Abstract
TCGA has identified predominant somatic copy number alterations (SCNA) affecting numerous genes in HGSOC. To identify cancer-driver genes from the regions of SCNA, we have devised a scoring system that integrates information from different genetic alterations. Applying this scoring system to the TCGA-HGSOC dataset (n = 316) we have identified several well-known and novel putative cancer genes in HGSOC. We functionally validated the roles of two previously unknown genes, RNF144B and PPP2R2A. RNF144B, an E3 ubiquitin-ligase is amplified and overexpressed in 16% of HGSOC (TCGA). Overexpression of RNF144B in ovarian cancer cells increased cell proliferation, colony formation, and migration. RNF144B was significantly overexpressed in 50% of primary tumors from patients with HGSOC compared to the ovary. Further, it had significantly reduced expression in tumors after chemotherapy. PPP2R2A, the regulatory subunit of PP2A is deleted and downregulated in 38% of HGSOCs (TCGA). Overexpression of PPP2R2A inhibited cell proliferation, colony-formation, migration, and invasion in ovarian cancer cells. In OVCAR-5, which expresses low levels of PPP2R2A, Niraparib inhibited cell proliferation. PPP2R2A was not expressed in 72% of HGSOCs. This report demonstrates this approach to identifying genes from the TCGA data. Further experiments are required to conclusively prove the role of these genes in the pathogenesis of ovarian cancer.
Similar content being viewed by others
Introduction
Ovarian cancer is one of the leading causes of gynecological cancer-related deaths among women globally. Globally in 2020, there were approximately 313,959 incident ovarian cancer cases and 207,252 deaths1. High-grade serous ovarian cancer (HGSOC) is the most common type and contributes to 80% of all deaths from ovarian cancer2. Despite advances in treatment, the five-year survival rates have shown only modest improvement. For patients diagnosed with advanced-stage disease (III & IV), the 5–year survival ranges between 30–40% globally3. Understanding the genetic and epigenetic causes of ovarian cancer will provide insights into the discovery of targets for drug development and for improving diagnosis and prevention strategies.
The Cancer Genome Atlas (TCGA) by NCI has comprehensively analyzed more than 500 ovarian tumor genomes at genetic, epigenetic, transcriptome, and protein levels4. The vast amount of data generated was deposited in the cancer genome portal (https://portal.gdc.cancer.gov/). The analysis of data from the TCGA of HGSOC has shown that p53 is mutated in nearly 100% of tumors. Most other genes are mutated at less than 5%. However, SCNA alterations were the most frequent in HGSOC compared to other tumors types4. SCNA-causing focal changes are less complex and may contain driver genes that recur in different tumor types5. SCNA results in loss of heterozygosity/deletion and gain/amplification of a gene or a chromosomal segment involving hundreds of genes. Though these affect numerous genes, only some of those genes are ‘Drivers’ provide a selective advantage, and assist tumor growth. Other genes that do not contribute to tumorigenesis are ‘Passengers’. Thus, the challenge is to identify the driver genes from passenger events6. If amplification (> 2N) or overexpression of a gene occurs within an SCNA, then it is likely to be an oncogene. In contrast, if deletion or loss of expression of a gene is detected then it is likely to be a tumor suppressor. Both these mechanisms significantly alter the expression of the gene. Previous strategies used to identify the driver gene were based on the frequency of CNA, mapping the minimal region of aberration and correlating the alteration with expression (positional cloning). Genes that are identified through these methods were further functionally validated either by silencing or by over-expressing in ovarian cancer cell lines. Oncogenes such as PAX8, ID4, GAB2, and BRD4 were identified through high throughput screening of more than a hundred genes7. However, this method is time-consuming, expensive, and requires additional validations to verify the relevance of a gene to cancer. Hence, several in-silico computational and statistical methods are being developed to identify driver genes from the regions of SCNA8,9,10.
In this study, we have addressed the challenge of identifying oncogenes and tumor suppressors from the regions of SCNA by using an integrated approach. It is based on the fact that a driver gene may exhibit different forms of alteration. For example, KRAS is found to be amplified in 10.7%, over-expressed in 8.86%, and mutated in 0.6% of TCGA HGSOC patients (n = 316). Similarly, the tumor suppressor BRCA1 is mutated in 3% and methylated in 11% of HGSOC patients11. Hence to identify the driver genes in SCNA regions, we integrated the information from various factors such as frequency of amplification or deletion, size of the amplicon or deleted segment, the number of genes within the amplicon or deletion, level of expression in the tumor compared to normal (ovary), presence of mutation, the effect of gene alteration on overall survival. We have identified numerous known cancer driver genes and novel putative genes using this approach whose function remain undetermined in ovarian cancer. We validated the expression and function of two new potential driver genes, RNF144B, an amplified gene, and PPP2R2A, a homozygously deleted gene in HGSOC cell lines and tumors.
Materials and methods
Data source
TCGA data (Nature, 2011) for 316 HGSOC’s were retrieved from different websites. The SCNA data was downloaded from FIREHOSE, Broad Institute https://gdac.broadinstitute.org). cBioportal http://www.cbioportal.org/publicportal/ was used to analyze mutation, SCNA, expression, and clinical data.
Scoring system
Scoring for each gene was computed separately by integrating data from different types of genetic analysis as detailed below and illustrated in Figs. 1a, b (for details see supplementary methods).
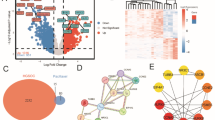
Scoring System and Integrated Approach for identification of cancer driver genes in HGSOC. Weighted scoring system by incorporating 8 different parameters to score and rank the (a) amplified and (b) deleted genes. Genes fulfilling all criteria received a maximum score of 24. (c) Framework for identification of driver genes. Focal SCNA data for the TCGA HGSOC dataset were downloaded from the Broad Institute FIREHOSE portal. Genes were shortlisted based on the significance of alteration p < 0.05 and score ≥ 5 (d) Oncoprint showing alterations of RNF144B in TCGA HGSOC dataset e Oncoprint of PPP2R2A alterations in TCGA HGSOC dataset.
Identification of amplified putative oncogenes
At the DNA level
Frequency of gene amplification
Driver genes are frequently amplified in cancer. To identify these genes, we analyzed amplification frequencies in HGSOC tumors. Genes were assigned scores from 1 to 7 based on frequency of amplification to assess their potential as driver genes. The frequency of amplification in TCGA ranged from <5% to 35%. Based on this we created scores corresponding to increasing intervals of 5% up to 35%.
The average size of the aberration
Focal amplifications are more likely to identify driver genes as they encompass fewer genes than larger, broad amplicons. Amplified chromosomal regions ranged in size from 0.024Mb to 1.18Mb with an average size of 0.4Mb. Therefore, genes within a 0.4Mb region were assigned a score of ‘1’ and others ′0′.
Number of genes in the aberration
Single-gene amplifications often indicate a driver role in tumorigenesis12. We assigned a maximum score of ‘3’ to regions of 0.4Mb containing a single gene. However, the scores were ‘2’ and ‘1’ if there were 2 or ≥ 3 genes respectively in the amplicon.
Frequency of somatic mutations
Missense mutations within oncogenes can result in aberrant activation, thereby contributing to tumorigenesis13. We considered the frequency of gain-of-function mutation when scoring a gene as a putative oncogene. Since driver genes were mutated in only a small subset of HGSOCs, we assigned a score of ‘1’ to genes mutated in >1% of tumors and ‘0’ to genes without such mutations.
At the RNA level
Frequency of gene overexpression
Overexpression at the mRNA level, independent of underlying genetic alterations, can signify oncogenic potential14. Genes that were overexpressed were assigned scores ranging from 1 to 7 depending on their frequency at the mRNA level.
Fold change
Fold change (FC) represents the relative expression of a gene in tumor samples compared to normal tissue. In the data, FC values ranged from 0 to 3-fold. Scores were assigned on a scale of 1-3 based on the magnitude of the increase in expression. Genes exhibiting a fold change greater than 3 were assigned a score of ′3′.
Other parameters
Impact on overall survival
Genetic alterations associated with patient survival outcomes are indicative of their biological significance15. A score of ′1′ was designated to genes whose aberrations had a significant impact (p≤0.05) on overall survival (OS) if there was no significant correlation then it was ‘0’. This analysis was performed using the cBioportal database, which employs the Kaplan-Meier statistics test to identify genes associated with the outcome. The patient cohort was divided into two groups; based on the presence or absence of alterations in amplification, mutation, or overexpression of a specific gene.
Validation of genes in other cancer types
Through literature curation (PubMed search), we assigned a score of ‘1’ to genes previously shown to have a driver role in other cancer types. This criterion helped us in identifying novel targets whose function is unknown in cancer.
Identification of deleted putative tumor suppressors
We used the same parameters for scoring homozygously deleted genes as those used for scoring the amplified genes, with the exceptions as shown below.
Frequency of gene deletion
To identify the putative tumor suppressors, we analyzed their frequency of homozygous deletions and assigned scores from ‘1’ to ‘7’ depending on their alteration frequencies.
The average size of the aberration
Homozygously deleted chromosomal regions ranged in size from 0.18Mb to 3.7Mb, with an average size of ≤1.4Mb. Consequently, genes located within a 1.4Mb were given a score of ‘1’
Number of genes in the aberration
We assigned a maximum score ‘of 3’ to regions of 0.4Mb containing one gene. Regions of the same size with 2 genes received a score of ‘2’, while those with 3 or more genes were assigned a score of ‘1’.
Frequency of somatic mutations
Nonsense somatic mutations within genes can result in gene silencing or protein dysfunction, contributing to tumorigenesis12. To assess a gene’s potential as a tumor suppressor, we considered the frequency of nonsense mutations. Genes mutated in more than 1% of tumors were assigned a score of ‘1’, while those without such mutations were given a score of '0'.
At the RNA level
Frequency of gene expression
To assess the potential tumor suppressor function of genes, mRNA downregulation was evaluated. Genes exhibiting decreased mRNA expression were assigned scores from ‘1’ to ‘7’, correlating with increasing downregulation frequency within the tumor cohort. Downregulation frequency ranged from 5% to 35% across analyzed tumors.
Fold change
In the dataset, fold change values ranged from 0-3-fold. Scores were assigned on a scale of 1 to 3 based on the magnitude of the decrease in expression. Genes exhibiting a fold change of less than -3 were assigned a score of ‘3’.
Other parameters
Impact on overall survival
A score of ‘1’ was assigned to genes whose aberrations had a significant impact (P ≤ 0.05) on overall survival (OS). To perform this analysis, the patient cohort was divided into two groups based on the presence or absence of homozygous deletion, nonsense mutation, or downregulation in a specific gene.
Validation of genes in other cancer types
Genes previously shown to function as tumor suppressors in other cancer types were assigned a score of ‘1’.
Other validation datasets
Four independent datasets were selected from the Oncomine database for analyzing the expression of RNF144B and PPP2R2A (See supplementary methods).
Cell lines and cell culture
HEK293T was obtained from ATCC (https://www.atcc.org/). The immortalized cell line HOSE642 (Human ovarian surface epithelial cells) was obtained from Dr. Samuel C. Mok (University of Texas, MD Anderson Cancer Center, USA) and was maintained in M19916. FT33 cell line, procured from Dr. R.Drapkin, (Dana-Farber Cancer Institute, Harvard Medical School, Boston) was maintained in DMEM with ultroser G17 . Other commonly used ovarian cell lines obtained from ATCC were maintained and cultured in DMEM medium or RPMI-1640, supplemented with 10% FBS, 1% penicillin–streptomycin, and grown at 37 °C and 5% CO2 atmosphere. All cell lines were regularly screened for mycoplasma by PCR.
RNA extraction and real-time PCR (qPCR)
Total RNA was extracted using Trizol reagent (Sigma-ALDRICH) following the manufacturer’s protocol and 1 µg of RNA was used to generate cDNA. Relative levels of expression of genes were detected using ABI Quant studio Flex 12 k. qPCR cycling conditions used were 94 °C for 2 min, then followed by 40 cycles of 94 °C for 45 secs, 60 °C for 45secs, and 72 °C for 55secs. To confirm specific product amplification, disassociation curves were analyzed. All samples were run in triplicates, and gene expression data were normalized to β-actin levels. Relative quantification of gene expression was performed using the 2 −ΔΔCT method. The primers used are listed in Supplementary Table 1.
Vectors
The pCDNA4/TO/MYC-HIS-C with RNF144B CDS (NM_182757.4) was obtained from Dr. Berna Sayan, University of Manchester18. The pLA-CMV N Flag-PPP2R2A construct was a generous gift from Dr. Anna Sablina, VIB Laboratory for the Mechanism of cell transformation, KU Leuven19. The PPP2R2A CDS (NM_002717.4) was further cloned into pCDNA4/To/Myc/His-c vector (Invitrogen) (See supplementary methods for details).
RNF144b peptide antibody
A polyclonal antibody specific to human RNF144B was developed in collabration with Abgenex Pvt. Ltd (Bhubaneswar, Odisha, India). The C-terminal amino acid sequence of RNF144B from 224 to 243 (QNLDNDIFLRHYDKGPCRNK) was chosen using MacVector (https://macvector.com). Rabbits were immunized with KLH-conjugated RNF144B peptide and the immune sera collected on day 64 were purified using a peptide affinity column (see supplementary methods).
Immunoblotting and Urea SDS-PAGE
Protein lysates were prepared using a lysis buffer consisting of 500 mM NaCl, 1 mM Tris pH 8.0, 1 mM EDTA, 1% Triton X-100, protease & phosphatase inhibitors (NAF, sodium orthovanadate). Protein concentration was estimated using a BCA assay, separated on SDS-PAGE, and transferred to a PVDF membrane. The blots were probed with respective primary and secondary antibodies20. Protein bands were detected using an ECL-prime kit (G-Biosciences) on X-ray film. Image J was used to calculate the relative protein band intensity.
Urea (6 M) polyacrylamide gel electrophoresis was used for enhanced resolution of proteins. For preparing a 10% 6 M urea SDS-PAGE, the protocol as used for SDS-PAGE was followed20, except that 3.6 g of urea was added to the resolving SDS-PAGE mixture.
Immunoprecipitation
Immunoprecipitation was performed by incubating lysate with 1 µg of antibody for 1 h at 4 °C. This lysate was then added to protein A/G agarose beads and incubated at 4 °C overnight on a Rotospin at 8 rpm.
For RNF144B, unconjugated RNF144B peptide (1 mg/ml) was incubated with 1 µg of RNF144B antibody for 4 h on ice as a control. Protein lysate was added to the peptide mix, which was then added to the beads and incubated at 4 °C overnight. Immunoprecipitates were washed with 1XPBS and spun at 2000 rpm for 1 min. The beads were boiled at 95 °C for 5 min and the supernatant collected after centrifugation was resolved on SDS-PAGE20.
Stable and transient transfection
Cells were plated in a 6-well plate containing DMEM media with 10% FBS. Four hours before transfection, the cells were transfected with 2 µg of plasmid using a Jet-prime reagent (Polyplus). After 48 h of transfection, fresh media was added to the cells containing the antibiotic zeocin. The colonies that remained on the culture plate after 14–18 days were isolated, expanded, and screened.
All the transient transfections were performed using Poly ethyl imine (PEI). 1-2 µg of plasmid was dissolved in 100 µl of serum-free medium, added to the cells, and lysed after 48 h. The screening was confirmed by performing a Western blot.
Cell proliferation and colony formation assay
Cell Proliferation was determined using the MTT assay. MTT reagent (0.5 mg/ml) was added to cells (n = 4000) seeded in each well of 96 well plates and incubated for 4 h. The formazan crystals formed were dissolved by adding 100 μl of DMSO and the absorbance was measured at 570 nm.
For colony formation assay, cells (n = 1000) were plated in a 6-well dish containing DMEM (10%FBS ± zeocin) and cultured for 2 weeks. Colonies formed were stained and counted manually21.
Wound healing and invasion assay
Cell migration was determined by scratch assay. Cells were seeded in a 24-well plate and allowed to reach 90–100% confluence. Using a sterile pipette tip, a scratch was made vertically across each well. The cells were washed with 1XPBS, replenished with fresh medium, and incubated at 37 °C. The closure of the wound was imaged at different time intervals (0 h-72 h). The percentage of wound closure was calculated using Image J21.
Invasion assay was performed using trans-well inserts (pore size 8 µm, Thermo-fisher) coated with Matrigel. Cells were adjusted at a density of 1 × 105 cells/ml in serum-free DMEM and 200 µl of cell suspension was added to the top chamber. In the lower chamber, 600 µl of DMEM + 20%FBS was added, and the plate was incubated at 37 °C for 24 h. Cells that invaded through the Matrigel were fixed, stained, and counted manually using the grid method.
Immunohistochemistry
The clinical characteristics of patients included in this study are given in the supplementary methods. Immunohistochemistry was performed on control tissues including the ovary (n = 7), fallopian tube (n = 7), and HGSOC tissues (n = 100). Colon and prostate adenocarcinoma tissues were used as positive controls. IHC was performed on paraffin-embedded tissues of 4 µm thickness as described previously22. The slides were incubated with the respective primary antibodies (RNF144B at 1:100 and PPP2R2A at 1:50 dilution).
The stained slides were scored by a pathologist who was blinded to the clinical data. Scoring was based on the intensity and the percentage of tumor cells stained. A score of 0 to 3 was assigned for staining intensity: negative = 0, low = 1, moderate = 2, and high = 3. The percentage of cells stained was scored as follows, no stain = 0, 1–30% = 1, 30–60% = 2, and 60–100% = 3. The score was calculated by multiplying the percentage of cells stained by the intensity of staining. These final scores were further recoded as (0: Negative, 1: 1&2, 2: 3&4, and 3: 6&9)23.
Statistical methods
Statistical analysis was performed using GraphPad Prism (version 5.0), IBM SPSS Statistics 22.0 software, and Microsoft Excel. The data is represented as mean ± standard deviation from three independent experiments performed in triplicate. P values less than or equal to 0.05 were considered statistically significant (See supplementary methods).
Results
Insilico analysis
Somatic copy number alterations present in HGSOC tumors were analyzed using the TCGA data from different SNP array platforms. The GISTIC algorithm was employed to identify the chromosomal peaks that are significantly altered24. GISTIC differentiates between focal-level and arm-level alterations based on the amplitude of aberrations. This SCNA data was downloaded from the Broad Institute (FIREHOSE)24. We chose focal SCNAs (amplification and deletion) which involve fewer genes, compared to the arm-level alterations, where hundreds of genes are altered. Since focal SCNA contains few genes, they are likely enriched for driver genes. In addition, focal SCNAs are frequently observed in cancer cell lines with a low number of passages and in the xenograft genome5. In total, 32 focal amplified regions containing 679 genes and 38 homozygously deleted regions with 1694 genes were identified in the TCGA HGSOC dataset. From this, we choose to analyze significant (FDR < 0.05) focal-level aberration containing a limited number of genes (≤ 10). Using this criterion, we identified 17 amplified peaks containing 95 genes and 15 homozygously deleted peaks with 59 genes (Supplementary Tables 2&3). To these 95 amplified and 59 homozygously deleted genes, we calculated scores based on the frequency of CNA alteration, mutation, number of genes in the peak & size of the peak. In addition, the frequency of alteration at the RNA level, fold change, and impact of all these alterations on overall survival were also considered for the scoring. Finally, a literature search was conducted to verify whether these were known cancer genes (Fig. 1a, b). Overall, 38 amplified and 18 homozygously deleted genes with a score of ≥ 5 out of 24 were selected (Fig. 1c and Tables 1 , 2). Using this scoring method, we could identify several novel putative genes and well-known cancer genes (Supplementary Table 5–7). Genes such as CCNE1, MECOM, BRD4, MYC, and PVT1 were among the top-ranked known oncogenes. Tumor suppressors such as MAP2K4, NF1, RB1, and WWOX were ranked in order of highest among the homozygously deleted genes (Supplementary Tables 4–6).
RNF144B, a novel oncogene in ovarian cancer
RNF144B was ranked first with a score of 10/24 among other amplified genes with unknown functions in ovarian cancer (Table 1). RNF144B is located on chromosome 6q22.3, identified as the only gene in the amplicon (chr6:18466469–18508269) with a significant q-value of 4.0712e-16. Alteration of RNF144B was found in 52% of tumors, including amplification in 6.9% and gains in 43.35% (Fig. 1d). Among the fourteen members of the RBR (RING between RING) family, RNF144B, and RNF19A are altered at a higher frequency in HGSOC compared to the other members (Supplementary Fig. 1a). However, RNF19A was located within an arm-level amplification of 8q and was, therefore, not considered for further analysis. Other RBR family members such as RNF144A25 and PARK226 were shown to function as tumor suppressors. Patients with tumors exhibiting gain and amplification of RNF144B have a significantly higher expression at the RNA level compared to the diploid tumors (tumors without alteration in the relevant gene) (p < 0.0001)27 (Supplementary Fig. 2a).
Having documented RNF144B amplification and overexpression using TCGA data, the analysis was further validated using the “Lu Ovarian cancer” dataset from Oncomine28. RNF144B was significantly overexpressed in serous and endometroid ovarian tumors compared to the OSE cells (p = 0.004) (Supplementary Fig. 2b). Additionally, we also found that the expression of RNF144B was significantly higher in chemo-sensitive HGSOC tumors (TCGA) compared to chemo-resistant (p = 0.008) (Supplementary Fig. 2c). Outcome analysis revealed that alterations in RNF144B (gain, amplification, and overexpression) were significantly correlated with improved overall survival (p = 0.01), but not with progression-free survival in the TCGA HGSOC dataset (Supplementary Fig. 2d).
Expression of RNF144B in ovarian cancer
RNF144B mRNA expression was initially evaluated in ovarian cancer cell lines (n = 10), immortalized cell lines HOSE, FT33, and in the ovary and fallopian tube tissue by qPCR. As shown in Fig. 2a, ovarian cancer cell lines A2780, SKOV3, PEO1, and CAOV2 had higher RNF144B expression compared to the control tissue, immortalized cell lines. Other cell lines CAOV3, OVCAR3, OVCAR5, OAW28, OAW42, and PEO4 showed minimal expression of the gene.
Expression of RNF144B in ovarian cancer cell lines and preparation of a polyclonal antibody against human RNF144B. (a) RNF144B mRNA expression in cell lines and controls was evaluated using semi-quantitative PCR (qPCR). β-actin was used as a control. The expression is shown relative to normal ovarian tissue (b) Schematic diagram of RNF144B protein, including the RING finger, IBR: in between ring finger and TM: transmembrane domain. The peptide antibody was raised against the amino acid sequence from 224–243 (accession number: NP_877434.2) (c) HEK293T cells were transfected with RNF144B (pCDNA4 + MYC-RNF144B plasmid) and the lysates were immunoprecipitated with an anti-MYC antibody and blotted with RNF144B specific polyclonal antibody (1:5000); lane 1-Untransfected, lane 2-transfected (d) The lysate was also immunoprecipitated with RNF144B and blotted with an anti-MYC antibody (1:1000 dilution); lane 1-Antibody was incubated with a peptide corresponding to the epitope, lane 2- antibody alone (1:5000 dilution). (e) RNF144B protein expression levels in normal tissues (ovary, fallopian tube), immortalized cell lines (HOSE, FT33), and ovarian cancer cell lines (n = 10) were examined by Western blot. RNF144B protein band intensity was measured using ImageJ software and normalized to GAPDH. Western blot analysis was performed to confirm the stable transfection of the RNF144B gene in different clones of (f) PEO4 and (g) OVCAR3 cells. Empty vector pCDNA4/To/Myc/His-c was transfected into these cells as a control. Data values represent the relative RNF144B protein expression intensity normalized to that of GAPDH.
Furthermore, we examined the endogenous expression of RNF144B at the protein level using Western blot analysis. We generated a polyclonal peptide antibody in rabbits against an epitope from the C-terminus (224-243a.a) of RNF144B (Fig. 2b). Immunoprecipitation with anti-RNF144B and anti-Myc antibodies in the presence or absence of cognate peptide from extracts of HEK293T cells transfected with MYC-tagged RNF144B showed the presence of a single band of approximately 35KDa (Fig. 2c, d). This was also confirmed by Western blotting of lysates. RNF144B was highly expressed in A2780, SKOV3, PEO1, and CAOV2. Other cell lines including OVCAR3, OVCAR5, PEO4, OAW42, and OAW28 had minimal expression of this protein. The Western blot results were consistent with qPCR findings, except for OAW28 which had an absence of expression at the protein level (Fig. 2e).
Functional role of RNF144B in ovarian cancer
To investigate the biological role of RNF144B in ovarian cancer, stable transfectants of PEO4 and OVCAR3 expressing RNF144B were established. Clone-3 from PEO4 and clone-1 from OVCAR3 cells were identified to have maximal overexpression of RNF144B compared to the other clones and vector-transfected cells. These clones were selected for subsequent experiments (Fig. 2f, g). MTT assay showed that the proliferative rate is significantly higher in RNF144B over expressed PEO4 (p = 0.0003) & OVCAR3 (p = 0.04) cells at 72 h compared to the control cells (Fig. 3a, b).
RNF144B promotes ovarian cancer cell proliferation, colony formation, and migration. Cell viability assays were performed on (a) PEO4 and (b) OVCAR3 cells stably overexpressing RNF144B compared to mock and parental cells. The number of viable cells was measured using an MTT reagent and spectrophotometric absorbance was taken at 570 nm, p = 0.0003 & 0.04. Ovarian cancer cells (n = 1000) (c) PEO4 (clone-3) and (d) OVCAR3 (clone-1) stably expressing RNF144B were plated in 6-well plates. Cells transfected with empty vector pCDNA4 were served as controls. After two weeks, the colonies formed were stained and counted using the grid method, p = 0.001 and < 0.0001. A wound healing assay was performed in (e) PEO4 (clone-3) and (f) OVCAR3 (clone-1) cells stably expressing RNF144B. Wound closure was captured at 0, 24, and 48 h at 4 × magnification. Wound closure was calculated by measuring the distance covered by the cells relative to the initial wound area determined at 0 h using image J software. The wound closure data is expressed in percentages, p = 0.0002 & 0.001. A transwell Matrigel invasion assay was performed on (g) PEO4 (clone-3) cells expressing RNF144B or empty vector. Cells were added to the upper chamber of the transwell insert. The cells that invaded through the Matrigel were fixed and stained. The average number of migrated cells was compared between RNF144B transfected and control, p = 0.09. All experiments were performed in triplicate, with three independent replicates for each. Statistical significance was calculated using the two-tailed student’s t-test, and the results are represented as mean ± SD.
Next, we assessed the colony-forming ability of cells overexpressing RNF144B. As shown in Fig. 3c&d, the relative colony-formation efficiency was significantly increased in PEO4 cells (p = 0.001) and OVCAR3 cells (p < 0.0001), compared to the control cells. Furthermore, a wound-healing assay was performed to determine whether RNF144B increases cell motility. Figure 3e&f shows that the migratory ability of cells (PEO4 and OVCAR3) transfected with RNF144B was significantly increased and, the complete wound closure was achieved at 48 h (p = 0.0002 & 0.001) in both the cell lines compared with that the control cells. A transwell Matrigel invasion assay demonstrated an increase in the number of invading cells in RNF144B compared to the controls. The average number of cells invaded was 429 for PEO4 cells transfected with vector, whereas for RNF144B transfected it was 585 (P = 0.09) (Fig. 3g). Taking all the results together, therefore, it suggests that RNF144B may have an important role in promoting tumorigenesis of ovarian cancer cells. Future experiments can be performed by overexpression of RNF144B in HOSE16 and FT3317 cell lines, which represent immortalized normal human surface epithelium of the ovary and fallopian tube.
Clinical data
A total of 100 patients with HGSOC diagnosed between the years 2014 and 2016 were included in this study. The median age of the cohort was 53 years (range 30 to 72 years). Thirty-one percent of patients were premenopausal and 69% were postmenopausal. The majority of patients had advanced disease: stage III (72%) and IV (27%). Histologically, the tumors were classified as grade II in 15% of patients, and high-grade (III) in 85% of patients. Only 5% of patients underwent primary surgery, while 69% of patients had optimal (total abdominal hysterectomy (TAH) with bilateral salpingo-oophorectomy (BSO), omentectomy with no macroscopic residual tumor) interval debulking surgery after 3–4 cycles of chemotherapy, 26% were inoperable and 5% refused to undergo surgery. Detailed patient characteristics and clinicopathological factors of the 100 patients are summarized in Supplementary Table 7.
Univariate and multivariate analyses were performed to evaluate the prognostic significance of the different clinicopathological factors. In univariate analysis, age (p = 0.012), comorbidities (p = 0.05), and surgery (Inoperable, p < 0.0001, and refused surgery, p < 0.0001) were significantly associated with decreased overall survival of patients. In multivariate analysis, only comorbidities (p = 0.01) and surgery (inoperable; p < 0.001, refused surgery; p < 0.0001) were identified to be strong prognostic factors that correlated with the overall survival of the patients (Supplementary Tables 8 & 9).
Expression of RNF144B in HGSOC tumors
The expression of RNF144B was evaluated by IHC in 100 primary HGSOC tumors (diagnostic biopsies), as well as in ovaries (n = 7) and fallopian tubes (n = 7). We initially tested the antibody in the positive control human colon tissue, which exhibits moderate to high expression of this protein. RNF144B antibody at dilution 1:100 demonstrated specific staining in the cytoplasm (Supplementary Fig. 4a). The expression of the RNF144B protein was predominantly located in the cytoplasm of the cells (both in normal and tumor cells) (Fig. 4a). RNF144B is expressed at low to moderate levels in the epithelial cells of the ovary and fallopian tube. Among the 100 HGSOC samples, RNF144B was strongly (score 3) expressed in 24 tumors (24%) and moderately expressed (score 2) in 16 (16%) of tumors. Low expression (score 1) was observed in 33 (33%) and 27 tumors (27%) had no expression of RNF144B (Fig. 4b). There was no significant correlation between the expression of RNF144B with the outcome (Supplementary Fig. 5a). To further understand the significance of RNF144B expression in response to chemotherapy, we have also examined samples obtained after neoadjuvant chemotherapy. Of the 100 pre-chemotherapy biopsies, matched paired tumor samples (resections following 3–4 cycles of chemotherapy) were available for 50 patients. Late recurrence of the disease was documented in 33 patients (66%), while 17 patients (34%) remained in complete remission A significant reduction in the expression of RNF144B was observed after adjuvant chemotherapy (p = 0.0001). The expression of RNF144B was observed in 26% (13/50) of tumors, with only 10% of tumors (5/50) showing immunostain intensity scores of 2 or greater after treatment compared to 40% before chemotherapy (Fig. 4c,d). However, the magnitude of the difference in the expression of RNF144B did not significantly correlate with the probability of relapse or remaining in complete remission (Fig. 4e).
Expression of RNF144B in tumors from patients with HGSOC. (a) Representative IHC staining of RNF144B in ovarian surface epithelium (OSE), fallopian tube (FT), and HGSOC tumors (n = 100) using anti-RNF144B antibody at concentration 1:100. (b) RNF144B protein expression levels in control and HGSOC tissues, p = 0.03. (c) RNF144B immunoreactivity in a panel of matched pair samples (n = 50). Tissues were collected from the same patient at the time of diagnostic biopsy and during surgery following 3–4 cycles of chemotherapy. (d) Plot showing the distribution of RNF144B intensity scores in matched pair samples, analyzed using two-tailed Student’s t-test, p < 0.0001. (e) Expression of RNF144B in patients who recurred (n = 33) compared to those in complete remission (n = 17). The bar graph represents the mean value of the difference in IHC scores between pre- and post-treated tumors obtained from patients who had either recurrence or complete remission, p = 0.821. The P-value was calculated using the Wilcoxon signed rank test. Magnification 40x (Microscope Nikon E200 Japan).
PPP2R2A, a novel tumor suppressor in ovarian cancer
PPP2R2A emerged as the top-ranked novel deleted gene (score 11/24) out of the selected 18 genes (Table 2). Loss of Heterozygosity (LOH) and homozygous Deletion (HD) of PPP2R2A were observed in 70.8% of tumors (n = 316) (Fig. 1e). Homozygous deletion of PPP2R2A was observed in 4.74% of these tumors, which provided initial evidence that it could function as a tumor suppressor since homozygous deletions are very rare. The expression of PPP2R2A was significantly lower in tumors with a single allele or loss of both alleles compared with diploid tumors (p = < 0.0001) (Supplementary Fig. 3a)11. A similar trend of reduced expression was also observed in three independent ovarian cancer datasets (Supplementary Fig. 3b). Furthermore, we also found that among different subunits of the PP2A holoenzyme, the expression of PPP2R2A is absent at a higher frequency in this tumor type (Supplementary Fig. 1b).
Expression of PPP2R2A in ovarian cancer
The expression of PPP2R2A was determined by qPCR in ovarian cancer cell lines (n = 9) and control samples (ovary, fallopian tube tissue & HOSE, FT33). Higher expression levels were observed in fallopian tube tissue and HOSE cells. Reduced expression of this gene was observed in four cell lines i.e., OVCAR3, OVCAR5, OAW28, and PEO4. The highest expression was noticed in SKOV3 followed by A2780. The other cell lines had a moderate expression of PPP2R2A (Fig. 5a). To confirm the results obtained by qPCR on the protein level, lysates from both the tissues and cell lines were subjected to Western blot analysis. We first examined the specificity of the antibody using the protein lysate isolated from HEK293T cells overexpressing PPP2R2A. Consistent with the qPCR results, downregulation of the expression of PPP2R2A was observed in four out of nine ovarian cancer cell lines (OVCAR3, OVCAR5, OAW28, and PEO4) (Fig. 5b).
PPP2R2A inhibits ovarian cancer cell proliferation, colony formation, migration, and invasion. (a) Relative mRNA levels of PPP2R2A in ovarian cancer cell lines (n = 9), immortalized cell lines (HOSE & FT33), and control tissue (ovary and FT) were evaluated by qPCR (qPCR). β-actin was used as a control. (b) the protein expression levels of PPP2R2A in these cell lines were also examined by Western blot using an anti-PPP2R2A antibody (1:1000 dilution). Protein band intensity was measured using ImageJ software and normalized to GAPDH. Western blot analysis of PPP2R2A expression in (c) OVCAR3 & (d) OVCAR5 cells after transient transfection with pcDNA4/To/MYC-His-A-Flag-PPP2R2A or the empty vector. MTT assays were performed on (e) OVCAR3 and (f) OVCAR5 cells 48 h after transient transfection with PPP2R2A plasmid. Cell proliferation rates were expressed as absorbance at 570 nm, p = 0.003 & 0.01. Colony formation assays were performed on (g) OVCAR3 and (h) OVCAR5 cells (n = 1000) plated in 6-well plates 24 h after transfection. Colonies formed were counted after 2 weeks, p = 0.001 and 0.0001. Wound healing migration assays were performed on (i) OVCAR3 and (j) OVCAR5 cells transiently expressing PPP2R2A. Wound closure was captured at 0, 24, and 48 h at 4 × magnification. Wound closure was quantified using Image J software and results are expressed as percentages, p = 0.001 & 0.01. A transwell Matrigel invasion assay was performed on (k) OVCAR5 cells transiently expressing PPP2R2A or the empty vector. Cells that invaded through the Matrigel were counted p = 0.01. Three independent experiments were performed, each in triplicates. Statistical significance was calculated using the two-tailed Student’s t-test, and results are presented as mean ± SD.
Functional role of PPP2R2A in ovarian cancer
We next examined the effect of ectopic expression of PPP2R2A in ovarian cancer cells through in-vitro experiments. Despite several experiments, we were unable to establish stable transfectants of PPP2R2A in cancer cell lines due to cell death. Consequently, all experiments were performed with transient transfection. First, the transfection of PPP2R2A plasmid into ovarian cancer cell lines (OVCAR3 and OVCAR5) was confirmed by Western blot (Fig. 5c, d). Next, we evaluated the effect of PPP2R2A overexpression on cell proliferation. The growth curves obtained from the MTT assay revealed that PPP2R2A significantly inhibited cell proliferation in both the cell lines (p = 0.003 & 0.01) at 72 h compared to the parental cells and cells transfected with an empty vector (Fig. 5e,f).
Consistent with the cell proliferation assay, the colony formation assay also demonstrated that the cells transfected with PPP2R2A had a reduced number of colonies compared to the controls (p = 0.001 &0.0001) (Fig. 5g,h). In addition, we also examined the effect of PPP2R2A overexpression on the migration and invasion of ovarian cancer cells. A wound-healing assay demonstrated that the migratory ability of cells was significantly reduced in ovarian cancer cells upon overexpression of PPP2R2A. The wound closure was about 88% in the control cells, whereas the PPP2R2A transfected OVCAR3 cells had a wound closure of 54% (p = 0.001). Similar results were obtained with OVCAR5 cells (p = 0.01) (Fig. 5i,j). In addition, the Matrigel invasion assay demonstrated that the number of cells invaded was markedly decreased in PPP2R2A transfected OVCAR5 cells compared to the control (p = 0.01) (Fig. 5k). These results confirm that PPP2R2A upon overexpression suppresses the growth of tumor cells in vitro.
Based on the previous reports we examined the sensitivity of cancer cells with reduced expression of PPP2R2A to PARP inhibitors28. To validate these findings, ovarian cancer cells SKOV3 and OVCAR5 were treated with the PARP inhibitor, Niraparib, at a concentration of 50 µM. Niraparib significantly inhibited the cell viability in both SKOV3 cells, which express high levels of PPP2R2A, and OVCAR5 cells, which express low levels of PPP2R2A (p 0.0004 and 5.27071E-05 respectively) compared to control cells. In addition, we identified that OVCAR5 cells were relatively more sensitive to Niraparib than SKOV3 cells (p = 0.0001) (Fig. 6a). While it is possible that the response to the PARP inhibitors is differentially regulated by the expression of PPP2R2A, further validation to demonstrate that increased sensitivity to PARP inhibitors is exclusively due to the depletion of PPP2R2A.
Sensitivity to PARP inhibitors and expression of PPP2R2A in patients with HGSOC. (a) Ovarian cancer cell lines with differential expression of PPP2R2A were treated with Niraparib (50 µm) and cell proliferation assays were performed using an MTT reagent. Two independent experiments were performed in triplicate. Statistical significance was calculated using the two-tailed Student’s t-test; p = 0.0001. Immunohistochemical control staining of human PPP2R2A. Representative immunostaining of PPP2R2A in ovarian surface epithelium (OSE) (b) fallopian tube, and HGSOC tumors. (c) PPP2R2A protein expression levels in control and HGSOC tissues. p < 0.0001 Magnification 40X.
Expression of PPP2R2A in HGSOC
To investigate the expression and clinicopathological significance of PPP2R2A in ovarian cancer, immunohistochemistry was performed on 100 HGSOC tumors and control samples from the ovary and fallopian tubes (n = 7 each). Prostate adenocarcinoma tissue was used as a positive control for IHC (Supplementary Fig. 4b). Immunoreactivity for PPP2R2A was observed in the cytoplasm of both normal and cancer cells (Fig. 6b). Ovarian and fallopian tube epithelial cells showed low to moderate cytoplasmic expression of PPP2R2A (score = 1&2). In contrast, the majority of HGSOC tissues exhibited low PPP2R2A expression. Out of 100 HGSOC samples, 72 tumors (72%) had no PPP2R2A immunoreactivity (score 0), and 10 tumors (10%) exhibited low expression (score 1) of this gene. The remaining, 18 cases had moderate (9%) and high expression (9%) of PPP2R2A (scores 2 & 3). Hence, from these results, it is clear that two-thirds of the HGSOC tumors had a loss of PPP2R2A expression (Fig. 6c). This data aligns with the loss of PPP2R2A in the TCGA HGSOC samples11. However, we did not find any correlation between the lack of PPP2R2A expression and overall survival (Supplementary Fig. 5b).
Discussion
Somatic copy number alteration results in activation of oncogenes and inactivation of tumor suppressors. SCNA’s are a major genetic alteration in HGSOC and are thought to contribute to its development29,30. However, the critical challenge in the analysis of SCNAs is to identify cancer-driver genes from other numerous random alterations that do not contribute to tumorigenesis. To address this, we have developed and validated a scoring system, to identify and prioritize cancer driver genes from SCNA’s. We implemented this method on HGSOC data from TCGA and identified 38 putative oncogenes and 18 putative tumor suppressors. This list includes well-known cancer genes such as CCNE1, BRD4, MYC, MAP2K4, NF1, and RB1, as well as novel putative cancer genes that may contribute to HGSOC. Among the novel genes identified, RNF144B (amplified gene), and PPP2R2A, (homozygously deleted gene) displayed a high score of 10 and 11 respectively. This is the first study to evaluate the expression and function of these two genes in HGSOC tumors.
The role of RNF144B, in the development of cancer, is emerging. This gene is significantly overexpressed in HNSCC18, endometrial cancer31, and chordoma32. Silencing of RNF144B results in decreased cell proliferation, colony formation, migration, invasion, and induced cell apoptosis in these tumor types18,31,32. Notably, the amplification of RNF144B is frequent in HGSOC (8.74%) next to uterine carcinosarcoma (12.5%) and is not frequently seen in other types of cancer based on the analysis of TCGA data11. In this study, using TCGA data we have shown that RNF144B is amplified and correlated with increased expression in HGSOC. Single allele gain was also noticed in a significant proportion of the tumors. Ectopic expression of RNF144B significantly increased cell proliferation, colony formation, and the migratory ability of the cells. E3 ubiquitin ligases function as oncogenes by targeting tumor suppressors such as p53, PTEN, and p21 for degradation18,33. Thus, it is reasonable to assume that RNF144B promotes ovarian tumorigenesis by targeting critical tumor suppressors. Further investigation into the selection of the substrates and the pathways could lead to the development of inhibitors. Several proteasomal inhibitors have shown success in targeting these E3 ubiquitin ligases and restoring the tumor suppressor function34. We also validated the overexpression of RNF144B in a significant proportion (50%) of HGSOC tumors using immunohistochemistry. Additionally, we, observed that the expression of RNF144B was significantly downregulated in tumors from patients who had received chemotherapy.
Homozygous deletion (HD) and Loss of heterozygosity (LOH) of chromosomal regions are common and critical in tumor progression. Several chromosomal regions demonstrating a high frequency of LOH such as 17p, 17q, 22q, 18q, 8p, 6q, and 13q have been identified in ovarian cancer35,36. TCGA identified significant loss of heterozygosity and deletion of chromosomal region 8p21.2 in HGSOC tumors4. PPP2R2A is likely to be the candidate tumor suppressor in this chromosomal region as the neighboring gene BNIP3L is not linked with ovarian cancer37. Loss of expression of PPP2R2A has been frequently observed in several tumor types including prostate38, bladder39, lung19, thyroid40, and breast cancer41. In HGSOC tumors, PPP2R2A is frequently lost due to homozygous and heterozygous deletions. However, heterozygous deletions are significantly common occurring in 65.1%, compared to the homozygous deletions which are observed only in 4.7% of tumors27. Similar results have been observed in prostate tumors38. PPP2R2A has been shown to function as a haploinsufficient tumor suppressor in prostate cancer where complete loss of this gene has been shown to have deleterious consequences. Loss of one allele of PPP2R2A correlates with poor prognosis in patients with prostate cancer42. Given the prevalence of hemizygous loss of PPP2R2A in HGSOC, it is crucial to investigate whether PPP2R2A functions as a haploinsufficient tumor suppressor in this tumor type. Future studies involving selective deletion of a single allele of PPP2R2A in this tumor type may provide valuable insights. Silencing PPP2R2A has been shown to result in cell cycle arrest at the G1-S phase and affects cell proliferation, colony formation, and tumor growth19,41,43. In this study, we demonstrated that overexpression of PPP2R2A exerts an inhibitory effect on the growth of HGSOC cells. This is supported by our findings that PPP2R2A overexpression reduces cell proliferation, colony formation, migration, and invasion in ovarian cancer cells. These results supported the hypothesis that PPP2R2A functions as a tumor suppressor gene in HGSOC. A recent study using a non-tumorigenic model demonstrated that homozygous deletion of PPP2R2A caused embryonic lethality. Hence PPP2R2A is an essential gene both in embryonic development and tumorigenesis. PPP2R2A was identified to exert its tumor suppressor function by targeting several substrates for dephosphorylation. This includes PI3K/AKT, Cyclin-dependent kinase B-Cdk1 and 4, β-catenin, and C-Myc44,45. Therefore, it is worthwhile to comprehensively investigate the substrates and the signaling pathways associated with PPP2R2A. We next, investigated the association of PPP2R2A with response to PARP inhibitors. PPP2R2A has been implicated to play a role in the DNA repair pathway by dephosphorylating the DNA repair gene ATM. Silencing PPP2R2A results in a defective HR pathway and sensitizes the cells to PARP inhibitors19. We found that OVCAR5 cells, which express low levels of PPP2R2A, are sensitive to Niraparib compared to SKOV3 cells, which have high expression of PPP2R2A. Although this experiment was performed in only two cell lines as an initial experiment, it suggests that PPP2R2A expression may mediate sensitivity to PARP inhibitors Further investigation involving a large panel of ovarian cancer cell lines and tumors, with PPP2R2A depletion, is needed to confirm this observation. Given the emerging evidence of PP2A modulators as therapeutic agents, particularly in PARP inhibitor-resistant patients, PP2A and its associated pathways represent a promising target for future therapeutic strategies in HGSOC46. Finally, the immunohistochemical analysis revealed that the expression of PPP2R2A is lost in the majority of HGSOC tumors.
Collectively these data suggest that the scoring system effectively identifies both novel and established cancer genes, supporting the robustness of this approach. The two novel cancer driver genes RNF144B and PPP2R2A have been shown to play a role in the tumorigenesis of HGSOC at the in vitro level. Although we have not yet demonstrated that either gene overexpression or loss correlates with long-term patient outcomes, such an analysis would require a larger sample size of patients treated with primary surgery. Ideally, we should compare expression from cognate sections of normal ovary and fallopian tube from the same patient which was not feasible always. Interestingly overexpression of RNF144B correlated with better outcomes with TCGA data (Supplementary Fig. 2c) for a trend observed with other oncogenes like GAB247. Future studies investigating the role of these two genes in relationship with the signaling pathways that regulate ovarian cancer cell proliferation, growth, and motility are warranted. Ultimately using a Cre-recombinase-dependent Cas9 ovarian cancer mouse model with sgRNA targeting these two genes RNF144B and PPP2R2A will ultimately substantiate the role of these two genes in HGSOC.
During the review of this manuscript, we noted a recently published study that also identified RNF144B as a potential oncogene in ovarian cancer. Their findings, highlighting RNF144B-mediated degradation of p21 regulated by HDAC348, further support the role of RNF144B in promoting ovarian cancer growth and metastasis. These complementary results reinforce the significance of RNF144B as a critical player in ovarian tumorigenesis and underscore the value of our approach in identifying cancer driver genes. Our study, focusing on the amplification and functional validation of RNF144B in HGSOC, adds a novel dimension and provides additional evidence for the oncogenic potential of RNF144B.
Data availability
All data generated or analysed during this study are included in this published article [and its supplementary information files].
Abbreviations
- EOC:
-
Epithelial ovarian cancer
- HGSOC:
-
High grade serous ovarian cancer
- OSE:
-
Ovarian surface epithelium
- HOSE:
-
Human ovarian surface epithelium
- FTSEC:
-
Fallopian tube secretory epithelial cells
- TCGA:
-
The cancer genome atlas
- SCNA:
-
Somatic copy number alteration
- CNA:
-
Copy number alteration
- Chr:
-
Chromosome
- FC:
-
Fold change
- GISTIC:
-
Genomic identification of significant targets in cancer
- FDR:
-
False discovery rate
- PARP:
-
Poly (ADP-ribose) polymerase
- PFS:
-
Progression-free survival
- OS:
-
Overall survival
References
Sung, H. et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 71 (3):209-249 (2021).
Bowtell, D. D. et al. Rethinking ovarian cancer II: reducing mortality from high-grade serous ovarian cancer. Nat. Rev. Cancer 15(11), 668–679 (2015).
Allemani, C. et al. Global surveillance of cancer survival 1995–2009: analysis of individual data for 25,676,887 patients from 279 population-based registries in 67 countries (CONCORD-2). Lancet 385(9972), 977–1010 (2015).
Cancer Genome Atlas Research N. Integrated genomic analyses of ovarian carcinoma. Nature 474(7353), 609–615s (2011).
Krijgsman, O., Carvalho, B., Meijer, G. A., Steenbergen, R. D. & Ylstra, B. Focal chromosomal copy number aberrations in cancer-Needles in a genome haystack. Biochem. Biophys. Acta. 1843(11), 2698–2704 (2014).
Grobner, S. N. et al. The landscape of genomic alterations across childhood cancers. Nature 555(7696), 321–327 (2018).
Manasa, P., Sidhanth, C., Krishnapriya, S., Vasudevan, S. & Ganesan, T. S. Oncogenes in high grade serous adenocarcinoma of the ovary. Genes Cancer 11(3–4), 122–136 (2020).
Zhou, W. et al. Identification of driver copy number alterations in diverse cancer types and application in drug repositioning. Mol. Oncol. 11(10), 1459–1474 (2017).
Graf, R. P., Eskander, R., Brueggeman, L. & Stupack, D. G. Association of copy number variation signature and survival in patients with serous ovarian cancer. JAMA Netw. Open 4(6), e2114162 (2021).
Sudhakar, M., Rengaswamy, R. & Raman, K. Multi-omic data improve prediction of personalized tumor suppressors and oncogenes. Front. Genet. 13, 854190 (2022).
Cerami, E. et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2(5), 401–404 (2012).
Vogelstein, B. et al. Cancer genome landscapes. Science 339(6127), 1546–1558 (2013).
Wood, L. D. et al. The genomic landscapes of human breast and colorectal cancers. Science 318(5853), 1108–1113 (2007).
Perron, G. et al. Pan-cancer analysis of mRNA stability for decoding tumour post-transcriptional programs. Commun. Biol. 5(1), 851 (2022).
Wang, P. W., Su, Y. H., Chou, P. H., Huang, M. Y. & Chen, T. W. Survival-related genes are diversified across cancers but generally enriched in cancer hallmark pathways. BMC Genomics 22(Suppl 5), 918 (2022).
Tsao, S. W. et al. Characterization of human ovarian surface epithelial cells immortalized by human papilloma viral oncogenes (HPV-E6E7 ORFs). Exp. Cell Res. 218(2), 499–507 (1995).
Karst, A. M. & Drapkin, R. Primary culture and immortalization of human fallopian tube secretory epithelial cells. Nat. Protoc. 7(9), 1755–1764 (2012).
Conforti, F. et al. PIR2/Rnf144B regulates epithelial homeostasis by mediating degradation of p21WAF1 and p63. Oncogene 32(40), 4758–4765 (2013).
Kalev, P. et al. Loss of PPP2R2A inhibits homologous recombination DNA repair and predicts tumor sensitivity to PARP inhibition. Cancer Res. 72(24), 6414–6424 (2012).
Sidhanth, C. et al. LASP-1 interacts with ErbB2 in ovarian cancer cells. Biochem. J. 479(1), 23–38 (2022).
Sneha, S. et al. The hedgehog pathway regulates cancer stem cells in serous adenocarcinoma of the ovary. Cell. Oncol. 43(4), 601–616 (2020).
Krishna Priya, S. et al. Expression of a novel endothelial marker, C-type lectin 14A, in epithelial ovarian cancer and its prognostic significance. Int. J. Clin. Oncol. 22(1), 107–117 (2017).
Nagare, R. P. et al. Expression of cancer stem cell markers CD24, EPHA1 and CD9 and their correlation with clinical outcome in epithelial ovarian tumours. Cancer Biomark. Sect. A Dis. Markers 28(3), 397–408 (2020).
Mermel, C. H. et al. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 12(4), 41 (2011).
Li, Y. et al. RNF144A suppresses ovarian cancer stem cell properties and tumor progression through regulation of LIN28B degradation via the ubiquitin-proteasome pathway. Cell Biol. Toxicol. 38(5), 809–824 (2022).
Wang, F. et al. PARK2 suppresses the proliferation of high-grade serous ovarian carcinoma via inducing the proteasomal degradation of ZNF703. Med Oncol 41(8), 207 (2024).
Gao, J. et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 6(269), pl1 (2013).
Rhodes, D. R. et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia 6(1), 1–6 (2004).
Margolis, B. et al. CCNE1 amplification among metastatic sites in patients with gynecologic high-grade serous carcinoma. Gynecol. Oncol. Rep. 37, 100850 (2021).
Despierre, E. et al. Somatic copy number alterations predict response to platinum therapy in epithelial ovarian cancer. Gynecol. Oncol. 135(3), 415–422 (2014).
Zhou, Q. et al. Pir2/Rnf144b is a potential endometrial cancer biomarker that promotes cell proliferation. Cell death Dis. 9(5), 504 (2018).
Wang, C. B., Wang, Y., Wang, J. J. & Guo, X. L. LINC00662 triggers malignant progression of chordoma by the activation of RNF144B via targeting miR-16-5p. Eur. Rev. Med. Pharmacol. Sci. 24(3), 1007–1022 (2020).
Deng, L., Meng, T., Chen, L., Wei, W. & Wang, P. The role of ubiquitination in tumorigenesis and targeted drug discovery. Sig. Transduct. Target. Ther. 5(1), 11 (2020).
Wang, D., Ma, L., Wang, B., Liu, J. & Wei, W. E3 ubiquitin ligases in cancer and implications for therapies. Cancer Metastasis Rev. 36(4), 683–702 (2017).
Shelling, A. N., Cooke, I. E. & Ganesan, T. S. The genetic analysis of ovarian cancer. Br. J. Cancer 72(3), 521–527 (1995).
Pribill, I. et al. High frequency of allelic imbalance at regions of chromosome arm 8p in ovarian carcinoma. Cancer Genet. Cytogenet. 129(1), 23–29 (2001).
Lai, J., Flanagan, J., Phillips, W. A., Chenevix-Trench, G. & Arnold, J. Analysis of the candidate 8p21 tumour suppressor, BNIP3L, in breast and ovarian cancer. Br. J. Cancer 88(2), 270–276 (2003).
Cheng, Y. et al. Evaluation of PPP2R2A as a prostate cancer susceptibility gene: a comprehensive germline and somatic study. Cancer Genet. 204(7), 375–381 (2011).
Zeng, L. P., Hu, Z. M., Li, K. & Xia, K. miR-222 attenuates cisplatin-induced cell death by targeting the PPP2R2A/Akt/mTOR Axis in bladder cancer cells. J. Cell. Mol. Med. 20(3), 559–567 (2016).
Huang, Y. et al. MicroRNA-222 promotes invasion and metastasis of papillary thyroid cancer through targeting protein phosphatase 2 regulatory subunit B alpha expression. Thyroid : Off. J. Am. Thyroid Assoc. 28(9), 1162–1173 (2018).
Di Conza, G., Trusso Cafarello, S., Zheng, X., Zhang, Q. & Mazzone, M. PHD2 targeting overcomes breast cancer cell death upon glucose starvation in a PP2A/B55alpha-mediated manner. Cell Rep. 18(12), 2836–2844 (2017).
Zhao, Z. et al. PPP2R2A prostate cancer haploinsufficiency is associated with worse prognosis and a high vulnerability to B55alpha/PP2A reconstitution that triggers centrosome destabilization. Oncogenesis 8(12), 72 (2019).
Cristobal, I. et al. PP2A inhibition is a common event in colorectal cancer and its restoration using FTY720 shows promising therapeutic potential. Mol. Cancer Ther. 13(4), 938–947 (2014).
Wlodarchak, N. & Xing, Y. PP2A as a master regulator of the cell cycle. Crit. Rev. Biochem. Mol. Boil. 51(3), 162–184 (2016).
Qiu, Z. et al. A genome-wide pooled shRNA screen identifies PPP2R2A as a predictive biomarker for the response to ATR and CHK1 inhibitors. Cancer Res. 80(16), 3305–3318 (2020).
Avelar, R. A. et al. Small-molecule-mediated stabilization of PP2A modulates the homologous recombination pathway and potentiates DNA damage-induced cell death. Mol. Cancer Ther. 22(5), 599–615 (2023).
Davis, S. J. et al. Enhanced GAB2 expression is associated with improved survival in high-grade serous ovarian cancer and sensitivity to PI3K inhibition. Mol. cancer Ther. 14(6), 1495–1503 (2015).
Zhuang, H., Zhang, Z., Wang, W. & Qu, H. RNF144B-mediated p21 degradation regulated by HDAC3 contribute to enhancing ovarian cancer growth and metastasis. Tissue Cell 86, 102277 (2024).
Acknowledgements
We would like to acknowledge the Department of Science and Technology-INSPIRE, Indian Council of Medical Research, and the Department of Biotechnology New Delhi, India for financial support. We are grateful to the staff of the Department of Pathology and Division of Epidemiology and Cancer Registry, Cancer Institute (WIA).
Funding
INSPIRE,Department of Science and Technology,New Delhi,India,Department of Biotechnology,India
Author information
Authors and Affiliations
Contributions
Manasa.P; Performed data analysis, invitro experiments, and prepared the manuscript. S.Krishnapriya; Investigation and manuscript editing, C.Sidhanth; Investigation, S.Vasudevan; Investigation, Kanchan Murhekar; Evaluated the histopathological staining and T.S.Ganesan; Designed the study, supervision, funding acquisition, critical revision & final approval.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Manasa, P., Krishnapriya, S., Sidhanth, C. et al. Characterization of RNF144B and PPP2R2A identified by a novel approach using TCGA data in ovarian cancer. Sci Rep 15, 5414 (2025). https://doi.org/10.1038/s41598-024-76801-3
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-024-76801-3