Abstract
Kidney organoids are connected to the host circulation and mature after transplantation. However, they are still immature compared to the adult kidneys, and their precise maturation stages remain unclear. By transplanting the mouse embryonic kidney as a model system for organoid transplantation, we report here the maturation defects of the graft, especially in the medulla. Single cell profiling of the developing kidneys in vivo identified gene sets associated with the maturation of the collecting duct epithelium and medullary stroma. These data revealed an upregulation of genes associated with channel/transporter functions and immune defense, as well as a downregulation of neuronal genes. Using these marker genes, we found that the maturation of the collecting duct and medullary stroma in the grafts barely corresponds to the perinatal stage, which was confirmed histologically by using representative genes. Thus, the gene sets obtained serve as maturation coordinates for the renal medulla and will be helpful in analyzing its maturation defects after transplantation. They will also provide a useful basis for further maturation of transplanted kidney organoids.
Similar content being viewed by others
Introduction
The kidney is derived from at least three types of precursors: the nephron progenitor, ureteric bud, and stromal progenitor1,2. The nephron progenitors give rise to nephrons, including glomerular podocytes, proximal tubules, loops of Henle (LoH), and distal tubules, while the ureteric bud branches extensively to form the collecting ducts (CD) and ureter. These differentiated components align to form the route for urinary flow. The stromal progenitors differentiate into cortical and medullary interstitial cells, as well as mesangial cells and renin cells that are located within or close to glomeruli, respectively. At embryonic day (E) 15.5, the overall nephron segments are specified, and urine starts to flow approximately at E16.5–E17.53. Meanwhile, the medulla of the kidney elongates, reflecting the elongation of the collecting ducts and possibly stromal expansion surrounding the duct epithelia. This medullary development continues after the day of birth (P0: postnatal day 0), while nephron progenitors cease self-renewal within a few days after birth, leading to the cessation of new nephron generation4.
Recent advances in stem cell research have enabled the generation of kidney organoids from pluripotent stem cells in vitro5,6,7,8,9,10 However, it has been noted that the organoids remain at an immature stage and may correspond to kidneys at approximately E14.5 in mice10 and at approximately 16 weeks of gestation in humans11. We and others have developed methods to transplant kidney organoids into immunodeficient mice, allowing for organoid vascularization and connection to the host circulation, leading to further maturation of the organoids12,13,14,15,16. Despite these advances, the maturation stage of transplanted kidney organoids, even those derived from mouse pluripotent stem cells (PSCs), has not yet reached that of adult kidneys17. Accurate determination of their maturation stage requires comprehensive information on gene expression at each stage of the developing kidneys. Single-cell RNA sequencing (scRNA-seq) analysis is a powerful tool to examine gene expression in complex and heterogeneous organs, such as the kidney. In our previous report18, we performed scRNA-seq analysis of the developing mouse kidney from E15.5 to newborn (P0). We also used the E12.5 embryonic kidney as a model system for organoid transplantation and found that most cell types, except for the collecting ducts, exhibited maturation similar to that of neonatal kidneys in vivo, revealing non-synchronous maturation across cell lineages18. Indeed, the glomeruli in the transplanted organoids are well vascularized and to some extent mature17, but the maturation status of the renal medulla remains unclear. Therefore, in the present study, we extend our scRNA-seq analysis to 7 days after birth (P7) and focus mainly on the maturation of the renal medulla, namely the collecting ducts and medullary stroma. We identified many maturation-dependent genes in these cell types in the renal medulla in vivo, and application of these maturation coordinates to the transplanted embryonic kidneys revealed maturation defects in the medulla. We also selected representative gene markers for histologic evaluation at a high resolution. Using these tools, we investigated the possible causes of medullary maturation defects after transplantation.
Results
Morphologic characteristics of the graft do not match those of the neonatal kidney
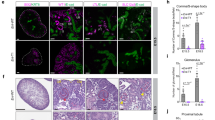
When E12.5 kidneys were transplanted under the kidney capsule of adult mice, the medulla did not develop well, and the corticomedullary junction in the grafts was unclear (Fig. S1a). This may be due to limited subcapsular space leading to compression against the growing grafts. In contrast, when transplanted near the blood vessels in the peri-testicular fat, as we previously reported18, the graft was clearly vascularized by the nearby vessels and grew well (Fig. 1a). Therefore, in this study, we mainly performed transplantation into the peri-testicular fat, unless otherwise indicated. The major and minor axes of the grafts at day 12 post-transplantation were larger than those of the E15.5 kidneys in vivo, although the grafts were still smaller than the neonatal (P0) kidneys (Fig. 1b). Histologically, the cortex and medulla were clearly visible both in the P0 kidneys and grafts. However, the renal medulla of the grafts showed less elongation toward the pelvis than the P0 kidney (Fig. 1c). KRT8+/CDH1+ CD in the grafts appeared thinner than in the P0 kidney, and SLC12A1+/CDH1+ LoH were less abundant in the grafts (Fig. 1c), which may underscore the poor medullary elongation in the graft. In contrast, PECAM1+ vascular endothelial cells (ECs) were present in both the cortex and medulla of the day 12 grafts (Fig. S1b). At day 14 post-transplantation, we observed hydronephrosis (Fig. S1c), suggesting the onset of blood filtration leading to urine production. At this stage, the renal parenchyma was compressed by the retained urine, which prevented a fair assessment of the maturation status. Therefore, the evaluation was mainly performed at day 12 post-transplantation, when no obvious hydronephrosis was present.
Morphologic characteristics of the graft do not match those of the neonatal kidney. (a) Transplantation of E12.5 kidneys into the peri-testicular fat. The left panel shows a planned transplant site (dotted circle). The middle and right panels show the transplanted embryonic kidneys at day 12. In the middle panel, the surface membrane of the graft was removed to visualize the overall appearance of the graft. In the right panel, the surface membrane was left intact to visualize the host blood vessel extending toward the graft. Arrows: a blood vessel from the host. Scale bars: 1.0 mm. (b) Size of the grafts and embryonic kidneys. The major and minor axis lengths of the transplanted embryonic kidneys (at day 12) were compared with those of the E15.5 and P0 kidneys (n = 15, 6, and 6, respectively). Multiple comparisons were performed using Dunnett’s T3 test. Fifteen grafts from eight independent transplantation experiments were analyzed. (c) Histologic analysis of the E15.5 and P0 kidneys and day 12 grafts. 1st column: Hematoxylin and eosin (H&E) staining. Scale bars: 500 μm; 2nd column: Immunostaining of SLC12A1 (loops of Henle), KRT8 (collecting ducts), and CDH1 (collecting ducts and nephrons). Scale bars: 500 μm; 3rd and 4th columns: magnified images of the 2nd column. Scale bars: 100 μm. Three grafts from three independent transplantation experiments were analyzed.
Identification of maturation coordinates of the renal medulla in vivo
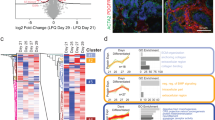
To analyze gene expression profiles at the single cell level, we performed scRNA-seq analysis on developing mouse kidneys at E15.5, P0, and 7 days after birth (P7) and transplanted E12.5 embryonic kidneys (8 and 12 days post-transplantation, Graft Day8 and Graft Day12 in Fig. 2a, respectively). In addition, another day 12 sample (Graft Day12(*)) from the previous report18 was added for reproducibility. E12.5 kidneys with ureter and cloaca were used for all three grafts in this scRNA-seq analysis.
Identification of maturation coordinates of the renal medulla in vivo. (a) scRNA-seq analysis of transplanted embryonic kidneys (day 8 and 12) and developing kidneys (E15.5, P0, P7). The day 12 grafts include the sample (*) from our previous report18. The UMAP plots are shown. NP: nephron progenitor; Pod: podocyte; PT: proximal tubule; LoH: loop of Henle; DT: distal tubule; UB: ureteric bud; CD: collecting duct; EC: endothelial cell; Mφ: macrophage; Ren: renin cell; Mes: mesangial cell; SP: stromal progenitor; iMS: inner medullary stroma; oMS: outer medullary stroma. (b) Volcano plots showing the differentially expressed genes between the renal medulla (CD, iMS, oMS) at E15.5 and P7. Red circles: representative genes that are more abundantly expressed at E15.5. Blue circles: representative genes that are more abundantly expressed at P7. (c) CNET plots showing the differentially expressed genes between the renal medulla (CD, iMS, oMS) at E15.5 and P7. Representative genes are shown in circles. (d) Heatmaps of the developing kidneys (E15.5, 17.5, P0, and P7) using the top 200 maturation coordinates. The representative genes are listed on the right side of the heatmaps for each cell type.
The UMAP plots showed the presence of the nephrogenic niche, namely nephron progenitor (NP, clusters 8, 17, 20), ureteric bud tip (UB, cluster 21), and stromal progenitor (SP, cluster 13), up to the P0 kidney and day 8 transplant, but these niche clusters disappeared at P7 in vivo and day 12 transplants (Fig. 2a), consistent with the transient nature of the nephrogenic niche. The clusters for differentiating nephrons, collecting ducts, and stromal cells overlapped at all developmental stages and in all grafts. However, proliferation-related genes (Mki67, Ccnd1, Top2a) were downregulated in the day 12 graft compared to the P7 kidney, especially in the proximal tubules (Fig. S2a). In contrast, there was little difference in the expression of apoptosis-suppressing genes (Bcl-2) and apoptosis-promoting genes (Bax and Bok) compared to the P7 kidney (Fig. S2b). Therefore, the decreased proliferation of differentiating nephrons, together with the disappearance of the nephrogenic niche, may at least partially explain the reduced graft size.
To assess the maturation stage of the grafts, we adopted a strategy of first identifying gene sets that can serve as maturation coordinates using the developing kidney in vivo, and then applying these coordinates to the grafts. Since we have previously studied the maturation of the renal cortex18, in this study we focused on the maturation of the renal medulla, namely the collecting duct (CD, cluster 10), the inner medullary stroma (iMS, cluster 24), and the outer medullary stroma (oMS, cluster 22) (Fig. 2a, S2c). We selected genes enriched in each cell type and picked up the differentially expressed genes between the E15.5 and P7 kidneys, as shown as volcano plots (Fig. 2b, Table S1). These genes were subjected to Gene Set Enrichment Analysis (GSEA), which highlighted “immune response” in all three cell types (Fig. S3). “Regulation of body fluid levels” increased in the P7 CD (Fig. S3a), while the “nervous system” decreased in the P7 iMS and oMS (Fig. S3b, c). The gene concept network (CNET) plots were then generated to visually display the genes in representative categories, showing upregulation of transporter/channel genes (e.g., Aqp2, Aqp4) in the CD and downregulation of neuronal genes (e.g., Gria1, Cntn6) in the medullary stroma (Fig. 2c). Genes related to the immune/defense response (e.g. Defb1, Jak2, Socs3, Il6, Ptgs2) were upregulated in all three cell types (Fig. 2C, Fig. S2d). Furthermore, the heatmaps constructed with the top 200 differentially expressed genes in the CD, iMS, and oMS showed a reasonable alignment of the in vivo kidneys from E15.5 to P7 (Fig. 2d, Table S2). Therefore, these gene sets will serve as promising coordinates for assessing the maturation stages of the renal medulla of developing kidneys in vivo and possibly of transplants.
Maturation coordinates place the graft medulla in the perinatal stage
We next applied these maturation coordinates to the transplants. Using the top 200 genes from the heatmaps as maturation coordinates, we included the day 8 and day 12 grafts and performed the hierarchical clustering analysis, which placed the grafts between the P0 and P7 kidneys (Fig. 3a, S4, S5, and S6). Heatmap analysis showed the progressive changes in maturation marker expression from day 8 to day 12 grafts (Fig. 3a). We also selected differentially expressed genes between day 8 and day 12 grafts (Fig. S7a). These gene sets partially overlapped with the maturation coordinates. For example, transporter/channel genes (e.g., Aqp2, Aqp4) in the CD were higher in day 12 grafts (Fig. 3c, Fig. S7b), suggesting that the grafts follow the maturation process, at least partially, into the days after transplantation. In contrast, genes involved in the immune/defense response (e.g. Jak2, Socs3, Il6, Ptgs2) were higher in the day 8 grafts (Fig. 3a, S7c), which may be due to the acute inflammation induced by transplantation. Neuronal genes (e.g., Gria1, Cntn6) in the medullary stroma were higher in day 12 grafts (Fig. S7d), which may reflect transplant-specific gene expression.
Maturation coordinates place the graft medulla in the perinatal stage. (a) Heatmaps of the developing kidneys and grafts (day 8 and 12) using the top 200 maturation coordinates. Results for each medullary cell type are shown (CD, iMS, oMS). (b) Volcano plots showing the differentially expressed genes between the day 12 graft and P7 kidney. Red circles: representative genes that are more abundantly expressed in the graft. Blue circles: representative genes that are more abundantly expressed in the P7 kidney. (c) UMAP plots of the representative maturation coordinates for the renal medulla. Blue arrowheads: CD; red arrowheads: innermost MS; purple arrowheads: oMS; green arrowheads: iMS.
The comparison between the day 12 graft and the P7 kidney in vivo still showed the insufficient expression of maturation markers, including water channel genes (Aqp2, Aqp4) in the CD and immune response genes (Defb1 in the CD, Ptgs2, IL6, Socs3 in the iMS), while neuronal genes (e.g., Gria1, Cntn6) remained high in the MS (Fig. 3b, Table S3), suggesting the immaturity of the grafts compared to the P7 kidney. At day 12, one of the grafts (*) appeared to be slightly closer to P7 than the other (Fig. 3a), but this difference may be due to different times at which transplantation was performed or to strain differences in the grafts and hosts (C57BL/6 kidneys in C57BL/6 mice for Graft day 12(*) versus ICR kidneys in immunodeficient mice for Graft day 12). In addition, the expression of Hif1a and its downstream targets Hk2 and Pfkl were lower in the iMS of the graft than in the P7 kidney (Fig. 3b). The UMAP plots confirmed these differences (Fig. 3c, S2e). In particular, Ptgs2 and Hif1a were detected in the cluster corresponding to the innermost MS of the P7 kidney but not of the grafts. Ptgs2 encodes the enzyme Cox-2, which catalyzes prostaglandin synthesis from arachidonic acid19,20, and Hif1a is involved in the hypoxia response21,22. It would be intriguing if prostaglandins and hypoxia were related to medullary elongation and maturation, although their functional roles in the iMS remain elucidated. Taken together, the use of maturation coordinates suggests that the day 12 grafts may correspond to the perinatal stage.
Histologic evaluation using maturation markers validates the graft medulla as a perinatal stage
To validate the usefulness of these maturation marker genes by histological examination, we used the highly sensitive in situ hybridization technology23.
The expression of Aqp2 and Aqp4 in the medullary CD increased as development progressed from E15.5 to P7, while Wnt7b appeared to decrease (Fig. 4a), which is consistent with the scRNA-seq data. (Fig. S7b). In the transplants, the expression levels of Aqp4 and Aqp2 and the thickness of Aqp2/Aqp4 + CD were lower than those in the P0 kidney. (Fig. 4b). No significant differences in Wnt7b expression were observed, although Wnt7 + CDs were thinner (Fig. 4b).
Histologic evaluation using maturation markers validates the graft medulla as perinatal stage. (a) In situ hybridization of maturation markers for the CD (Aqp4 and Aqp2) at E15.5, P0, and P7. Wnt7b is used as a constant control. Scale bars: 500 μm. (b) In situ hybridization of the CD maturation markers in the P0 kidney and the day 12 graft. Scale bars: 500 μm. 2nd and 3rd rows: magnified images of the 1st row. Scale bars: 100 μm. Two grafts from two independent transplantation experiments were analyzed. (c) In situ hybridization of maturation markers for the iMS (Hif1a and Ptgs2) at E15.5, P0, and P7. Scale bars: 500 μm. 2nd row: magnified images of the medulla in the 1st row. Scale bars: 100 μm. 3rd row: magnified images of the cortex in the 1st row. Ptgs2 is also expressed in the macula densa of the cortex. Scale bars: 100 μm. (d) In situ hybridization of the iMS maturation markers in the P0 kidney and the day 12 graft. Scale bars: 500 μm. 2nd row: magnified images of the medulla in the 1st row. Scale bars: 100 μm. 3rd row: magnified images of the cortex in the 1st row. Ptgs2 is also expressed in the macula densa of the cortex. Scale bars: 100 μm. Two grafts from two independent transplantation experiments were analyzed. (e) In situ hybridization of neuronal genes at E15.5, P0, and P7. Scale bars: 500 μm. 2nd row: magnified images of the medulla and cortex in the 1st row. Cntn6 is expressed in the oMS at E15.5. Scale bars: 100 μm; 3rd row: magnified images of the cortex in the 1st row. Gria1 is expressed in stromal progenitor at E15.5 and P0. Scale bars: 100 μm. (f) In situ hybridization of neuronal genes in the E15.5 kidney and the day 12 graft. Scale bars: 500 μm. 2nd row: magnified images of the medulla and cortex in the 1st row. In the graft, Cntn6 is expressed in the oMS, and Gria1 is expressed in the iMS and oMS. Scale bars: 100 μm; 3rd row: magnified images of the cortex in the 1st row. Scale bars: 100 μm. Two grafts from two independent transplantation experiments were analyzed.
The expression of Ptgs2 and Hif1a in the iMS increased with development (Fig. 4c), but their expression in the transplants was lower than in the P0 kidney (Fig. 4d). Ptgs2 is also known as a representative marker for the macula densa in the renal cortex19. Therefore, the expression of Ptgs2 in the cortex remained relatively constant and was also detected in the transplants.
The expression of neuronal genes was highest at E15.5 in normal development: Gria1 was expressed in stromal progenitors and Cntn6 in the oMS (Fig. 4e). Gria1 encodes the glutamate ionotropic receptor AMPA type subunit 1 involved in excitatory neurotransmission in the central nervous system24, and Cntn6 encodes Contactin 6, a member of the immunoglobulin superfamily that functions as a cell adhesion protein and promotes neurite outgrowth and synapse formation25,26. These genes were almost undetectable at P7. However, in the transplants, Gria1 and Cntn6 were robustly detected in the oMS (Fig. 4f).
Therefore, these maturation markers are useful for assessing the maturation status of the kidneys in vivo and the transplants histologically, and our data validated that renal medulla (CD, iMS, oMS) in the grafts barely reached the P0 stage. Relatively unaffected Wnt7b expression in the grafts likely rules out possible involvement of this gene in the medullary defects in the grafts, although Wnt7b is known to be essential for medullary development and elongation27,28.
Administration of HIF1 activator or transplantation into female mice does not ameliorate medullary maturation defects
Next, we searched for the causes of medullary maturation defects after transplantation. Embryonic kidneys are nourished by a low-oxygen blood stream from the placenta. However, embryonic kidneys transplanted into adult mice are likely to receive the high oxygen blood flow directly. As described above, the expression of Hif1a and its target glycolytic genes, Hk2 and Pfkl, was lower in the grafts than in the P7 iMS. The UMAP plots of the P7 kidney showed that these genes were abundantly expressed in the deeper region of the medullary stroma (Fig. 3c, S2e), suggesting a possible role of hypoxia in the medullary stroma surrounding the elongating collecting duct at this postnatal stage. Therefore, we administered Roxadustat, a HIF1 protein activator29, to the host animals daily from day 2 to day 11 after transplantation of E12.5 kidneys (10 mg or 25 mg/kg/day). The dose was determined on the basis of previous reports30,31. Indeed, administration of 25 mg/kg/day resulted in an increase in blood hemoglobin levels (Fig. S8), suggesting that the drug sufficiently activates the HIF pathway. The medullary maturation markers were then used to assess maturation. However, there was no clear improvement in the CD, iMS, or oMS after treatment (Fig. 5a-d).
Administration of the HIF1 activator does not ameliorate medullary maturation defects. The treated and untreated grafts transplanted into the peri-testicular fat were compared histologically at day 12 after transplantation of E12.5 kidneys. Two independent transplantation experiments were performed (two grafts in each condition per experiment). (a) H&E staining of the grafts with and without Roxadustat treatment. Scale bars: 500 μm. (b) In situ hybridization of the CD maturation markers (Aqp4 and Aqp2) in the grafts with and without Roxadustat treatment. Scale bars: 500 μm. 2nd row: magnified images of the 1st row. Scale bars: 100 μm. (c) In situ hybridization of the iMS maturation markers (Hif1a and Ptgs2) in the grafts with and without Roxadustat treatment. Scale bars: 500 μm. 2nd row: magnified images of the 1st row. Scale bars: 100 μm. (d) In situ hybridization of the neuronal genes (Cntn6 and Gria1) in the grafts with and without Roxadustat treatment. Scale bars: 500 μm. 2nd row: magnified images of the 1st row. Scale bars: 100 μm.
We also speculated that there may be differences in circulating hormones between male and female hosts. Therefore, E12.5 kidneys were transplanted into the peri-ovarian fat of adult female mice (Fig. S9a) and compared with grafts in male peri-testicular fat. However, no obvious differences in the expression of maturation marker genes were observed in the CD, iMS, or oMS. (Fig. S9b-e). Nevertheless, the markers we selected served as useful coordinates for assessing the maturation status of the grafts.
Discussion
Organ maturation is an important aspect of developmental biology. In the field of stem cell biology, transplantation is a common approach to facilitate the maturation of pluripotent stem cell-derived organoids, including kidney organoids. However, currently available kidney organoids remain immature in vitro and even after transplantation. In this study, we used embryonic kidney transplantation as a model system for organoid transplantation. First, we identified maturation coordinates for the renal medulla (collecting ducts and medullary stroma) and used them to assess the maturation status of the transplanted embryonic kidneys.
We performed scRNA-seq analysis of the kidney at different developmental stages, from E15.5 to 7 days after birth. Including data at P7 was critical for our analysis because we found that dramatic gene expression changes occur after birth, and many of the differentially expressed genes turned out to serve as maturation coordinates. This was achieved by our technique of dissociating the postnatal kidney to make it suitable for single “cell” RNA-seq analysis, which allowed a fair comparison between pre- and postnatal stages, whereas single “nucleus” RNA-seq is often used for the postnatal kidney. By comparing E15.5 and P7 kidneys, we found channels/transporters that regulate body fluid homeostasis, including aquaporins, are upregulated in the collecting duct after birth. Interestingly, genes related to the immune/defense response are also upregulated after birth in the collecting duct (Defb1, etc.) and medullary stroma (Ptgs2, Il6, etc.), which may be caused by postnatal exposure to the external environment. It remains to be determined whether these immune responses are simply a consequence of the urinary tract infection or whether they contribute to further maturation of the renal medulla. Another interesting finding is the relatively high expression of neuronal genes at E15.5 and their postnatal decrease in the medullary stroma. Although no obvious renal phenotypes are reported in mice lacking these genes24,25,26, there may be some redundant roles in immature stromal cells. At least a decrease of such neuronal genes clearly correlates with stromal maturation.
These maturation coordinates allowed us to assess the maturation status not only of the kidney in vivo but also of the transplanted embryonic kidney. We also selected several key markers and confirmed this conclusion by high-resolution in situ hybridization. This type of histological assessment bypasses costly scRNA-seq analysis and provides a user-friendly method to roughly assess the maturation state of the in vivo kidney and the transplant. Based on these assays, we conclude that the renal medulla in the grafts barely reached the P0 stage. By further comparing gene expression between the graft and P7 kidney, we found lower expression of hypoxia-related genes, including Hif1a, Hk2, and Pfkl, in the grafts. Therefore, we administered the HIF1 activator to the transplanted hosts, but no apparent effects were observed. Genetic activation of the HIF pathway, such as deletion of Vhl, which encodes the protein that degrades HIF, would be required to address this issue unequivocally. We also transplanted the embryonic kidney into female hosts because we suspected a role for female hormones in the maturation of the grafts. However, there was no significant improvement compared to transplantation into male mice. The development of transplantation technique in pregnant female mice will be necessary to elucidate the role of the embryonic environment in kidney maturation, e.g. maternal hormones and metabolites secreted by the placenta or transferred from the mother across the placenta. Another possibility is insufficient urine flow in the grafts. In this study, we mainly used the isolated kidneys as donors but also the kidney with ureter and cloaca in some of the experiments, including scRNA-seq analysis (Figs. 2 and 3). The presence of the ureter and cloaca, the latter of which develops into the urinary bladder, allows urine to flow until the bladder is filled with urine32,33. Even in this situation, our scRNA-seq data showed the poor medullary maturation after transplantation, and there is limited histologic benefit in the grafts with cloaca (Fig. S1d). Nevertheless, a slight increase in intraluminal pressure due to urinary retention may have affected medullary maturation, although we did not observe obvious hydronephrosis at harvest. It has been reported in rats and pigs that the connection of a donor-derived bladder to the host ureter suppressed hydronephrosis and preserved nephron structures32. It would be informative to see the medullary development/maturation in these rats and pigs, as a similar surgical procedure in mice is technically difficult due to their small size.
The mouse kidney organoids, consisting of nephrons, collecting ducts, and stroma, have already been generated17, but they still lack the ureter and bladder. Even if this is achieved, our data suggest that there are other hurdles to overcome, including medullary maturation, which is critical for urine concentration after birth. Elucidation of the underlying mechanisms of medullary maturation will be necessary for the further maturation of kidney organoids. In the human organoids, even the assembly of nephrons and collecting ducts into the organotypic higher-order structure has not been reported. After the generation of human kidney organoids with ureter and bladder, maturation hurdles should again be overcome. It remains unclear whether maturation coordinates are conserved between mice and humans, and the upstream factors that promote maturation need to be identified. These may include maternal hormones and metabolites, urine flow, milk and food intake, gut and urinary microbiome, and oxygen concentration1. These non-genetic parameters, as well as genetic cascades, should be identified to generate mature kidney organoids.
In conclusion, we have identified the maturation coordinates of the renal medulla and the impaired maturation of the transplanted embryonic kidney. These coordinates, as well as our transplantation technique, will serve as a basis to elucidate the mechanism of maturation and to generate functionally mature kidney organoids.
Materials and methods
Animals
C57BL/6J mice and ICR mice were purchased from Japan SLC, Inc., and immunodeficient mice (NOD.CB17-Prkdcscid /J) were purchased from KBT Oriental, Inc. The mice were housed in a specific pathogen-free animal facility. All animal experiments were performed in accordance with our institutional ethical guidelines and approved by the licensing committee of Kumamoto University (approval numbers: A2021-008, A2023–009, and A2024-073). We also complied with ARRIVE guidelines. Mice were euthanized by intraperitoneal injection of 0.75 mg/kg medetomidine, 4.0 mg/kg midazolam, and 5.0 mg/kg butorphanol followed by cervical dislocation.
Transplantation of mouse embryonic kidneys
E12.5 mouse kidneys were used for all transplantation experiments. C57BL/6J embryos as the donors and C57BL/6J mice (8–10 weeks old) as the hosts were used in most of the transplantation experiments. The embryonic kidneys were transplanted into the peri-testicular fat unless indicated otherwise12,18,34. For transplantation into the peri-ovarian fat, the embryonic kidneys were transplanted near the blood vessels in the peri-ovarian fat. In this experiment, ICR embryos as the donors and NOD.CB17-Prkdcscid /J mice (8–10 weeks old) as the hosts were used. Host mice were anesthetized by intraperitoneal injection of 0.75 mg/kg medetomidine, 4.0 mg/kg midazolam, and 5.0 mg/kg butorphanol. After surgery, atipamezole was administered intraperitoneally as an anesthetic antagonist. The grafts were harvested at day 12 after transplantation. Fifteen grafts from eight independent transplantation experiments were analyzed for Fig. 1b and three samples were analyzed for Fig. 1c. Two grafts from two independent transplantation experiments were analyzed for Fig. 4. Two independent transplantation experiments (two grafts in each condition per experiment) were performed for Fig. 5. Two grafts from two independent transplantation experiments were analyzed for Fig S1a, S1c, S1d. Two independent transplantation experiments (two grafts in each condition per experiment) were performed for Fig. S9.
scRNA-seq analysis
scRNA-seq data of E15.5, E17.5, and P0 kidneys and the transplanted embryonic kidneys with a cloaca in the C57BL/6J background (transplanted at E12.5 and harvested at day 12 after transplantation) was described previously18. For the additional transplantation experiments, E12.5 embryonic kidneys with a cloaca from the ICR pregnant mice were transplanted into the testicular fat of the male NOD.CB17-Prkdcscid /J mice (8- week-old), and harvested at day 8 and 12 after transplantation.
Dissociation of E15.5, E17.5, and P0 kidneys were described previously18. Grafts harvested on days 8 and 12 were dissociated using a similar method for the P0 kidney. Specifically, the grafts were digested with dissociation buffer comprising 2 mg/ml collagenase (Sigma; Cat#9407), 2.4 U/ml dispase II (Roche; Cat# 04942078001), 2 mM CaCl2 (Wako; Cat# 031–00435), 50 µg/ml DNase I (Worthington #LS002139), and 10% fetal calf serum (FCS) (Sigma; Cat# 172012) in Dulbecco’s modified Eagle’s medium (DMEM) (Sigma; Cat# D5796) for 20 min at 37 °C, washed with phosphate-buffered saline (PBS), and treated with 0.25% trypsin/EDTA 37 °C for 10 min. For the postnatal kidney (P7), the kidney was digested with dissociation buffer comprising 300U/ml collagenase Type I (Worthington; Cat# LS004194), 1 mg/ml Pronase E (Sigma; Cat# P8811), 50 U/ml DNase I (Worthington #LS002139) in Hanks’ balanced salt solution (HBSS) (Thermo; Cat# 14185-052) for 20 min at 37 °C, followed by treatment with 0.25% trypsin/EDTA at 37 °C for 10 min.
Aliquots containing 5000 dissociated cells from each sample were applied to a Chromium Controller (10× Genomics). A Chromium Single Cell 3′ Library & Gel Beads Kit v2 or v3 (10× Genomics) was used to generate cDNA libraries, which were then sequenced by an Illumina HiSeq X (757,548,299 reads for E15.5; 259; 180,950 reads for E17.5; 955,496,397 reads for P0; 370,196,520 reads for P7; 374,558,242 reads for the graft at day 8; 384,179,897 reads for the graft at day 12; 401,965,326 reads for the graft at day 12 (*)18. The Q30 base RNA reads (Q-scores indicating sequencing quality) of the samples were 86.2% for E15.5, 63.8% for E17.5, 93.6% for P0; 91.9% for P7, 91.9% for the graft at day 8, 92.2% for the graft at day 12, and 91.2% for the graft at day 12 (*).
The raw sequence data were processed using the cell ranger count command in Cell Ranger version 7.1.0 (10× Genomics), for E15.5, E17.5, P0, P7, for the graft at day 8, for the graft at day 12, and for the graft at day 12 (*) to generate count tables of unique molecular identifiers (UMIs) for each gene per cell. At this point, each dataset contained 32,749, 31,333, 33,951 34,048 35,486 35,115, and 30,347 genes, 3,969, 5,358 10,356 5,875 5,423 4,912, and 7,699 cells for E15.5, E17.5, P0, P7, the graft at day 8, the graft at day 12, and the graft at day 12 (*), respectively. All of these individually generated datasets were integrated using the cell ranger aggr command (10× Genomics). All subsequent analyses were performed in the R statistical programming language version 4.3.135, and the RStudio software36. Seurat package (version 4.3.0 and 5.0.3) was used for analyses including quality control, data normalization, data scaling, visualization, and the differential analyses37,38. For quality control, cells that expressed < 500 genes, > 35% of mitochondrial genes, > 1% of hemoglobin genes, were filtered out. Potential doublets were also removed by using DoubletFinger (2.0.4) before the further analysis. The final dataset contained 27,421 genes and 34,024 cells. A principal component analysis was used for dimension reduction with a dimension value of 95 determined by the JackStrawPlot function39. Normalization was performed by SCTransform with the top 3000 highly variable genes selected by the SelectIntegrationFeatures function. Integrated data was generated by IntegrateData with using anchor genes create by FindIntegrationAnchors function, followed by scaling the data with regressing out cell cycle, high mitochondrial, ribosomal, and hemogrobin ratio. Cluster segmentation was performed using a resolution value of 2.0. The FindClusters command generated a total of 50 clusters that were easily distinguished with cluster-specific marker genes obtained with the FindMarkers function of the Seurat package. Uniform Manifold Approximation and Projection for Dimension Reduction (UMAP) plots were generated using the uwot package39. The UMAP coordinates, Seurat cluster coordinates, and cluster-specific markers obtained were exported as csv files for confirmation analysis using Loupe Cell Browser software (10x Genomics).
Selection of maturation stage-dependent genes
To select maturation stage-dependent gene candidates in each cluster, we first used the FindMarkers function to pick up genes that were differentially expressed between P7 and E15.5 (e.g. comparison between cluster 10 at P7 and cluster 10 at E15.5 for collecting ducts). To determine the genes enriched in each cell type, we used the FindMarkers function to compare the target cluster with all other clusters and selected the genes with low p-values (p < 0.05). After selecting the overlapping genes, we verified them in UMAP plots to finalize the cell type-enriched stage-dependent genes. These gene lists were used for volcano plots using the EnhancedVolcano function40. For GSEA analysis, the same gene lists were processed using the gseGO function41, and major pathways involved in each cell type were visualized using the dotplot function in DOSE package42. For CNET plots, part of the GSEA result were plotted using the cnetplot function in the DOSE package42.
Heatmap
We first utilized the AverageExpression function to add the average gene expression data for each cluster at each stage (log-fold change > 1 and p-value < 0.05). The top 200 genes were selected from the maturation stage-dependent gene lists described above. The RNAseqChef program43 was used to obtain the heatmap, and pheatmap function44 was also used to obtain a heatmap with unbiased hierarchal clustering analysis.
Immunohistochemistry
Paraffin sections were subjected to antigen retrieval in 10 mM citrate buffer (pH 6.0) after the deparaffinization. The sections were washed three times with PBS and blocked by incubation with 1% BSA in PBS for 1 h at room temperature. The sections were incubated with primary antibodies at 4 °C overnight, followed by incubation with secondary antibodies conjugated with Alexa Fluor 488, 568, or 633 for 90 min at room temperature. Cell nuclei were counterstained with 4,6-diamidino-2-phenylindole (Roche). The following primary antibodies were used: mouse anti-E-cadherin (CDH1) (BD; 610181; 1:100); rabbit anti-NKCC2 (SLC12A1) (StressMarq Bioscience; SPC-401D; 1:100); rat anti-CK8 (KRT8) (DSHB; 11562-1-AP; 1:20). Fluorescence images were captured with a confocal microscope (TSC SP8; Leica).
Whole-mount immunostaining
Whole-mount immunostaining was performed previously described with some minor modifications45. The graft or P0 kidney was fixed in 25%, 50%, and 75% methanol/ H2O for 10 min each at room temperature and then in methanol with 20% DMSO overnight on a shaker at 4℃. The samples were rehydrated in 75%, 50%, and 25% methanol/ H2O for 60 min each at room temperature and then washed three times with 1% Triton X-100 in PBS for 5 min at room temperature. The washed samples were placed in PBS containing 0.3 M glycine, 1% TritonX-100, and 20% DMSO for 3 h at room temperature and then blocked in PBS containing 10% goat serum (Nippon Bio-Test Laboratories), 1% BSA (Sigma Life Science; A4503), 1% Triton X-100, 0.2% dry skim milk, and 10% DMSO overnight on a shaker at room temperature. After washing in PBS containing 1% TritonX-100 and 10 µg/ mL heparin (PTrH) twice for 60 min at room temperature, the tissues were incubated with primary antibodies conjugated with rabbit anti-E-cadherin (Cell Signaling; 3195; 1:50) and rat anti-CD31 (BD; 557355; 1:20) in PBS containing 10% goat serum, 1% BSA, 1% Triton X-100, 0.2% dry skim milk, and 5% DMSO overnight on a shaker at 4 °C. The samples were washed with PTrH for 1 day at 4 °C and then incubated with secondary antibodies conjugated with Alexa Fluor 488 and 568 (1:100) overnight at 4 °C. The tissues were washed with PTrH for two days at 4 °C. The samples were serially dehydrated with 50%, 70%, and 100% ethanol for 60 min each at 4 °C and cleared with ethyl cinnamate (Sigma-Aldrich) for more than 30 min at room temperature. Three-dimensional fluorescence images were captured using a confocal microscope (TSC SP8; Leica) and reconstructed using LAS X (Leica) software and Imaris (Bitplane).
In situ hybridization
RNAscope analysis23 of 10% formalin-fixed paraffin sections was performed using an RNAscope Multiplex Fluorescent Reagent Kit v2 (Advanced Cell Diagnostics; Cat# 323100). Signal amplification was performed with TSA plus fluorophores (Thermo Fisher Scientific). The following RNAscope probes were used: Wnt7b (401131), Aqp4 (417161-C2), Aqp2 (452411-C3), Hif1a (313821), Ptgs2 (316621-C2), Gria1(426241-C2), and Cntn6 (836461).
Roxadustat treatment
E12.5 kidneys were transplanted into the peri-testicular fat. Roxadustat (FG4592) was dissolved in DMSO at 50 mg/ml and further diluted in 5% DMSO with 40% PEG300, 5% Tween 80, and 50% H2O to 2.5 mg/ml30,31. The host mice received daily intraperitoneal injections of 10 or 25 mg/kg/day Roxadustat from day 2 to day 11 after transplantation. The grafts at day 12 were compared histologically between those with and without Roxadustat treatment.
Blood analysis
8-week-old male mice received daily intraperitoneal injections of vehicle alone or 25 mg/kg/day Roxadustat for 10 days (n = 5 and 3, respectively). Mice were anesthetized by intraperitoneal injection of 0.75 mg/kg medetomidine, 4.0 mg/kg midazolam, and 5.0 mg/kg butorphanol. Blood samples were collected from the inferior vena cava. Blood hemoglobin levels were analyzed using an i-STAT1 analyzer with an EG6 + cartridge (Abbott, Princeton, NJ, USA). Student’s t-test was performed.
Statistical analysis
We assumed that our sample data were normally distributed for the analysis of the three groups. Then, Bartlett’s test was performed to determine the equality of variances among the three groups. If the variances were not significantly equal, Welch’s ANOVA was performed. If the ANOVA was significant, multiple comparisons were performed using Dunnett’s T3 test. Differences with values of p<0.05 were considered statistically significant. The asterisk (*) represents the p-value of the statistical test. One asterisk indicates that the p-value is less than 0.05. Four asterisks indicate that the p-value is less than 0.0001. This analysis was performed using GraphPad Prism 10 (GraphPad Software, San Diego, CA).
Data availability
All data supporting the conclusions are present in the paper and the supplementary materials. Figures 2 and 3 have associated scRNA-seq raw data, which have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus (GSE271515 [https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE271515]). The scRNA-seq data of E15.5, E17.5, and P0 embryonic kidneys, and one of the day 12 grafts were described previously18 (GSE149134 [https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE149134]).
References
Nishinakamura, R. Advances and challenges toward developing kidney organoids for clinical applications. Cell. Stem Cell. 30, 1017–1027 (2023).
Nishinakamura, R. Human kidney organoids: Progress and remaining challenges. Nat. Rev. Nephrol. 15, 613–624 (2019).
Rasouly, H. M. & Lu, W. Lower urinary tract development and disease. Wiley Interdiscip Rev. Syst. Biol. Med. 5, 307–342 (2013).
Hartman, H. A., Lai, H. L. & Patterson, L. T. Cessation of renal morphogenesis in mice. Dev. Biol. 310, 379–387 (2007).
Taguchi, A. et al. Redefining the in vivo origin of metanephric nephron progenitors enables generation of complex kidney structures from pluripotent stem cells. Cell. Stem Cell. 14, 53–67 (2014).
Takasato, M. et al. Directing human embryonic stem cell differentiation towards a renal lineage generates a self-organizing kidney. Nat. Cell. Biol. 16, 118–126 (2014).
Takasato, M. et al. Kidney organoids from human iPS cells contain multiple lineages and model human nephrogenesis. Nature 526, 564–568 (2015).
Morizane, R. et al. Nephron organoids derived from human pluripotent stem cells model kidney development and injury. Nat. Biotechnol. 33, 1193–1200 (2015).
Freedman, B. S. et al. Modelling kidney disease with CRISPR-mutant kidney organoids derived from human pluripotent epiblast spheroids. Nat. Commun. 6, 8715 (2015).
Taguchi, A. & Nishinakamura, R. Higher-order kidney organogenesis from pluripotent stem cells. Cell. Stem Cell. 21, 730–746 (2017).
Wu, H. et al. Comparative analysis and refinement of human PSC-derived kidney organoid differentiation with single-cell transcriptomics. Cell. Stem Cell. 23, 869–881e8 (2018).
Sharmin, S. et al. Human induced pluripotent stem cell-derived podocytes mature into vascularized glomeruli upon experimental transplantation. J. Am. Soc. Nephrol. 27, 1778–1791 (2016).
Bantounas, I. et al. Generation of functioning nephrons by implanting human pluripotent stem cell-derived kidney progenitors. Stem Cell. Rep. 10, 766–779 (2018).
van den Berg, C. W. et al. Renal subcapsular transplantation of PSC-derived kidney organoids induces neo-vasculogenesis and significant glomerular and tubular maturation in vivo. Stem Cell. Rep. 10, 751–765 (2018).
Freedman, B. S. & Dekel, B. Engraftment of kidney organoids in vivo. Curr. Transpl. Rep. 10, 29–39 (2023).
Fujimoto, T. et al. Generation of human renal vesicles in mouse organ niche using nephron progenitor cell replacement system. Cell. Rep. 32, 108130 (2020).
Tanigawa, S. et al. Generation of the organotypic kidney structure by integrating pluripotent stem cell-derived renal stroma. Nat. Commun. 13, 611 (2022).
Naganuma, H. et al. Molecular detection of maturation stages in the developing kidney. Dev. Biol. 470, 62–73 (2021).
Harris, R. C., Wang, J. L., Cheng, H. F., Zhang, M. Z. & McKanna, J. A. Prostaglandins in macula densa function. Kidney Int. 54, S49–S52 (1998).
Norwood, V. F., Morham, S. G. & Smithies, O. Postnatal development and progression of renal dysplasia in cyclooxygenase-2 null mice. Kidney Int. 58, 2291–2300 (2000).
Kaelin, W. G. & Ratcliffe, P. J. Oxygen sensing by metazoans: The Central role of the HIF hydroxylase pathway. Mol. Cell. 30, 393–402 (2008).
Taylor, C. T. & Scholz, C. C. The effect of HIF on metabolism and immunity. Nat. Rev. Nephrol. 18, 573–587 (2022).
Wang, F. et al. RNAscope: A novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J. Mol. Diagnostics. 14, 22–29 (2012).
Granger, A. J., Shi, Y., Lu, W., Cerpas, M. & Nicoll, R. A. LTP requires a reserve pool of glutamate receptors independent of subunit type. Nature 493, 495–500 (2013).
Takeda, Y. et al. Impaired motor coordination in mice lacking neural recognition molecule NB-3 of the contactin/F3 subgroup. J. Neurobiol. 56, 252–265 (2003).
Ye, H. et al. Neural recognition molecules CHL1 and NB-3 regulate apical dendrite orientation in the neocortex via PTPα. EMBO J. 27, 188–200 (2008).
Yu, J. et al. A Wnt7b-dependent pathway regulates the orientation of epithelial cell division and establishes the cortico-medullary axis of the mammalian kidney. Development 136, 161–171 (2009).
Roker, L. A., Nemri, K. & Yu, J. Wnt7b signaling from the ureteric bud epithelium regulates medullary capillary development. J. Am. Soc. Nephrol. 28, 250–259 (2017).
Provenzano, R. et al. Oral hypoxia–inducible factor prolyl hydroxylase inhibitor roxadustat (FG-4592) for the treatment of anemia in patients with CKD. Clin. J. Am. Soc. Nephrol. 11, 982–991 (2016).
Su, X. et al. HIF-α activation by the prolyl hydroxylase inhibitor roxadustat suppresses chemoresistant glioblastoma growth by inducing ferroptosis. Cell. Death Dis. 13, 861 (2022).
Yan, P. et al. Hypoxia-inducible factor upregulation by roxadustat attenuates drug reward by altering brain iron homoeostasis. Signal. Transduct. Target. Ther. 8, 355 (2023).
Saito, Y. et al. Generation of functional chimeric kidney containing exogenous progenitor-derived stroma and nephron via a conditional empty niche. Cell. Rep. 39, 110933 (2022).
Yamanaka, S. et al. Generation of interspecies limited chimeric nephrons using a conditional nephron progenitor cell replacement system. Nat. Commun. 8, 1719 (2017).
Tanigawa, S. et al. Organoids from nephrotic disease-derived iPSCs identify impaired NEPHRIN localization and slit diaphragm formation in kidney podocytes. Stem Cell. Rep. 11, 727–740 (2018).
R Core Team. R: A language and environment for statistical computing. R Foundation for statistical computing, Vienna, Austria. (2018).
RStudio Team. RStudio: Integrated Development for R. RStudio (PBC, 2020).
Butler, A., Hoffman, P., Smibert, P., Papalexi, E. & Satija, R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420 (2018).
Stuart, T. et al. Comprehensive integration of single-cell data. Cell 177, 1888–1902e21 (2019).
Chung, N. C. & Storey, J. D. Statistical significance of variables driving systematic variation in high-dimensional data. Bioinformatics 31, 545–554 (2015).
Blighe, K., Rana, S. & EnhancedVolcano, L. M. Publication-ready volcano plots with enhanced colouring and labeling. R package version 1.22.0. (2024).
Korotkevich, G., Sukhov, V., Budin, N., Atryomov, M. N. & Sergushichev, A. Fast gene set Enrich. Anal. bioRxiv 060012 doi:https://doi.org/10.1101/060012 (2021).
Yu, G., Wang, L. G., Yan, G. R. & He, Q. Y. DOSE: An R/Bioconductor package for disease ontology semantic and enrichment analysis. Bioinformatics 31, 608–609 (2015).
Etoh, K. & Nakao, M. A web-based integrative transcriptome analysis, RNAseqChef, uncovers the cell/tissue type-dependent action of sulforaphane. J. Biol. Chem. 299, 104810 (2023).
Kolde, R. pheatmap: Pretty Heatmaps. R package version 1.0.12. (2019).
Renier, N. et al. IDISCO: A simple, rapid method to immunolabel large tissue samples for volume imaging. Cell. 159. , 896–910 (2014).
Acknowledgements
We thank Prof. H. Masutani, Tenri University, and S. Fujimura, S. Usuki, and K. Yasunaga, members of the Liaison Laboratory Research Promotion Center, Institute of Molecular Embryology and Genetics, Kumamoto University, for experimental assistance. We also thank all members of the Nishinakamura laboratory for helpful discussions.
Author information
Authors and Affiliations
Contributions
All authors participated in the design, interpretation of the studies and analysis of the data and review of the manuscript. Conceptualization, R.N.; methodology, H.I., K.M., and R.N; investigation, H.I. (transplantation, scRNA-seq, histologic analysis), K.M. (scRNA-seq), T.O. (in situ hybridization), K.M. (blood analysis), and Y.I. (blood analysis), S.T. (scRNA-seq); resources, H.I. and R.N.; writing-original draft, H.I., K.M., and R.N.; writing-review & editing, R.N.; funding acquisition, R.N.; project administration and supervision, R.N.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Ide, H., Miike, K., Ohmori, T. et al. Mouse embryonic kidney transplantation identifies maturation defects in the medulla. Sci Rep 14, 30293 (2024). https://doi.org/10.1038/s41598-024-81984-w
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-024-81984-w
Keywords
This article is cited by
-
In vitro generation of a ureteral organoid from pluripotent stem cells
Nature Communications (2025)