a deep learning model to diagnose covid-19 x-ray images. Curr. Med. Imaging 20, 1–11 (2024).
Alsaaidah, B., Mustafa, Z., Al-Hadidi, M. & Alharbi, L. A. Automated identification and categorization of covid-19 via X-ray imagery leveraging roi segmentation and cart model. Traitement Signal 40, 2259–2265 (2023).
Chen, Y. et al. Bgsnet: A cascaded framework of boundary guided semantic for covid-19 infection segmentation. Biomed. Signal Process. Control 90, 105824 (2024).
Zhou, T., Lian, B., Wu, C., Chen, H. & Chen, M. U-former: Covid-19 lung infection segmentation based on convolutional neural network and transformer. J. Electron. Imaging 33, 013041 (2024).
Zhao, S. et al. Scoat-net: A novel network for segmenting covid-19 lung opacification from ct images. Pattern Recogn. 119, 108109 (2021).
Devi, M., Singh, S. & Tiwari, S. Covlis–Munet segmentation model for covid-19 lung infection regions in ct images. Neural Comput. Appl. 36, 7265–7278 (2024).
Liu, S., Cai, T., Tang, X. & Wang, C. Mrl-net: Multi-scale representation learning network for covid-19 lung ct image segmentation. IEEE J. Biomed. Health Inform. 27, 4317–4328 (2023).
Saha, S., Dutta, S., Goswami, B. & Nandi, D. Adu-net: An attention dense u-net based deep supervised dnn for automated lesion segmentation of covid-19 from chest ct images. Biomed. Signal Process. Control 85, 104974 (2023).
Fan, D.-P. et al. Inf-net: Automatic covid-19 lung infection segmentation from ct images. IEEE Trans. Med. Imaging 39, 2626–2637 (2020).
Li, X., Song, J., Jiao, W. & Zheng, Y. Minet: Multi-scale input network for fundus microvascular segmentation. Comput. Biol. Med. 154, 106608 (2023).
Yin, P., Cai, H. & Wu, Q. Df-net: Deep fusion network for multi-source vessel segmentation. Inf. Fusion 78, 199–208 (2022).
Szegedy, C. et al. Going deeper with convolutions. In Proceedings of the IEEE Conference o Computer Vision and Pattern Recognition 1–9 (2015).
Tang, H. et al. Htc-net: A hybrid cnn-transformer framework for medical image segmentation. Biomed. Signal Process. Control 88, 105605 (2024).
Wu, R. et al. Mhorunet: High-order spatial interaction unet for skin lesion segmentation. Biomed. Signal Process. Control 88, 105517 (2024).
Tahir, A. M. et al. Covid-19 infection localization and severity grading from chest X-ray images. Comput. Biol. Med. 139, 105002 (2021).
Yamac, M. et al. Convolutional sparse support estimator-based covid-19 recognition from X-ray images. IEEE Trans. Neural Netw. Learn. Syst. 32, 1810–1820 (2021).
Selvaraj, A. & Nithiyaraj, E. Cedrnn: A convolutional encoder-decoder residual neural network for liver tumour segmentation. Neural Process. Lett. 55, 1605–1624 (2023).
Trinh, M.-N. et al. An efficientnet-encoder u-net joint residual refinement module with Tversky–Kahneman Baroni–Urbani–Buser loss for biomedical image segmentation. Biomed. Signal Process. Control 83, 104631 (2023).
Li, J. et al. Class-aware attention network for infectious keratitis diagnosis using corneal photographs. Comput. Biol. Med. 151, 106301 (2022).
Oulefki, A., Agaian, S., Trongtirakul, T. & Laouar, A. K. Automatic covid-19 lung infected region segmentation and measurement using ct-scans images. Pattern Recogn. 114, 107747 (2021).
Hu, K. et al. Dsc-net: A novel interactive two-stream network by combining transformer and cnn for ultrasound image segmentation. IEEE Trans. Instrum. Meas. 72, 5030012 (2023).
Yu, Z., Yu, L., Zheng, W. & Wang, S. Eiu-net: Enhanced feature extraction and improved skip connections in u-net for skin lesion segmentation. Comput. Biol. Med. 162, 107081 (2023).
Zhao, P., Wang, W., Zhang, G. & Lu, Y. Alleviating pseudo-touching in attention u-net-based binarization approach for the historical Tibetan document images. Neural Comput. Appl. 35, 13791–13802 (2023).
Muhammad, Z.-U.-D., Huang, Z., Gu, N. & Muhammad, U. Dcanet: Deep context attention network for automatic polyp segmentation. Vis. Comput. 39, 5513–5525 (2023).
Fu, H. et al. Joint optic disc and cup segmentation based on multi-label deep network and polar transformation. IEEE Trans. Med. Imaging 37, 1597–1605 (2018).
Xu, Q., Ma, Z., Na, H. & Duan, W. Dcsau-net: A deeper and more compact split-attention u-net for medical image segmentation. Comput. Biol. Med. 154, 106626 (2023).
Zhou, W. et al. Dual-path multi-scale context dense aggregation network for retinal vessel segmentation. Comput. Biol. Med. 164, 107269 (2023).
Wu, H., Zhao, Z. & Wang, Z. Meta-unet: Multi-scale efficient transformer attention unet for fast and high-accuracy polyp segmentation. In IEEE Transactions on Automation Science and Engineering (2023).
Wu, C. et al. Msraformer: Multiscale spatial reverse attention network for polyp segmentation. Comput. Biol. Med. 151, 106274 (2022).
Liu, Z. et al. Swin transformer v2: Scaling up capacity and resolution. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition 12009–12019 (2022).
Li, G. et al. Mcafnet: Multiscale cross-layer attention fusion network for honeycomb lung lesion segmentation. Med. Biol. Eng. Comput. 62, 1121–1137 (2024).
Liu, Y., Yao, S., Wang, X., Chen, J. & Li, X. Md-unet: A medical image segmentation network based on mixed depthwise convolution. Med. Biol. Eng. Comput. 62, 1201–1212 (2024).
Doc, Y. Z. & Doc, S. W. Duala-net: A generalizable and adaptive network with dual-branch encoder for medical image segmentation. Comput. Methods Progr. Biomed. 243, 107877 (2024).
Acknowledgements
Thanks to the editorial team and all the anonymous reviewers who helped us improve the quality of this paper.
Author information
Authors and Affiliations
Contributions
Conceptualization, methodology, and writing by X.L.; validation and experiments by X.L. and W.J.; writing review and editing, X.L. and W.J. Both authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Lan, X., Jin, W. Multi-scale input layers and dense decoder aggregation network for COVID-19 lesion segmentation from CT scans. Sci Rep 14, 23729 (2024). https://doi.org/10.1038/s41598-024-74701-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-74701-0