Abstract
Despite all prevention programs, many cases of colorectal cancer (CRC) are diagnosed when they have already metastasized. Herein, chemotherapy is required, and combination of 5-fluorouracil, irinotecan, and leucovorin (FOLFIRI) is one of the first-line treatments chosen. However, it is so toxic that compromises patient outcomes. Thus, with the aim of improving FOLFIRI pharmacokinetics while reducing its side effects, the three compounds that make it up were simultaneously absorbed in this work into polydopamine nanoparticles (PDA NPs), also loaded with an antibody to target CRC cells overexpressing the epithermal growth factor receptor (EGFR). All adsorptions, which were successfully executed without toxic solvents, were electrostatic in nature according to the calorimetry results obtained. Otherwise, based on the experiments done, 5-flurouracil, irinotecan, and leucovorin release from PDA NPs followed a burst-like pattern, which was possibly mediated by Fickian diffusion mechanisms. Finally, the assays performed with two EGFR-overexpressing CRC cell lines showed that the uptake of the nanosystem was rapid, and that its therapeutic effect was very significant. It managed to greatly reduce the viability of these cells to 22–30% after 72 h of incubation. Furthermore, when tumor spheroids were developed and treated with PDA NPs loaded with FOLFIRI and the anti-EGFR antibody (FOLFIRI-CTX@PDA NPs), these demonstrated to continue to have very marked therapeutic activity. In addition, FOLFIRI-CTX@PDA NPs affected to a lesser extent the survival rate of stromal cells, with which viability experiments were also done. Therefore, the novel developed PDA nanocarrier could be a promising strategy to enhance metastatic CRC therapy hereafter.
Similar content being viewed by others
Introduction
Among the different types of cancer, colorectal cancer (CRC) ranks the third most diagnosed, and it represents the second leading cause of cancer-related deaths worldwide1,2,3. When detected in early stages, CRC can be cured by surgical resection and shows a 5-year survival rate near to 90%. However, although its early detection has been improved by colonoscopy screening, over 50% of CRC cases are still diagnosed at late stage, when it has already metastasized and its 5-year survival rate is lower than 25% on average. In these cases, chemotherapy is required after surgical intervention to try to improve therapeutic outcomes1,4.
5-fluorouracil (5FU), which interferes with DNA and RNA synthesis by inhibiting the thymidylate synthetase, has been the first choice for CRC treatment since the 1950s2,5. Nevertheless, its hydrophilic nature and rapid metabolism make it necessary to continuously administer 5FU in high doses, which cause severe toxic effects3. Besides, nearly half of the patients with CRC become resistant to 5FU-based chemotherapy after a while2,5, and it is required to combine this drug with other cytotoxic agents and targeted therapies (like monoclonal antibodies (mAbs) or small-molecule tyrosine kinase inhibitors (TKIs)) to achieve a rescue therapy3,6. Thus, nowadays, the first-line chemotherapy for metastatic colorectal cancer (mCRC) is FOLFOX or FOLFIRI, in which 5FU is combined with leucovorin (LV) and oxaliplatin (FOLFOX regime), or with LV and irinotecan (CPT11) (FOLFIRI regime)1. LV is not a chemotherapeutic drug as such, but it is often administered with 5FU because it boosts its antitumor activity by augmenting 5FU inhibitory effect to thymidylate synthetase7. Otherwise, CPT11, which is also administered in the FOLFIRI regime, is a topoisomerase I inhibitor that blocks DNA replication and transcription8,9. In addition to 5FU, CPT11 is often combined with other cytotoxic drugs for the treatment of other types of cancer, but it is very poorly soluble in water and has such a complex pharmacological profile that the dose-dependent toxicity that it causes limits its administration6,8,9. Therefore, although the combination of 5FU, LV and CPT11 has been shown in several clinical trials to be very beneficial for patients with mCRC10,11, it is associated with poor tolerance. Its toxicity compromises the life quality of patients and treatment outcomes, so new approaches are still needed1.
In this context, Nanotechnology has shed some light with the development of drug delivery systems (DDSs) that have improved 5FU and CPT11 pharmacokinetic profile and therapeutic efficacy by reducing their premature metabolization and non-targeted uptake1,5,9. For instance, different liposomes carrying 5FU or CPT11, often also conjugated with mAbs, can be found in the literature as an attempt to achieve more effective therapies against CRC6,12,13,14,15. Similarly, different types of polymeric nanoparticles have already been loaded with one of these two antitumor drugs7,12,16,17, but very few authors have developed DDS in which both cytotoxic agents are transported simultaneously. Herein, a couple of 5FU and CPT11 nanocarriers have already been synthesized, but for the treatment of other digestive cancers other than CRC18,19.
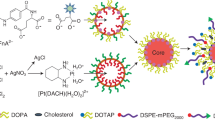
For this reason, the aim of this work was to develop polydopamine nanoparticles (PDA NPs) for the transport of not only 5FU and CPT11, but also LV and a mAb that would allow targeting the epidermal growth factor receptor (EGFR), a valuable blank in the treatment of CRC metastases20. Due to their excellent properties, such as the capacity of loading drugs without the need of employing toxic solvents, PDA NPs have been used in recent years by many authors to develop new DDSs21,22, in which they also provide an added intrinsic antiproliferative effect23. Likewise, PDA has also been used to coat other 5FU nanocarriers to enhance their properties24. However, to date, PDA NPs had not been simultaneously loaded with all FOLFIRI compounds in addition to a mAb to improve CRC therapy, and this is precisely what is described in this manuscript. Following an already stablished protocol25, ~ 170 nm PDA NPs were synthesized, characterized, and easily co-loaded with 5FU, CPT11, LV and cetuximab, one of the anti-EGFR mAbs whose administration is recommended for mCRC patients with wild-type KRAS tumors, but whose effectiveness is limited by the appearance of resistances26. Later, after determining the incorporation efficiency of all the compounds and verifying it by Fourier Transform-Infrared Spectroscopy (FT-IR), loaded NPs were again characterized. Isothermal titration calorimetry (ITC) assays were performed to study PDA interaction with the different drugs, and further experiments were carried out to analyze their pH-dependent release behavior. Moreover, the selective cytotoxicity of the PDA NPs loaded with all FOLFIRI compounds and with cetuximab was in vitro determined using two EGFR-overexpressing CRC cell lines, as well as stromal cells. Ultimately, their antitumor capabilities were also validated in 3D CRC spheroids, which were developed from the carcinoma cell lines used for the viability assays (Scheme 1).
Schematic illustration summarizing this work: PDA NPs were produced by oxidative polymerization and jointly loaded with 5FU, CPT11, LV, and an anti-EGFR antibody (cetuximab). Later, after complete physicochemical characterization of the nanosystem developed, its therapeutic activity was in vitro analyzed using conventional cell cultures and tumor spheroids. In the scheme, DLS refers to dynamic light scattering, SEM to scanning electron microscopy, UHPLC to ultra-high-performance liquid chromatography, and CLSM to confocal laser scanning microscopy. This figure was created with BioRender.com.
Experimental
Chemicals
Dopamine hydrochloride (DA), ammonium hydroxide (NH4OH, 28–30%, ACS reagent), calcium folinate (leucovorin calcium, LV), 5-fluorouracil (5FU, 99%), Trizma base, sodium acetate (anhydrous), phosphate buffered saline (PBS, 0.01 M), potassium bromide (FT-IR grade), pyrocatechol (99%), Dulbecco´s Modified Eagle´s Medium (DMEM), fetal bovine serum (FBS, USA origin), and methyl thiazolyltetrazolium bromide (MTT) were obtained from Sigma Aldrich (St. Louis, MO, USA). Irinotecan hydrochloride (CPT11) was supplied by MedChemExpress. The Pierce™ BCA protein assay kit, penicillin-streptomycin (5,000 U/mL), trypsin-EDTA (0.25%), dimethyl sulfoxide (DMSO, 99.9%), calcein AM, propidium iodide (PI, 1 mg/mL), and CellTracker™ Deep Red were purchased at ThermoFisher Scientific (Waltham, MA, USA). Absolute ethanol (EtOH, 99.5%) was supplied by VWR Chemicals (Radnor, PA, USA). Purified anti-human EGFR antibody (clone AY13), labeled or not with Alexa Fluor 488 (A488), was obtained from Inmunostep (Salamanca, Spain).
PDA NP synthesis
PDA NPs were synthesized in a water (90 mL) / EtOH (40 mL) mixture containing NH4OH (3.01% (v/v)), which was kept under magnetic stirring at 25 °C for 30 min. After this time, DA (0.5 g) was dissolved in deionized water (10 ml) and added directly to the previous solution. Polymerization reaction was allowed to proceed for 3 h25, and PDA NPs obtained were isolated by centrifugation. Next, they were purified through at least four centrifugation-redispersion cycles in deionized water. To determine DA conversion rate, NP suspension was dried in an oven at 105 °C until constant weight was achieved.
Physicochemical characterization of PDA NPs
PDA NP hydrodynamic diameter, polydispersity index (PDI), and zeta potential were determined by dynamic light scattering (DLS) (Nano ZS90, Malvern Instruments, Hertfordshire, UK). To perform diameter measurements, since they are more stable in alkaline media, NPs were diluted to a concentration lower than 0.01% (WT) in Trizma buffer (50 mM, pH 10.0). The correlation functions that were obtained were analyzed using the cumulant method. Otherwise, zeta potential of PDA NPs was determined after re-dispersing them in two more media in addition to Trizma buffer: (i) acetate buffer (pH 4.5), and (ii) PBS (pH 7.4). Both, size and zeta potential measurements, were performed in triplicate at 25 °C.
The morphology and sphericity of PDA NPs was observed by scanning electron microscopy (SEM) (FESEM Ultra plus, ZEISS, Oberkochen, Germany). Again, samples were prepared by diluting the NPs to a concentration lower than 0.01% (WT) in deionized water. Drops of the resulting dispersion were placed on a crystalline silicon wafer coated with silicon nitride, and they were left to dry for 24 h. Then, samples were metallized with thin iridium films (5 nm) to prevent their surface from electrical charging, and images were taken using a 3 kV acceleration. A secondary electron detector (SE/InLens) was used to observe NP surface. Once images were captured, size distribution histograms were created to determine the mean diameter of the dried PDA NPs. This last analysis was carried out with the ImageJ® software, with which more than 300 NPs were measured to obtain statistically significant results.
Preparation and characterization of FOLFIRI-CTX@PDA NPs
5FU, CPT11, and LV were loaded in PDA NPs both independently and in combination to build adsorption isotherms. 100 µg/mL stock solutions of each drug were prepared in PBS/DMSO mixtures (9:1 (v/v)) to later make standards ranging from 1 to 100 µg/mL. Next, PDA NPs (2.5 mg/ml, 1 ml), previously washed five times with PBS, were centrifuged so that their supernatant could be exchanged by the same volume of the different drug standards prepared. NPs were allowed to incubate with the drugs at room temperature for 15 h, maintaining 100 rpm orbital shaking. Then, they were again centrifuged to quantify the amount of the drugs existing in the supernatant and, thus, the amount of the drugs loaded. Such quantification was carried out by ultra-high-performance liquid chromatography (UHPLC)/mass spectrometry (Vanquish/ Q Exactive Focus, Thermo Fisher Scientific, Waltham, MA, USA), using two different C18 columns (Ascentis® Express (2.7 μm, 150 × 4.6 mm) for 5FU and CPT11 quantification, and Avantor® ACE® (3 μm) for LV determination) that were maintained at 30 °C. In all cases, the mobile phase consisted of a mixture of (A) deionized water/formic acid (99.9/0.1 (v/v)) and (B) acetonitrile/ formic acid (99.9/0.1 (v/v)), and the flow rate was set at 0.3 ml/min. Retention times of 5FU, CPT11, and LV were 0.96, 3.99 and 2.82 min, respectively27,28,29.
Once PDA NP adsorption capacity of the three drugs was examined, adsorption experiments were anew conducted to characterize the physicochemical properties of the NPs obtained and perform in vitro assays with them. The initial concentrations of the drugs used for these experiments were 14 µg/ml (5FU), 50 µg/ml (CPT11), and 80 µg/ml (LV), which were chosen after determining their LD50 in the cell lines mentioned below. Furthermore, a solution containing the three drugs was also prepared to assess their effect in combination. PDA NPs were also incubated with the anti-EGFR mAb (CTX) to incorporate it onto their surface. In this way, both bare PDA NPs and PDA NPs loaded with 5FU, CPT11 and LV were incubated with 38 µg/ml CTX at room temperature (100 rpm) once 3 h of drug loading were elapsed. After 12 h, all NPs were centrifuged, and the amount of the three drugs adsorbed, as well as the amount of the mAb loaded, were quantified (Eqs. 1 and 2)30. Determination of the amount of mAb existing in NP supernatants was done with the commercial Pierce™ BCA protein assay kit.
Finally, NPs loaded with all the drugs independently and jointly were characterized by the same techniques as bare PDA NPs (DLS and SEM). In addition, IR absorption spectra of all NPs were obtained in the 4000–400 cm− 1 wavelength range (PerkinElmer SpectrumTwo™) after preparing the corresponding sample pellets with KBr.
ITC analysis
Interactions between PDA catechol groups, 5FU, CPT11, LV, and the anti-EGFR mAb were investigated at 25 °C by ITC (Nano-ITC system, TA Instruments, Waters Corporation, Milford, MA, USA). Prior to titration, samples underwent a degassing process to eliminate potential bubbles that could influence the measurements. Then, pyrocatechol dissolved in PBS (pH 7.4, 75.3 mM, 1.5 ml) was introduced in the reaction cell, while fresh PBS (pH 7.4) was placed in the reference cell. In all cases, titration was conducted using a 250 µl syringe filled with the respective drug or antibody (5FU: 2.15 mM, CPT11: 1.7 mM, LV: 3.4 mM, and CTX: 0.005 mM) by performing 15 injections (17.4 µl) every 200 s and at a stirring speed of 250 rpm. After the measurements, integrated heats of the reference titrations were subtracted from the integrated heats of the experimental titrations, achieving normalized heats for each titration step. Finally, normalized heats were fitted according to an independent binding model (assuming that there were no cooperative effects between the drugs) to obtain the corresponding association constant (Ka), reaction enthalpy (ΔH°), free energy (ΔG°) and entropy (ΔS°) values31.
FOLFIRI release study
5FU, CPT11, and LV release from PDA NPs was analyzed using the dialysis bag method. PDA NPs (1 ml) loaded with all the drugs (4.9 µg 5FU/mg, 18.4 µg CPT11/mg, and 29 µg LV/mg) were dispersed within a dialysis bag that was subsequently placed in a beaker containing 20 ml of PBS (pH 7.4) or acidic PBS (pH 4.85). The beaker was kept in orbital shaking (40 rpm) at 37 °C for 72 h, and aliquots were collected at specified time intervals. The subtracted volume (1 ml) was replaced by fresh PBS or acidic PBS, and the amount of 5FU, CPT11, and LV existing in the aliquots was determined by UV-Vis spectroscopy at 266 nm, 256 nm, and 286 nm, respectively32,33,34. Then, release data were analyzed by applying four different kinetic models: the zero-order, first-order, Higuchi, and Korsmeyer-Peppas models. Detailed explanation regarding these models can be found in the Supplementary Information.
Cellular uptake assay
EGFR-overexpressing colon carcinoma cells (HTC116 and HT29 cells) were seeded on glass-bottom dishes (35 mm) (Ibidi, Gräfelfing, Germany) at a density of 50,000 cells/dish and cultivated overnight in DMEM supplemented with 10% (v/v) FBS. Next day, cells were stained with CellTracker™ Deep Red (20 \(\:{\upmu\:}\)M) following manufacturer’s instructions. Also, culture medium was removed and replaced with fresh medium containing FOLFIRI-CTX-A488@PDA NPs (30 \(\:{\upmu\:}\)g/ml), which were synthesized as previously mentioned, but using an anti-EGFR antibody labeled with A488 (CTX-A488). After 0.5, 6, and 24 h of incubation, the medium was again removed. Cells were washed with PBS for 3 times and observed under confocal microscope (LSM510, Zeiss, Oberkochen, Germany).
Cytotoxicity evaluation
HTC116, HT29 and stromal (HS5) cells were used to assess the cytotoxicity, by the MTT method, of PDA NPs, PDA NPs loaded independently with each drug or the anti-EGFR mAb (5FU@PDA NPs, CPT11@PDA NPs, LV@PDA NPs, and CTX@PDA NPs), PDA NPs loaded simultaneously with all drugs (FOLFIRI@PDA NPs), and PDA NPs loaded with all drugs plus cetuximab (FOLFIRI-CTX@PDA NPs). In brief, all cells were inoculated at a density of 12,000 cells/well and cultivated overnight in 24-well plates in supplemented DMEM. Subsequently, after determining the LD50 of 5FU, CPT11, and LV in HTC116 and HT29 cells (data not shown), different concentrations of all PDA NPs were added to the wells, ranging from 10 to 50 \(\:{\upmu\:}\)g/ml. Control cells were subjected to treatment with PBS. After a further incubation for either 24, 48, or 72 h, solution containing 5 mg/ml MTT was added to each well to a volume of 110 \(\:{\upmu\:}\)l and allowed to incubate for a further 1 h. Thereafter, culture medium was discarded and exchanged with DMSO (500 \(\:{\upmu\:}\)l/well). The absorbance of each well at 550 and 700 nm was determined using a microplate reader23. Three parallel tests were carried out for every concentration.
Analysis of particle penetration into multicellular tumor spheroids (MCTS)
EGFR-overexpressing spheroid models were obtained after culturing HCT116 and HT29 cells for 72 h in U-shaped bottom 96-well plates (Nunclon™ Sphera™ 96-well plates) at an initial density of 2,000 cells/well. Then, spheroids were transferred to glass-bottom dishes (4–5 spheroids/dish) and incubated with PDA NPs and FOLFIRI-CTX@PDA NPs (30 \(\:{\upmu\:}\)g/ml) for further 48 h. After this time, cells were stained with calcein AM and PI (1.3 \(\:{\upmu\:}\)M) and observed 30 min later under confocal laser scanning microscopy (CLSM). Also, some of the spheroids were not stained, but rather observed by SEM (QUANTA 200 FEG, FEI, Hillsboro, OR, USA) after being coated with gold.
Statistical analysis
All data, in both the manuscript and Supplementary Information, are presented as the mean \(\:\pm\:\) standard deviation (SD). One-way and two-way ANOVA and t-tests were used to determine statistical significance. The significance thresholds for differences were set at *p < 0.05, **p < 0.01, and ***p < 0.001.
Results and discussion
Characterization of PDA NP size, zeta potential and morphology
As mentioned, PDA NPs were synthesized through DA oxidative polymerization in an alkaline medium containing EtOH in the presence of atmospheric oxygen35,36. Generally, most authors let this polymerization reaction take place for 24 h. Nevertheless, it has found that it is possible to obtain PDA NPs with the same physicochemical properties after 3 h in previous work25, and the reaction was only allowed to pass this time. Herein, PDA NP hydrodynamic diameter after 3 h of reaction was 172.7 ± 53.6 nm (PDI: 0.074), while their zeta potential was − 39.9 mV at pH 10.0, -17.4 mV at pH 7.4, and − 7.1 mV at pH 4.5. These last values were like other previously reported, and indicated that PDA nanocolloids had minor stability in acidic environments25.
After loading PDA NPs with the different drugs and with the mAb, both their size and surface charge changed, as can be seen in Fig. 1A-B. On one hand, NP diameter increased after all loading processes. In this manner, PDA NPs reached 210.4 ± 72.9 nm (PDI: 0.130) after 5FU loading (3.9 µg/mg), while their diameter increased up to 216.6 ± 80.9 nm (PDI: 0.116) after CPT11 loading (17 µg/mg). Likewise, once LV was incorporated into PDA NPs (32 ug/mg), their diameter turned out to be 205.3 ± 67.7 nm (PDI: 0.101), therefore being this increase somewhat less remarkable than in the previous cases. Otherwise, when 5FU, CPT11 and LV were all loaded together in PDA NPs, their hydrodynamic diameter increased to 231.6 ± 81.9 nm (PDI: 0.139). Moreover, interestingly, while the incorporation efficiency (IE) of LV was like that obtained after individual adsorption of the drugs (29 µg/mg), 5FU and CPT11 IEs were higher (4.9 µg/mg and 18.4 µg/mg, respectively) when the three drugs were adsorbed together, which may indicate that there was not significant competition for PDA binding sites among 5FU, CPT11, and LV. Besides, when PDA NPs were loaded with all the FOLFIRI compounds and CTX, their diameter augmented even further, reaching 297.3 ± 102.3 nm (PDI: 0.193). Since the increase was quite large, it was also decided to determine the size of the NPs loaded only with the mAb, finding that it was about 258.8 ± 97.7 nm (PDI: 0.100). Consequently, CTX incorporation significantly modified PDA NP diameter. In addition, 5FU and LV IEs were remarkably reduced (to 1.7 µg/mg and 3 µg/mg, respectively) after the incorporation of the mAb into the nanosystem, which was also lower than when CTX was adsorbed in isolation (3.2 µg/mg vs. 5.2 µg/mg). The only IE that did not decrease much after CTX loading was that of CPT11 (18.4 µg/mg). To facilitate the comprehension of the work, the IEs obtained after all adsorption processes, expressed both in ug/mg and in percentage, are collected in Table S1.
(A) DLS intensity distributions of the different PDA NPs dispersions in Trizma buffer (pH 10.0, 50 mM; (B) Hydrodynamic diameter (nm) of PDA NPs before and after loading them with the different drugs and the antibody specified under each bar; (C) Zeta potential values of the different PDA-based nanosystems, as well as of free 5FU, CPT11 and LV, at pH 4.5, 7.4, and 10.0.
On the other hand, PDA NP zeta potential became more negative after the loading of three drugs, either individually or in isolation, both at pH 7.4 and 4.5. This behavior, which can be seen in Fig. 1C, could be indicative that drugs were being internalized into PDA NP structure, as also shown by the SEM images that were taken (Fig. 2). In these images, morphology of bare PDA NPs (Fig. 2A) and of those loaded with 5FU, CPT11, and LV (Fig. 2B-E) did not differ significantly despite the increase in the size of the latter. However, when CTX was adsorbed, NP zeta potential notably acquired more positive values, regardless of whether they were previously conjugated with FOLFIRI drugs. This fact, which had already been reported by El Hallal et al.37, could show that the mAb was retained on PDA NP surface. Besides, this fact would explain why the size of CTX@PDA NPs and FOLFIRI-CTX@PDA NPs increased so much after the incorporation of the antibody, and why their morphology differed somewhat more from that of the rest of the PDA nanosystems (Fig. 2F).
Drug-loading characterization by FT-IR
With the aim of assessing 5FU, CPT11, and LV incorporation into PDA NPs, FT-IR assays were performed to compare the spectra of PDA NPs, 5FU, CPT11, LV, 5FU@PDA NPs, CPT11@PDA NPs, and LV@PDA NPs.
Starting with 5FU loading, some of the bands existing in 5FU spectrum could also be observed in that of 5FU@PDA NPs (Fig. 3A). Some of them were, for instance, the intensity band at 420 cm− 1 corresponding to bending of the halogen bond within the ring of the aromatic fluorinated compound; the absorption band at 550 cm− 1 that could be attributed to C-F deformations31; and the absorption bands observed at 752 and 870 cm− 1, which may correspond to C–H out of the plane vibrations at –CF = CH–38. Likewise, the absorption band at 1180 cm− 1, which could be assigned to C-O vibrations31; the absorption band at 1429 cm− 1 that could be assigned to C-N stretching vibrations39; the absorption band at 1580 cm− 1 that may correspond to C-N and C-C ring stretching vibrations; the adsorption band at 1723 cm− 1 that could be related to C = O stretching frequency; and the adsorption band at 2930 cm− 1 that may be attributed to the -CH2 group could be differentiated in both spectra31, so 5FU loading was successfully performed.
Secondly and regarding the absorption of CPT11, it was also observed that some characteristic bands of this drug existed in CPT11@PDA NPs, so its loading was successful, too (Fig. 3B). Some of these bands were, for example, those at 1160 cm− 1 and 1230 cm− 1, which may correspond to the stretching vibration of the C–O ester and to the C = C and C–CH3 aromatic stretching vibrations, respectively; and those bands at 1660 and 1745 cm− 1, which may be assigned to the C = O group from the lactone, the carbamate, and the pyridine group40.
Finally, it could also be seen that LV and LV@PDA NPs shared some absorption bands (Fig. 3C), such as those existing at 765 cm− 1, 1190 cm− 1, and 1320 cm− 1, which may correspond to the C-H group, the C = O group, and aromatic ring of LV, respectively41.
Analysis of the interaction between PDA pyrocatechol, 5FU, CPT11, LV and CTX by ITC
Once 5FU, CPT11, LV, and CTX loading on PDA NPs was verified by FT-IR experiments, the interaction between the different compounds and the nanocarrier was analyzed by ITC. Nonetheless, assays were not carried out directly with the NPs, since PDA molecular weight is not accurately known. Instead, ITC assays were done with pyrocatechol, one primary monomer that is produced during DA oxidative polymerization, and which is responsible for PDA’s ability to load numerous compounds (drugs, peptides, proteins…)42,43,44. In this way, a concentration of monomer like that of PDA NPs employed was used to adsorb FOLFIRI compounds and CTX. Heat changes were detected when all drugs and the antibody interacted with pyrocatechol, and binding isotherms were built and fitted using an independent binding model (Figure S1). The calculated thermodynamic and binding parameters, including the association constants (Ka), the binding enthalpies (ΔH°), the binding entropies (ΔS°), and the binding free energies (ΔG°), can be seen in Table 1. According to the data obtained, it was verified that 5FU, CPT11, LV, and CTX interactions with pyrocatechol exhibited an exothermic behavior and were thermodynamically favorable. In general, there was a negative change of enthalpy and a positive change of entropy that favored such interactions, and the Ka values that were obtained may suggest that these bindings were electrostatic in nature. Besides, Ka value for CTX-pyrocatechol interaction was higher than for the rest of interactions, which could explain its slightly more negative ΔG° value45,46.
pH-responsive release of 5FU, CPT11, and LV from PDA NPs
Next, before performing cellular assays, the release rate of 5FU, CPT11, and LV from PDA NPs was evaluated in PBS at pH 4.85 and 7.40 to simulate lysosomal and physiological conditions, respectively. The drug release profiles, evaluated for 72 h, are depicted in Fig. 4. When analyzing the two graphs in that figure, it can be observed that, in general, the release of the three compounds displayed a burst-like pattern during the first hours, and that it was a little faster at acidic pH. More specifically and regarding 5FU, it should be noted that about 70% 5FU was released after 5 h in the acidic medium, while this release was somewhat less marked (54%) at the physiological pH after the same time. However, after 24 h, 5FU release profile at both pH values became more equal and, after 72 h, the amount of drug released in both media was similar and close to 95%. Regarding the release profile of CPT11, it was like that of 5FU. During the first 3 h of the assay, about 10% more drug was released at acidic pH than at the physiological pH but, after 5 h, CPT11 release was similar in both buffers, reaching values of 64–69% after 72 h. Finally, LV release also occurred more rapidly at acidic pH during the first hours of the experiment. Nevertheless, at the end of the assay, when the differences between both buffers were not so marked, the release of this compound was considerably lower than that of the two previous cytotoxic agents, reaching 44–53% after 72 h.
Globally, the release of greater percentages of drug at pH 4.85 may be attributed to the protonation of PDA amino groups in acidic environments, which could account for facilitating drugs’ release when NPs are endocyted in cells compared to physiological conditions47.
Otherwise, to study what mechanism could contribute to 5FU, CPT11, and LV release from PDA NPs under physiological conditions (diffusion, erosion, or swelling), four different mathematical models were implemented to evaluate the release kinetics of the drugs: zero-order, first-order, Higuchi and Korsmeyer-Peppas models. According to the results obtained, which can be found in the Supplementary Information along with the equations of the different models applied (Table S3), the Korsmeyer-Peppas model was found to be the best fitted to the drug release data, supported by the highest correlation coefficient (R2) values. In obedience to this model, the values of k, which is the release rate constant, were higher at pH 4.85, which could be explained by the fact that drug release took place faster at acidic pH. On the other hand, n is the diffusional exponent characteristic of the release mechanism and, as described in the literature, if it acquires values < 0.43, it will correspond to Fickian diffusion mechanisms, which may consequently govern the release of the FOLFIRI compounds from PDA NPs48,49,50.
Rapid cellular uptake of FOLFIRI-CTX@PDA NPs
Before accomplishing cytotoxicity studies, internalization of FOLFIRI-CTX-A488@PDA NPs in EGFR-overexpressing CRC cells was visualized under CLSM. Both HTC116 and HT29 human cell lines were used for this assessment51,52. As can be seen in Fig. 5, cellular uptake took place fast, since most NPs were distributed in the cellular perinuclear region after 30 min of incubation. Then, after 6 h of incubation, FOLFIRI-CTX-A488@PDA NPs could already be seen in the cytoplasm of the cells, especially in the HT29 cell line. In the HTC116 one, the complete internalization of the NPs, however, was much clear after 24 h. In any case, despite the anionic nature of their zeta potential, FOLFIRI-CTX-A488 NPs demonstrated excellent internalization, which is a fundamental characteristic that DDSs must have53,54. Possibly, thanks to the presence of the anti-EGFR antibody in the nanosystem, its internalization was propitiated by a receptor-mediated endocytosis process55.
Cytotoxicity of PDA NPs loaded with the different FOLFIRI compounds and the anti-EGFR mAb
As for uptake experiments, HTC116 and HT29 CRC cells were used to assess FOLFIRI-CTX@PDA NP therapeutic efficacy in vitro. Thus, both human carcinoma cell lines were treated for 72 h with concentrations of this nanosystem ranging from 10 to 50 \(\:{\upmu\:}\)g/ml. Furthermore, they were also treated with the same concentrations of bare PDA NPs, PDA NPs loaded with one of the compounds (5FU@PDA NPs, CPT11@PDA NPs, LV@PDA NPs, and CTX@PDA NPs), and PDA NPs loaded simultaneously with the three drugs (FOLFIRI@PDA NPs) to make appropriate comparisons. All the results obtained can be seen in the Supplementary Information (Figures S2-S4). In the manuscript, with the aim of summarizing the results, only those corresponding to the assays that were performed treating HTC116 and HT29 cells with 30 \(\:{\upmu\:}\)g/ml of the different PDA NPs can be found (Fig. 6A-B).
According to them, bare PDA NPs were able to reduce the viability of both tumor cells by more than 30% after 72 h, highlighting what had already been reported in several previous works23,56,57. Besides, LV@PDA NPs and CTX@PDA NPs had similar cytotoxic effect to PDA NPs, which could lead to the assumption that the concentrations of LV and the anti-EGFR antibody incorporated in PDA NPs did not have significant effect on CRC cell viability. On the contrary, those that had more marked cytotoxic effect were the NPs loaded with 5FU or CPT11, which, after 72 h of treatment, managed to reduce tumor cell viability by 50–60%. Moreover, it should be noted that 5FU@PDA NPs action was more remarkable during the first 24 h, possibly because, according to the release assays, 5FU was released quicklier from PDA NPs than CPT11. Finally, as can be noted in Fig. 6A-B, co-loading PDA NPs with 5FU, CPT11, and LV significantly contribute to augment their cytotoxicity, especially if the anti-EGFR mAb was also incorporated. In this manner, FOLFIRI@PDA NPs and FOLFIRI-CTX@PDA NPs managed to reduce the survival rate of HTC116 cells to 24% and 22% after 72 h, while they decreased HT29 viability rate to 33 and 30%, respectively. Thus, synergy among all the drugs, the antibody, and the PDA-based nanocarrier was evident.
Viability reduction (%) of (A) HTC116, (B) HT29 (B), and (C) HS5 cells caused by the different PDA-based nanosystems synthesized (30 \(\:{\upmu\:}\)g/ml). Each assay was repeated thrice, and p-values were obtained by comparing the cytotoxic effect of the different nanosystems with that of bare PDA NPs after 72 h.
On the other hand, in addition to studying the cytotoxicity of the different PDA-nanosystems for CRC cells, viability assays were also carried out with healthy stromal cells, which were treated with the same NP concentrations as tumor cells. In this case, as can be noted in Fig. 6C, all the synthesized NPs exhibited much lower cytotoxicity. Firstly, the intrinsic antiproliferative effect of bare PDA NPs was significantly smaller, only reducing the viability of stromal cells by 20% after 72 h. After this time, CTX@PDA NPs also reduced the viability of healthy cells by about 20%, while LV@PDA NPs did so by 30%, significant lower percentages than for CRC cell lines. Likewise, after 72 h days, 5FU@PDA NPs and CPT11@PDA NPs decreased stromal cell viability by 35%, while this reduction had been 50–55% for malignant cells. Also, FOLFIRI@PDA NPs and FOLFIRI-CTX@PDA NPs reduced HS5 cell viability 20% less than the viability of HTC116 and HT29 cells. Thereby, the lower toxicity of the nanosystems for healthy cells than for tumor cells was noticeable, a very relevant fact that could contribute to reduce the side effects of the carried drugs in the future.
Therapeutic efficacy of FOLFIRI-CTX@PDA NPs in EGFR-overexpressing MCTS
Given that 2D cell cultures often have limitations, it was decided to finish this work by studying the therapeutic efficacy of the developed nanosystem also in 3D cell cultures, since they better mimic tumor clinical behavior58. MCTS were developed from HTC116 and HT29 cells and, when they were incubated with PDA NPs and FOLFIRI-CTX@PDA NPs for 72 h, alterations appeared in their morphology. Firstly, it should be noted that bare PDA NPs, despite not having very marked effect, could reduce by themselves the size of both types of spheroids and could reduce their degree of compaction, as can be seen in Fig. 7 and S5. Thus, PDA NP intrinsic anticancer activity was also evident in 3D cell cultures. Otherwise, FOLFIRI-CTX@PDA NP therapeutic effect was very significant, since they managed to considerably diminish the size of the MCTS, which exhibited much less spherical shape after the elimination of the tumor cells that surrounded their necrotic core (Fig. 7). In this way, FOLFIRI-CTX@PDA NPs, by maintaining their effectiveness in these 3D models that better mimic the effect of the tumor microenvironment on drug response, could be an excellent candidate for carrying out in vivo assays in a near future.
Conclusions
In conclusion, in this study, PDA NPs were successfully synthesized and novelty loaded with all the drugs that are administered in the FOLFIRI regime. In addition, they were also loaded with an anti-EGFR mAb, and quite good adsorption efficiencies were achieved despite having four different compounds loaded in the nanocarrier. Adsorption of all the drugs and the antibody was verified by FT-IR assays, and the ITC experiments that were performed afterwards showed that the interaction between PDA NPs and the drugs could be electrostatic in nature. Likewise, the release assays that were later carried out showed that 5FU, CPT11, and LV release could be governed by Fickian diffusion mechanisms, and that such release was faster at the lysosomal pH than at the physiological pH. Otherwise, the PDA-based DDS developed, which was rapidly up-taken by EGFR-overexpressing CRC cells, exhibited marked therapeutic effect in vitro, since it notably reduced HTC116 and HT29 cell viability and caused significant alterations in the 3D models that were developed from these two cell lines. However, FOLFIRI-CTX@PDA NPs did not affect stromal cell viability as much, so they may constitute a great approach to enhance 5FU and CPT11 pharmacokinetics while reducing their side effects in the future. Overall, this work provides potential evidence for a promising therapeutic application of FOLFIRI-CTX@PDA NPs as a DDS that may improve mCRC treatment.
Data availability
The datasets used and/or analyzed during the current study available from the corresponding author on reasonable request.
References
Yu, Z., Li, X., Duan, J. & Yang, X-D. Targeted treatment of colon cancer with aptamer-guided albumin nanoparticles loaded with docetaxel. Int. J. Nanomed. 15, 6737–6748 (2020).
Sefidan Jadid, M. F., Jafari-Gharabaghlou, D., Bahrami, M. K., Bonabi, E. & Zarghami, N. Enhanced anti-cancer effect of curcumin loaded-niosomal nanoparticles in combination with heat-killed Saccharomyces cerevisiae against human colon cancer cells. J. Drug Deliv Sci. Tech. 80, 104167 (2023).
Smith, T. et al. Application of smart solid lipid nanoparticles to enhance the efficacy of 5-fluorouracil in the treatment of colorectal cancer. Sci. Rep. 10, 16989 (2020).
Liang, X., Chen, M., Bhattarai, P., Hameed, S. & Dai, Z. Perfluorocarbon@porphyrin nanoparticles for tumor hypoxia relief to enhance photodynamic therapy against liver metastasis of colon cancer. ACS Nano. 14, 13569–13583 (2020).
Mirzaghavami, P. S., Khoei, S., Khoee, S. & Shirvalilou, S. Folic acid-conjugated magnetic triblock copolymer nanoparticles for dual targeted delivery of 5-fluorouracil to colon cancer cells. Cancer Nanotechnol. 13, 12 (2022).
Santos Nunes, S. et al. pH-responsive and folate-coated liposomes encapsulating irinotecán as an alternative to improve efficacy of colorrectal cancer treatment. Biomed. Pharmacother. 144, 112317 (2021).
Ibrahim, B., Mady, D. Y., Tambuwala, M. M. & Haggag, Y. A. pH-sensitive nanoparticles containing 5-flurouracil and leucovorin as an improved anti-cancer option for colon cancer. Nanomedicine 17 (6), 367–381 (2022).
Giram, P. S. et al. Poly (D,L-lactide-co-glycolide) surface-anchored biotin-loaded irinotecan nanoparticles for active targeting of colon cancer. ACS Omega. 9, 3807–3826 (2024).
Bhaskaran, N. A., Jitta, S. R., Salwa, Cheruku, S., Kumar, N. & Kumar, L. Orally delivered solid lipid nanoparticles of irinotecan coupled with chitosan surface modification to treat colon cancer: Preparation, in-vitro and in-vivo evaluations. Int. J. Biol. Macromol. 211, 301–315 (2022).
Heinemann, V. et al. FOLFIRI plus cetuximab or bevacizumab for advanced colorectal cancer: final survival and per-protocol analysis of FIRE-3, a randomized clinical trial. Br. J. Cancer. 124, 587–594 (2021).
Fisher, L. E. et al. Efficacy of FOLFIRI plus Cetuximab vs. FOLFIRI plus Bevacizumab in 1st -line treatment of older patients with RAS wild-type metastatic colorectal cancer: An analysis of the randomized trial FIRE-3. Br. J. Cancer. 127, 836–843 (2022).
Khan, S. et al. Folate decorate lipid chitosan hybrid nanoparticles of 5-fluorouracil for enhanced anticancer efficacy against colon cancer. Int. J. Biol. Macromol. 222 (A), 497–508 (2022).
Wang, X. et al. Novel 5-fluorouracil carbonate-loaded liposome: Preparation, in vitro, and in vivo evaluation as an antitumor agent. Mol. Pharm. 19 (7), 2061–2076 (2022).
Liu, Y. et al. Targeted delivery of irinotecán to colon cancer cells using epidermal growth factor receptor-conjugated liposomes. Biomed. Eng. Online. 21, 53 (2022).
Li, Z. et al. Irinotecan/scFu co-loaded liposomes coaction on tumor cells and CAFs for enhanced colorectal cancer therapy. J. Nanobiotechnol. 19, 421 (2021).
Das, R. P. et al. Tuning the pharmacokinetics and efficacy of irinotecan (IRI) loaded gelatin nanoparticles through folate conjugation. Int. J. Pharm. 586, 119522 (2020).
Pai, F-T. & Lin, W. J. Synergistic cytotoxicity of irinotecan combined with polysaccharide-based nanoparticles for colorectal carcinoma. Biomater. Adv. 153, 213577 (2023).
Hong, J. & Feng, Z. Synergic fabrication of combination therapy of Irinotecan and 5-Fluorouracil encapsulated polymeric nanoparticles for the treatment of gastric cancer therapy. Process. Biochem. 106, 191–198 (2021).
Gao, Z., Li, Z., Yan, J. & Wang, P. Irinotecan and 5-fluorouracil-co-loaded hyaluronic acid-modified layer-by-layer nanoparticles for targeted gastric carcinoma therapy. Drug Des. Dev. Ther. 11, 2595–2604 (2017).
Janani, B. et al. EGFR-based targeted therapy for colorectal cancer – Promises and challenges. Vaccines 10 (4), 499 (2022).
Li, H. et al. Polydopamine-based nanomaterials and their potentials in advanced drug delivery and therapy. Colloid Surf. B Biointerfaces. 199, 111502 (2021).
Wang, Z., Duan, Y. & Duan, Y. Application of polydopamine in tumor targeted drug delivery systems and its drug release behavior. J. Control Release. 290, 56–74 (2018).
Nieto, C., Vega, M. A., Marcelo, G.& Martín Del Valle, E.M. Polydopamine nanoparticles kill cancer cells. RSC Adv. 8, 36201 (2018).
Singh, S. & Pal, K. Actively targeted gold-polydopamine (PDA@Au) nanocomplex for sequential drug release and combined synergistic chemo-photothermal therapeutic effects. Int. J. Pharm. 645, 123374 (2023).
Djermane, R., Nieto, C., Vargas, J. C., Vega, M. & Martín del Valle, E. M. Insight into the influence of the polymerization time of polydopamine nanoparticles on their size, surface properties and nanomedical applications. Polym. Chem. 13 (2), 235–244 (2022).
Zhou, J., Ji, Q. & Li, Q. Resistance to anti-EGFR therapies in metastatic colon cancer: Underlying mechanisms and reversal strategies. J. Exp. Clin. Cancer Res. 40, 328 (2021).
Duan, G. L., Zheng, L-X., Chen, J., Cheng, W-B. & Li, D. High-performance liquid chromatographic method for determination of leucovorin in plasma: Validation and application to a pharmacokinetic study in healthy volunteers. Biomed. Chromatogr. 16, 282–286 (2002).
Hanif, M. et al. Development and validation of a new HPLC method for the detection of 5-fluorouracil in mobile phase in plasma. Curr. Pharm. Anal. 14 (1), 3–7 (2018).
Shende, P. & Gaud, R. Validated RP-HPLC analysis of irinotecan HCl in the bulk material and in pharmaceutical formulations. Acta Chromatogr. 21 (1), 71–82 (2009).
Safwat, M. A., Soliman, G. M., Sayed, D. & Attia, M. A. Gold nanoparticles enhance 5-fluorouracil anticancer efficacy against colorectal cancer cells. Int. J. Pharm. 513, 648–658 (2016).
Singh, P., Tyagi, G., Mehrotra, R. & Bakhshi, A. K. Thermal stability studies of 5-fluorouracil using diffuse reflectance infrared spectroscopy. Drug Test. Anal. 1 (5), 240–244 (2009).
Yusefi, M. et al. The potential anticancer activity of 5-fluorouracil loaded in cellulose fibers isolated from rice straw. Int. J. Nanomed. 15, 5417–5432 (2020).
Hasan-Nasab, B. et al. A promising targeting system to enrich irinotecan antitumor efficacy: Folic acid targeted nanoparticles. J. Drug Deliv Sci. Technol. 63, 102543 (2021).
Jalal, N. R., Madrakian, T., Afkhami, A. & Ghoorchian, A. Graphene oxide nanoribbons/polypyrrole nanocomposite film: Controlled release of leucovorin by electrical stimulation. Electrochim. Acta. 370, 137806 (2021).
Liu, Y. et al. Dopamine-melanin colloidal nanospheres: An efficient near‐infrared photothermal therapeutic agent for in vivo cancer therapy. Adv. Mater. 25 (9), 1353–1359 (2013).
Vega, M. A., Nieto, C., Marcelo, G. & Martín del Valle, E. M. Cytotoxicity of paramagnetic cations - Loaded polydopamine nanoparticles. Colloids Surf. B. 167, 284–290 (2018).
El Hallal, R., Lyu, N. & Wang, Y. Effect of cetuximab-conjugated gold nanoparticles on the cytotoxicity and phenotypic evolution of colorectal cancer cells. Molecules 26 (3), 567 (2021).
Huang, L., Sui, W., Wang, Y. & Jiao, Q. Preparation of chitosan/chondroitin sulfate complex microcapsules and application in controlled release of 5-fluorouracil. Carbohydr. Polym. 80 (1), 168–173 (2010).
Selvaraj, V. & Alagar, M. Analytical detection and biological assay of antileukemic drug 5-fluorouracil using gold nanoparticles as probe. Int. J. Pharm. 337 (1–2), 275–281 (2007).
Iliescu, R. I. et al. Montmorillonite–alginate nanocomposite as a drug delivery system–incorporation and in vitro release of irinotecan. Int. J. Pharm. 463 (2), 184–192 (2014).
Li, P., Wang, Y., Peng, Z., She, F. & Kong, L. Development of chitosan nanoparticles as drug delivery systems for 5-fluorouracil and leucovorin blends. Carbohydr. Polym. 85 (3), 698–704 (2011).
Ball, V. Polydopamine nanomaterials: Recent advances in synthesis methods and applications. Front. Bioeng. Biotechnol. 6, 109 (2018).
Jin, A., Wang, Y., Lin, K. & Jiang, L. Nanoparticles modified by polydopamine: Working as drug carriers. Bioact Mater. 5 (3), 522–541 (2020).
Ryu, J. H., Messersmith, P. B. & Lee, H. Polydopamine surface chemistry: A decade of discovery. ACS Appl. Mater. Interfaces. 10 (9), 7523–7540 (2018).
Jodko-Piorecka, K. & Litwinienko, G. First experimental evidence of dopamine interactions with negatively charged model biomembranes. ACS Chem. Neurosci. 4 (7), 1114–1122 (2013).
Bhuiya, S., Pradhan, A. B., Haque, L. & Das, S. Molecular aspects of the interaction of iminium and alkanolamine forms of the anticancer alkaloid chelerythrine with plasma protein bovine serum albumin. J. Phys. Chem. B. 120 (1), 5–17 (2016).
Harini, G. et al. Antiproliferative and apoptotic effects of pH-responsive veratric acid-loaded polydopamine nanoparticles in human triple negative breast cancer cells. Chem. Biodivers. 20, e202201006 (2023).
Dash, S., Murthy, P. N., Nath, L. & Chowdhury, P. Kinetic modeling on drug release from controlled drug delivery systems. Acta Pol. Pharm. 67 (3), 217–223 (2010).
Bohrey, S., Chourasiya, V. & Pandey, A. Polymeric nanoparticles containing diazepam: Preparation, optimization, characterization, in-vitro drug release and release kinetic study. Nano Converg. 3 (1), 1–7 (2016).
Ritger, P. L. & Peppas, N. A. A simple equation for description of solute release I. Fickian and non-fickian release from non-swellable devices in the form of slabs, spheres, cylinders or discs. J. Control Release. 5 (1), 23–36 (1987).
Paiva, I. et al. Synthesis and analysis of 64Cu-labeled GE11-modified polymeric micellar nanoparticles for EGFR-targeted molecular imaging in a colorectal cancer model. Mol. Pharm. 17 (5), 14701481 (2020).
Zavieh, S. E. & Safari, F. The antitumor activity of hAMSCs secretome in HT-29 colon cancer cells throough downregulation of EGFR/c-Src/IRTKS expression and p38/ERK1/2 phosphorylation. Cell. Biochem. Biophys. 80, 395–402 (2022).
Liu, J. et al. Simple and tunable surface coatings via polydopamine for modulating pharmacokinetics, cell uptake and biodistribution of polymeric nanoparticles. RSC Adv. 7, 15864 (2017).
Zhang, P., Xu, Q., Du, J. & Wang, Y. Polydopamine-based nanoparticles with excellent biocompatibility for photothermally enhanced gene delivery. RSC Adv. 8, 34596 (2018).
Maya, S. et al. Cetuximab conjugated O-carboxymethyl chitosan nanoparticles for targeting EGFR overexpressing cancer cells. Carbohydr. Polym. 93 (2), 661–669 (2013).
Nieto, C., Vega, M. A., Enrique, J., Marcelo, G. & Martín Del Valle, E.M. Size matters in the cytotoxicity of polydopamine nanoparticles in different types of tumors. Cancers 11, 1679 (2019).
Nieto, C., Vega, M. A. & Martín Del Valle, E.M. Nature-inspired nanoparticles as paclitaxel targeted carrier for the treatment of HER2-positive breast cancer. Cancers 13, 2526 (2021).
Smith, T., Affram, Bulumko, E. & Agyare, E. Evaluation of in-vitro cytotoxicity effect of 5-FU loaded chitosan nanoparticles against spheroid models. J. Nat. Sci. 4 (10), e535 (2018).
Acknowledgements
Authors thanks the Service of Elemental Analysis, Chromatography, and Masses of the University of Salamanca for the UHPLC/mass spectrometry analysis, the Microscopy Unit of the Cancer Research Center of Salamanca for the CLSM images and the Microscopy unit of the University of Santiago de Compostela for the SEM images. Also, R.D. thanks to the Algerian government her predoctoral scholarship and M.V. thanks O. Acosta for her help.
Funding
This work was supported by grants from the University of Salamanca (PIC2-2021-19 and FS/5-2020) and the Spanish Ministry of Science and Innovation (PID2022-140599OB-I00).
Author information
Authors and Affiliations
Contributions
R.D. and M.V. were involved in nanoparticle synthesis and characterization, as well as in the release and ITC studies. C.N. conduced all in vitro assays and performed statistical analysis. C.N. and M.V. mainly designed the experiments, analyzed the results, and wrote and reviewed the manuscript. E.M., as supervisor, provided experimental and strategic guidance. All authors approved the final version of this manuscript. All authors agreed to publish this manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
Not applicable.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Djermane, R., Nieto, C., Vega, M.A. et al. EGFR-targeting polydopamine nanoparticles co-loaded with 5-fluorouracil, irinotecan, and leucovorin to potentially enhance metastatic colorectal cancer therapy. Sci Rep 14, 29265 (2024). https://doi.org/10.1038/s41598-024-80879-0
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-024-80879-0
Keywords
This article is cited by
-
Fabrication of size-tunable polydopamine nanoparticles: toxicity evaluation in cellular and animal models
Naunyn-Schmiedeberg's Archives of Pharmacology (2026)