Abstract
Stomach adenocarcinoma (STAD) is a prevalent malignancy that is highly aggressive and heterogeneous. Intratumor heterogeneity (ITH) showed strong link to tumor progression and metastasis. High ITH may promote tumor evolution. An ITH-related signature (IRS) was created using as integrative technique including 10 machine learning methods based on TCGA, GSE15459, GSE26253, GSE62254 and GSE84437 datasets. The relevance of IRS in predicting the advantages of immunotherapy was assessed using a number of prediction scores and three immunotherapy datasets (GSE78220, IMvigor210 and GSE91061). Vitro experiments were performed to verify the biological functions of AKR1B1. The RSF + Enet (alpha = 0.1) projected model was proposed as the ideal IRS because it had the highest average C-index. The IRS demonstrated a strong performance in serving as an independent risk factor for the clinical outcome of STAD patients. It performed exceptionally well in predicting the overall survival rate of STAD patients, as seen by the TCGA cohort’s AUC of 1-, 3-, and 5-year ROC curves, which were 0.689, 0.683, and 0.669, respectively. A low IRS score demonstrated a superior response to immunotherapy, as seen by a lower TIDE score, lower immune escape score, greater TMB score, higher PD1&CTLA4 immunophenoscore, higher response rate, and improved prognosis. Common chemotherapeutic and targeted treatment regimens had lower IC50 values in the group with higher IRS scores. Vitro experiment showed that AKR1B1 was upregulated in STAD and knockdown of AKR1B1 obviously suppressed tumor cell proliferation and migration. The present investigation produced the best IRS for STAD, which may be applied to prognostication, risk stratification, and therapy planning for STAD patients.
Similar content being viewed by others

Introduction
Gastric cancer is the three deadliest cancers in 2020 globally, with an estimated of 1,089,103 new diagnosed cases and an estimated of 768,793 deaths1. The most prevalent subtype of gastric cancer, known as stomach adenocarcinoma (STAD), has a high degree of invasiveness and significant heterogeneity2. Currently, surgery is no longer the only method of treating STAD; a multidisciplinary, all-encompassing approach that includes immunotherapy, targeted therapy, chemotherapy, and radiation is used instead3. However, many STAD patients still experience tumor progress and relapse after treatment, leading to treatment failure. The main reasons are the intratumor heterogeneity (ITH) and the complex mechanism of tumor development4. Furthermore, a small number of reliable markers may be able to forecast a STAD patient’s prognosis and therapy outcomes. These depressing findings underscore the critical need for biomarkers that can anticipate treatment outcomes and prognosis in STAD.
ITH refers to the uneven distribution of genomic diversity in a single tumor over space or time due to accumulated genetic mutations5. It is believed that the ITH is related to the randomness of gene mutation and environmental factors6. Research has demonstrated a strong correlation between ITH and tumor progression, suggesting that high ITH and metastasis may promote the evolution of tumors7. Moreover, low ITH indicates a better survival, lower tumor burden, and better benefits in anti-PD-1 immunotherapy8. ITH signature could indicate the prognosis and therapy response in patients with colon adenocarcinoma9, cholangiocarcinoma10 and cutaneous melanoma11. Considering the importance of tumor heterogeneity, it is necessary to comprehensively elucidate the genes mediating ITH in STAD and their role in forecasting the prognosis and therapeutic outcomes for STAD cases.
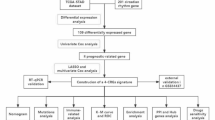
Using the information from the Cancer Genome Atlas (TCGA) and Gene Expression Omnibus (GEO), we identified those genes mediating ITH in STAD and developed a gene signature. In order to provide insights into prognosis prediction and the immunological landscape in STAD, we also investigated the relationship between the ITH-related signature (IRS) and immune infiltration, and medication sensitivity in STAD.
Materials and methods
Data acquisition and MATH score of STAD cases
From TCGA, the bulk RNA-seq data of STAD were extracted. To confirm our findings, we additionally examined four more datasets: GSE15459, GSE26253, GSE62254, and GSE84437. Three immunotherapy datasets were used to assess the contribution of IRS to the prediction of immunotherapy benefits: GSE91061, GSE78220, and IMvigor210. Those patients diagnosed with STAD with complete available overall survival information included in our study. Death within three months of diagnosis should be excluded. The ITH score for STAD cases were calculated by using DEPTH2 method, an algorithm for evaluating intratumor heterogeneity12. Next, we used the optimal cut-off to divide STAD patients into two cohorts: low DEPTH2 score and high DEPTH2 score. Using “limma” packages, differentially expressed genes (DEGs) between these two cohorts were found by setting a cutoff value of P-value < 0.05 and a |Log2FC| value > 1.5.
Integrative machine learning algorithms constructed an optimal IRS
After that, we used univariate Cox analysis to find probable predictive biomarkers (p-value < 0.05) among these DEGs in STAD. Then, in order to create a stable prognostic IRS, we subjected these putative prognostic biomarkers to an integrative machine learning analysis process. We created the IRS in four steps, following the methodology of the R scripts (https://github.com/Zaoqu-Liu/IRLS) of earlier research13,14, we developed the IRS in four steps: (1) Univariate Cox regression was conducted to identify prognostic biomarkers in the TCGA dataset; (2) Then, the prediction model of TCGA data set was fitted with 101 algorithms combinations; (3) All algorithms combinations were performed in GEO cohorts; (4) C-index was calculated across all cohorts. Furter details in machine learning algorithms were shown in Supplementary Methods. We used the “surv_cutpoint” function in the R package “survminer” to identify the optimal cutoff point, which allowed us to categorize STAD patients into high and low risk groups based on their risk score (IRS score).
The performance of IRS
The survival curves were produced using the Kaplan-Meier survival method. The “survivalROC” R package was utilized to generate the ROC and C-index curves. We then identified the risk factors for the prognosis of STAD patients by running univariate and multivariate Cox analyses. The clinical outcome of STAD patients was then predicted by building a predictive nomogram using the “nomogramEx” program based on IRS-based risk score and additional clinical characteristics. The calibration curve was used to show how real and expected survival differed.
Immune infiltration analysis
Every STAD case’s immunological score and ESTIMATE score were assessed using the ESTIMATE method15. Seven approaches were utilized to determine the quantity of immune cells in STAD cases16. We displayed the relative expression of immunological checkpoints and genes associated to the human leukocyte antigen (HLA) in various groups of STAD cases using the R package “ggplot” or “ggpubr.” Using the “GSVA” software and the ssGSEA approach, the gene set score that was connected with immune cells and immune-related activities or functions in STAD was found.
Drug sensitivity analysis
We employed multiple measures, such as tumor immune dysfunction and exclusion (TIDE) score17, immune escape score14,18, immunophenoscore19, to assess how well IRS predicted the immunotherapy response of STAD patients. Increased risk of immune escape and decreased efficacy of immunotherapy treatment are indicated by higher TIDE, higher tumor escape, lower immunophenoscore, and lower TMB scores. The “oncoPredict R” program was used to determine the IC50 of each drug in each STAD case based on data from the Genomics of Drug Sensitivity in Cancer. Reduced sensitivity was indicated by a higher IC50 value.
Cell lines and knockdown of AKR1B1
Gastric epithelial cell line (RGM-1) and STAD cell lines (NCI-N87, CRL-5822, RPMI1640, BGC-823, HGC-27) were purchased from Shanghai Institute of Biochemistry and Cell Biology (Shanghai, China). Cells were maintained in circumstances containing 5% CO2 and 95% saturated humidity at 37 °C using respective ATCC recommended medium. Fetal bovine serum (FBS; Gibco) and 1% penicillin-streptomycin (Sigma-Aldrich, St. Louis, USA) were added to the medium. BGC-823 and HGC-27 cells were transfected with AKR1B1 siRNA or scrambled negative control siRNA using Lipofectamine 3000 transfection reagent (Invitrogen, Thermo Fisher Scientific) based on the manufacturer’s instructions.
RT-qPCR, proliferation assay and wound healing assay
Using TRIzol (Takara Bio, Dalian, China), we extracted RNA from cells, which were reversely transcribed into cDNA using an oligo (dT) primer subsequently. Based on the ABI 7900HT detection system (Thermo Fisher Scientific Inc.), we then performed RT-qPCR with SYBR Premix Ex Taq (Takara Bio). Gene expression levels were normalized to the endogenous GAPDH. For proliferation assay, STAD cell lines were plated in 96-well plates (5,000 cells/well in triplicates). Cell Counting kit-8 (CCK-8; Beyotime) was added to cells at indicated times. Proliferation index was calculated as the ratio of OD value at the indicated time/OD value of the input cells. Cell migration ability was examined using wound healing assay. Transfected cells were allowed to grow to 80–90% confluence in 6-well plates. A wound was built afterwards across the cell monolayer using a bioclean 200 µl pipette tip. After rinsed with phosphate-buffered saline (PBS), the cells were incubated for another 24 h using serum-free medium. The images of wound closure were photographed (0 and 24 h) using a digital camera system (Olympus Corporation, Tokyo, Japan).
Results
The ITH score of STAD cases
Supplementary Table 1 displayed the STAD cases’ ITH scores. Higher ITH score was correlated with lower tumor grade, higher clinical stage, higher pT stage and distant metastasis (Fig. 1A). STAD cases were categorized into low and high ITH score. A higher ITH score was associated with a poorer overall survival (OS) rate (Fig. 1B, p = 0.041). Next, we looked at the DEGs in STAD between the groups with low and high ITH scores, with which we could identify genes mediating the ITH of STAD. As a result, we obtained 925 DEGs (Fig. 1C, p < 0.05). Twenty-one of these DEGs had a strong correlation with the clinical prognosis of STAD patients (Fig. 1D).
The intratumor heterogeneity score of STAD cases. A The correlation between intratumor heterogeneity score and the clinical characters of STAD patients. B Low intratumor heterogeneity score indicated a lower overall survival rate in STAD. C The different expressed genes between in high and low intratumor heterogeneity score. D Univariate cox analysis identified potential genes significantly correlated with the prognosis of STAD patients.
A prognostic IRS was created by machine learning
Then, in order to create an IRS, we fed these 21 genes into our machine learning-based integrative process. Using the LOOCV framework, we fitted 101 different types of prediction models in the TCGA cohort. We then computed the C-index for each model over all GEO cohorts (Fig. 2A). The C-index for each prediction model across all cohorts was displayed in Fig. 2A. With the greatest average C-index of 0.63, the IRS created using the RSF + Enet (alpha = 0.1) method was recommended as the ideal IRS (Fig. 2A). The IRS was created utilizing nine genes based on the RSF + Enet (alpha = 0.1) technique, and the IRS score (risk score) of STAD patients was computed using the formula below: risk score = (-0.2838) × DGKQexp + (0.1515) × SERPINE1exp + 0.1034 × PRTGexp + 0.0515 × CPNE8exp + 0.1614× NT5Eexp + 0.1919 × AKR1B1exp + 0.0493 × FGF1exp + (-0.0243) × SLITRK2exp + (-0.022) × ASPAexp. STAD cases were divided into groups with high and low IRS scores using the optimal cut-off. The study findings revealed that patients with STAD who had a high IRS score had a lower OS rate in the TCGA, GSE15459, GSE26253, GSE62254 and GSE84437 datasets(Fig. 2B and F), with 1-, 3-, and 5-year AUCs of 0.689, 0.683, and 0.669 in TCGA cohort (Fig. 2G); 0.670, 0.634, and 0.634 in GSE15459 cohort (Fig. 2H); 0.622, 0.601, and 0.693 in GSE26253 cohort (Fig. 2I); 0.669, 0.631, and 0.603 in GSE62254 cohort (Fig. 2J), 0.652, 0.686, and 0.636 in GSE84437 cohort (Fig. 2K), respectively (Fig. 2B and F).
Development of IRS by integrative machine learning algorithms. A IRS was evaluated using 101 machine learning combinations. The concordance index was calculated for each model of TCGA and GEO datasets. The survival curve of STAD patients with different IRS score in TCGA (B), GSE15459 (C), GSE26253 (D), GSE62254 (E) and GSE84437 (F) cohort. Time-dependent ROC curves for IRS in evaluating the 1-year (Red line), 3-year (Blue line), and 5-year (Green line) overall survival for the TCGA (G), GSE15459 (H), GSE26253 (I), GSE62254 (J) and GSE84437 (K) cohort.
An assessment of IRS’s performance
We also computed the C-index of IRS and these clinical characteristics in order to assess how well they performed in predicting the clinical outcome of STAD cases. The C-index of IRS was greater in all datasets than that of clinical characteristics, such as age, gender, tumor grade, and clinical stage, as Fig. 3A illustrates. In the TCGA, GSE15459, GSE26253, GSE62254, and GSE84437 datasets, additional univariate and multivariate cox regression analysis revealed IRS as an independent risk factor for the clinical outcome of STAD cases (Fig. 3B and C, all p < 0.05). We also created a nomogram to predict the clinical fate of STAD patients based on risk score and stage (Fig. 3D). Regarding the 1, 3, and 5-year survival rates in the TCGA cohort, the calibration plots revealed good agreement between the nomogram prediction and actual observation (Fig. 3E). These data revealed that the IRS is capable of accurately and consistently predicting the clinical result of STAD cases.
Evaluation the performance of IRS in predicting clinical outcome of STAD patients. A The C-index of IRS, age, gender and clinical stage for the performance in predicting the clinical outcome of STAD patients in TCGA and GEO datasets. B, C Univariate and multivariate cox regression analysis identified risk factors for the clinical outcome of STAD patients. D, E Predictive nomogram and calibration evaluating the overall survival rate of STAD patients.
Analysis of the relationship between IRS and the tumor immunological milieu
Figure 4A displayed the relationship between the IRS score and immune cell abundance. The number of CD8 + T cells, B cells, and dendritic cells was negatively correlated with the IRS score (Fig. 4B and D, p < 0.05). Additionally, a reduced amount of B cells, CD8 + T cells, macrophages, neutrophils, and TIL was suggested by a higher IRS score (Fig. 4E). A lower score was connected with cytolytic activity, T cell co-stimulation, and APC co-stimulation when the IRS score was higher (Fig. 4F). Additionally, we discovered that stromal, immune, and ESTIMAE scores were significantly lower in STAD patients with high IRS scores (Fig. 4G and I, all p < 0.001).
The association between IRS and immune infiltration in STAD. A Correlation atlas between IRS and immune infiltration in STAD based on seven state-of-the-art algorithms. B–D IRS score was negatively correlated with the abundance of CD8 + T cell, B cell and Dendritic cell. E, F ssGSEA analysis revealing the level of immune cells and immune related functions in different IRS score group. G–I The immune score, stroma score and ESTIMAE score in different IRS score group. *p < 0.05, **p < 0.01, ***p < 0.001.
IRS as a predictor of treatment outcomes in STAD
A greater immunophenotype and TMB score indicated a higher likelihood of benefiting from immunotherapy20. As shown in Fig. 5A and B, STAD patients with low IRS score had a higher TMB score and PD1&CTLA4 immunophenoscore. A low TIDE score indicated a better response to immunotherapy and a decreased risk of immune escape21,22. The data found a higher score of TIDE, T cell exclusion and dysfunction in higher IRS score group (Fig. 5C, all p < 0.05). A greater variety of antigen presentation was indicated by high expression of immunological checkpoints and HLA-related genes, which increased the possibility that immunotherapy might be beneficial23. The findings indicated that STAD patients with low IRS scores expressed more immunological checkpoints and genes associated to HLA (Fig. 5D and E, all p < 0.05). Consequently, patients with STAD and low IRS scores may benefit more from immunotherapy. We then computed the IRS score in immunotherapy patients to confirm the findings even more. In the IMvigor210 cohorts, non-responders had a higher IRS score than responders (p < 0.01), as illustrated in Fig. 5F. A higher IPS score (p = 0.02) was associated with a worse clinical outcome. Additionally, a lower IRS score was associated with a higher immunotherapy response rate (p < 0.01). It’s interesting to note that we saw comparable outcomes in the GSE78220 and GSE91061 dataset (Fig. 5G and H). We next investigate the IC50 value of STAD cases, taking into account the critical role that targeted therapy and standard chemotherapy play in the treatment of STAD. The information showed that STAD patients with high IRS scores had lower IC50 values for 5-Fluorouracil, Docetaxel, Oxaliplatin, Paclitaxel, Cytarabine, Gefitinib, Crizotinib, Erlotinib, and Osimertinib (Fig. 6A and B, all p < 0.05). This suggests that STAD patients with high IRS scores are more sensitive to chemotherapy and targeted therapy.
IRS acted as an indicator for predicting the immunotherapy response in STAD. The TMB score (A), PD1&CTLA4 immunophenoscore (B), TIDE score (C) in STAD patients with different IRS score. The level of HLA-related genes (D) and immune checkpoints (E) in different IRS score group. The immunotherapy response and overall rate in patients with high and low IRS score in IMvigor210 (F), GSE91061 (G) and GSE78220 (H) datasets. *p < 0.05, **p < 0.01, ***p < 0.001.
Analysis of the variations in functional enrichment across IRS score groups
The gene sets scoring for angiogenesis, DNA repair, EMT signaling, glycolysis, hypoxia, IL2-STAT5 signaling, mTORC1 signaling, NOTCH signaling, P53 pathway, and PIK-AKT-MTOR signaling were higher in STAD patients with high IRS scores, as Supplementary Fig. 1 illustrates (all p < 0.05).
Biological functions of the selected gene
To further verify the performance of IRS, we selected AKR1B1 that contributed the most to the IRS for further analysis. We explored the expression of AKR1B1 in STAD cell lines, demonstrating that the expression of AKR1B1 was higher in most of STAD cell lines (Supplementary Fig. 2A). In the follow-up study, the results of the CCK-8 assay proved that knockdown of AKR1B1 obviously inhibited the proliferation of BGC-823 and HGC-27 cells (Supplementary Fig. 2B-2 C, all p < 0.05). Moreover, knockdown of AKR1B1 obviously inhibited the migration of BGC-823 and HGC-27 cells (Supplementary Fig. 2D-2G, all p < 0.05).
Discussion
In this work, we used ten integrative machine learning techniques to build an IRS for STAD. The RSF + Enet (alpha = 0.1) projected model was proposed as the ideal IRS since it had the highest average C-index. IRS performed well in predicting the overall survival rate and functioned as an independent risk factor for the clinical outcome of STAD patients. A low IRS score was associated with a lower TIDE score, a lower immunological escape score, a higher PD1&CTLA4 immunophenoscore, a higher TMB score, a greater response rate, and a better prognosis, according to additional study. IRS was also proposed as an indicator for predicting the advantages of immunotherapy.
Our IRS was constructed using 9 potential genes, including DGKQ, SERPINE1, PRTG, CPNE8, NT5E, AKR1B1, FGF1, SLITRK2 and ASPA. SERPINE1 was a biomarker associated with the prognosis and immune infiltrates in STAD24. Upregulated of PRTG accelerated tumor progression via CGMP/PKG signaling pathway in gastric cancer25. CPNE8 favored STAD metastasis through modulating focal adhesion pathway26. Correlated with unfavorable prognosis, NT5E could regulates cell proliferation and motility in STAD27. AKR1B1 accelerated STAD progression via AKT-mTOR pathway and correlated with poor prognosis28. FGF1 functioned as a biological therapeutic target and predictive biomarker for STAD29.
We discovered that IRS had a strong predictive ability for the overall survival rate and functioned as an independent risk factor for the clinical outcome of STAD patients. It turns out that the intratumor heterogeneity signature may also be used to forecast how patients with other cancer types will fare clinically. IRS could forecast a colon cancer patient’s prognosis and responsiveness to chemotherapy9.
It has been suggested that immunotherapy, which includes STAD, is one of the most promising methods for treating cancer30. Our results showed that a low IRS score was linked to increased response rate, reduced TIDE, immunological escape, and TMB scores, as well as a better prognosis and higher PD1&CTLA4 immunophenoscore. Higher TMB and MSI scores were indicative of a better immunotherapy outcome31,32. A low TIDE score suggested a lower chance of immunological escape21. Intratumor heterogeneity has been demonstrated in prior research to be able to predict the response to immunotherapy8. According to this data, IRS may serve as a predictor of immunotherapy advantages for STAD patients; a lower IRS score was associated with greater immunotherapy benefits.
Additionally, we discovered a correlation between a high IRS score and higher gene sets scoring in angiogenesis, EMT signaling, glycolysis, and hypoxia—all characteristics associated with cancer. In STAD, angiogenesis was the main factor promoting tumor growth and metastasis33. An essential part of the onset and development of STAD is EMT signaling34. Targeting glycolysis is suggested as a viable approach in STAD, as STAD cells undergo metabolic reprogramming and switch to glycolysis to improve their survival and metastasis35. Therefore, IRS may use these cancer-related markers to mediate the development of STAD.
Some limitations could be found in our study. Training cohort and testing cohorts of IRS were obtained from different databases. Although we have normalized the data before the analysis, there may still be heterogeneity. We could see that the C-index of IRS in TCGA dataset was higher than all the GEO datasets. The heterogeneity between datasets from different databases may affect the performance of IRS. The IRS was not verified by an in-house clinical cohort. It would be better to further verify the functions of other genes or conduct experiments in more cell lines to ensure the robustness of our results. Further study could focus on the functions and potential mechanisms of IRS genes in STAD. In actual clinical settings, we can calculate the IRS score of STAD patients based on the expression of each gene in IRS, with which we could predict the potential effect in immunotherapy. However, the expression of certain genes in IRS may not be detected in STAD patients, which affects the calculation of IRS score and the clinical practice of IRS.
Conclusion
The present investigation produced the best IRS for STAD, which may be applied to prognostication, risk stratification, and therapy planning for STAD patients.
Data availability
Availability of data and materials from TCGA dataset (https://tcga-data.nci.nih.gov/tcga/) and GEO (https://www.ncbi.nlm.nih.gov/gds).
References
Sung, H. et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 71, 209–249 (2021).
Luo, L. et al. A necroptosis-related lncRNA-based signature to predict prognosis and probe molecular characteristics of stomach adenocarcinoma. Front. Genet. 13 (2022).
He, Y. et al. Comprehensive characterization of transforming growth factor beta receptor 1 in stomach adenocarcinoma identifies a prognostic signature for predicting clinical outcomes and immune infiltrates. Int. J. Gen. Med. 15, 3375–3391 (2022).
Xu, H., Jiao, D., Liu, A. & Wu, K. Tumor organoids: applications in cancer modeling and potentials in precision medicine. J. Hematol. Oncol. 15, 58 (2022).
Vitale, I., Shema, E., Loi, S. & Galluzzi, L. Intratumoral heterogeneity in cancer progression and response to immunotherapy. Nat. Med. 27, 212–224 (2021).
Andor, N. et al. Pan-cancer analysis of the extent and consequences of intratumor heterogeneity. Nat. Med. 22, 105–113 (2016).
Gerlinger, M. et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N. Engl. J. Med. 366, 883–892 (2012).
Li, J. et al. Tumor cell-intrinsic factors underlie heterogeneity of immune cell infiltration and response to immunotherapy. Immunity 49, 178–193.e7 (2018).
Liu, C. et al. An intratumor heterogeneity-related signature for predicting prognosis, immune landscape, and chemotherapy response in colon adenocarcinoma. Front. Med. (Lausanne) 9, 925661 (2022).
Chen, X. et al. Machine learning developed an intratumor heterogeneity signature for predicting prognosis and immunotherapy benefits in cholangiocarcinoma. Transl. Oncol. 43, 101905 (2024).
Zhang, W. & Wang, S. Machine learning developed an intratumor heterogeneity signature for predicting prognosis and immunotherapy benefits in skin cutaneous melanoma. Melanoma Res. 34, 215–224 (2024).
Song, D. & Wang, X. DEPTH2: an mRNA-based algorithm to evaluate intratumor heterogeneity without reference to normal controls. J. Transl. Med. 20, 150 (2022).
Liu, Z. et al. Machine learning-based integration develops an immune-derived lncRNA signature for improving outcomes in colorectal cancer. Nat. Commun. 13, 816 (2022).
Li, Z., Guo, M., Lin, W. & Huang, P. Machine learning-based integration develops a macrophage-related index for predicting prognosis and immunotherapy response in lung adenocarcinoma. Arch. Med. Res. 54, 102897 (2023).
Yoshihara, K. et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat. Commun. 4, 2612 (2013).
Li, T. et al. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 48, W509–w14 (2020).
Jiang, P. et al. Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat. Med. 24, 1550–1558 (2018).
Sun, Y. et al. Single-cell landscape of the ecosystem in early-relapse hepatocellular carcinoma. Cell 184, 404–421e16 (2021).
Charoentong, P. et al. Pan-cancer immunogenomic analyses reveal genotype-immunophenotype relationships and predictors of response to checkpoint blockade. Cell. Rep. 18, 248–262 (2017).
Palmeri, M. et al. Real-world application of tumor mutational burden-high (TMB-high) and microsatellite instability (MSI) confirms their utility as immunotherapy biomarkers. ESMO Open. 7, 100336 (2022).
Fu, J. et al. Large-scale public data reuse to model immunotherapy response and resistance. Genome Med. 12, 21 (2020).
Lin, A., Zhang, J. & Luo, P. Crosstalk between the MSI status and tumor microenvironment in colorectal cancer. Front. Immunol. 11, 2039 (2020).
Lin, A. & Yan, W. H. HLA-G/ILTs targeted solid cancer immunotherapy: opportunities and challenges. Front. Immunol. 12, 698677 (2021).
Feng, L., Li, G., Li, D., Duan, G. & Liu, J. Cuproptosis-related gene SERPINE1 is a prognostic biomarker and correlated with immune infiltrates in gastric cancer. J. Cancer Res. Clin. Oncol. 149, 10851–10865 (2023).
Xiang, T. et al. The novel ZEB1-upregulated protein PRTG induced by Helicobacter pylori infection promotes gastric carcinogenesis through the cGMP/PKG signaling pathway. Cell. Death Dis. 12, 150 (2021).
Zhang, P. et al. CPNE8 promotes gastric cancer metastasis by modulating focal adhesion pathway and tumor microenvironment. Int. J. Biol. Sci. 18, 4932–4949 (2022).
Hu, S., Meng, F., Yin, X., Cao, C. & Zhang, G. NT5E is associated with unfavorable prognosis and regulates cell proliferation and motility in gastric cancer. Biosci. Rep. 39 (2019).
Liu, L. et al. Aberrant expression of AKR1B1 indicates poor prognosis and promotes gastric cancer progression by regulating the AKT-mTOR pathway. Aging (Albany NY) 15, 9661–9675 (2023).
Liu, N. et al. Expression and clinical significance of fibroblast growth factor 1 in gastric adenocarcinoma. Onco Targets Ther. 8, 615–621 (2015).
Ruiz-Cordero, R. & Devine, W. P. Targeted therapy and checkpoint immunotherapy in lung cancer. Surg. Pathol. Clin. 13, 17–33 (2020).
Liu, L. et al. Combination of TMB and CNA stratifies prognostic and predictive responses to immunotherapy across metastatic cancer. Clin. Cancer Res. 25, 7413–7423 (2019).
Seo, I. et al. Neoadjuvant chemoradiation alters biomarkers of anticancer immunotherapy responses in locally advanced rectal cancer. J. Immunother Cancer 9 (2021).
Xu, P. et al. Identification and validation of a novel angiogenesis-related gene signature for predicting prognosis in gastric adenocarcinoma. Front. Oncol. 12, 965102 (2022).
Baj, J. et al. Mechanisms of the epithelial-mesenchymal transition and tumor microenvironment in Helicobacter pylori-induced gastric cancer. Cells 9 (2020).
Zheng, S. et al. The emerging role of glycolysis and immune evasion in gastric cancer. Cancer Cell. Int. 23, 317 (2023).
Acknowledgements
Not applicable.
Funding
This work was supported by Clinical Research Special Funds of Wu Jieping Medical Foundation (No. 320.6750.2022-01-16).
Author information
Authors and Affiliations
Contributions
Hongcai Chen: Writing- Original draft preparation, Investigation, Formal analysis and Funding acquisitionCui Yang: Data curation, Writing- Review and editingTingting Tang: Conceptualization, ResourcesYi Jiang: Methodology, Supervision, Software, ValidationWenwu Xue: Visualization, Writing- Review and editing, Project administration.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Chen, H., Zheng, Z., Yang, C. et al. Machine learning based intratumor heterogeneity signature for predicting prognosis and immunotherapy benefit in stomach adenocarcinoma. Sci Rep 14, 23328 (2024). https://doi.org/10.1038/s41598-024-74907-2
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-024-74907-2
Keywords
This article is cited by
-
Multi-omics analysis reveals different cholesterol metabolism subtypes in colorectal cancer
Discover Oncology (2025)