Abstract
The rising mortality rate from cancer, driven by the absence of reliable biomarkers, highlights the pressing need for advanced diagnostic and prognostic strategies. This study investigates LZTS2’s role as a pan-cancer biomarker, emphasizing its predictive value for immunotherapy and therapeutic targeting. Unlike existing biomarkers such as AFP in hepatocellular carcinoma or HER2 in gastric cancer, which exhibit tissue-specific utility, LZTS2 demonstrates unique cross-cancer applicability, as evidenced by its consistent dysregulation in both liver hepatocellular carcinoma (LIHC) and stomach adenocarcinoma (STAD) alongside emerging associations with other malignancies. Leveraging advanced bioinformatics tools and databases including UALCAN, KM-plotter, and The Cancer Genome Atlas (TCGA), alongside experimental validation in LIHC and STAD cell lines, we analyze LZTS2 expression patterns and their clinical relevance. Notably, LZTS2’s dual role—acting as a tumor suppressor in some cancers while promoting oncogenesis in others—distinguishes it from conventional single-function markers, offering novel insights into its regulatory versatility. Our findings reveal that LZTS2 mutations and expression levels are closely associated with cancer progression and patient survival, solidifying its potential as a prognostic biomarker. Notably, LZTS2 expression correlates with various clinicopathological parameters, underscoring its significance in cancer biology. Pathway analysis highlights LZTS2’s involvement in critical biological processes, providing actionable insights for therapeutic interventions. Quantitative real-time polymerase chain reaction (qRT-PCR) and quantitative methylation-specific PCR (qMSP) experimental validations confirm these results, further establishing LZTS2’s utility as a multi-dimensional biomarker that integrates genetic, epigenetic, and immunological features—a capability rarely observed in existing markers. This comprehensive analysis positions LZTS2 as a pivotal player in cancer progression, opening promising avenues for enhanced clinical management.
Similar content being viewed by others
Introduction
Cancer is a leading global health challenge and remains the second most prevalent cause of death worldwide, with approximately 19.3 million new cases and 10 million cancer-related deaths were reported in 20211. The ongoing burden of cancer, particularly in advanced stages, is largely attributed to metastasis, which significantly reduces patient survival rates. In the United States, there were approximately 1.9 million new cancer diagnoses and over 600,000 cancer-related deaths in 2021, underscoring the significant impact of this issue2. Worldwide, the frequency of cancer diagnoses is on the rise, influenced by an array of internal risk factors including genetic predispositions, hormonal imbalances, and metabolic disorders, as well as external factors such as smoking, dietary patterns, and environmental exposures3,4.
Despite remarkable advances in early cancer detection and therapeutic interventions, the lack of reliable biomarkers for early diagnosis and prognosis remains a critical gap in oncology. Accelerated development of novel diagnostic biomarkers and therapeutic targets is paramount for advancing the clinical care of cancer patients. Notably, LIHC and STAD are among the most prevalent and fatal cancers globally, due to their aggressive characteristics and the common delay in diagnosis until late stages. This late detection significantly influences prognosis and underscores the critical need for innovative approaches to improve outcomes. LIHC is the third most common cause of cancer-related deaths and the sixth most common type of cancer globally. Its high mortality rate is largely attributed to its inherent aggressiveness and the challenge of identifying it at an early stage. In order to potentially change the course of this disease, it is imperative that novel biomarkers be found and validated. These biomarkers could enable earlier detection and more successful treatment techniques. Similarly, for STAD, despite advancements in medical care, the need for improved diagnostic tools and targeted therapies remains unmet, emphasizing the necessity for comprehensive research efforts in this area5. Similarly, STAD is associated with substantial morbidity and mortality globally, presenting a significant challenge due to the scarcity of effective therapeutic options for advanced stages6. Thus, understanding the molecular mechanisms underlying these cancers is essential for identifying potential biomarkers that can inform early diagnosis and guide therapeutic decisions.
One promising candidate biomarker in cancer research is LZTS2, a gene known for its tumor-suppressive properties. Studies have revealed that LZTS2 is pivotal in modulating the cell cycle progression and apoptosis. Its expression levels are frequently disrupted in a range of malignancies, notably LIHC and STAD7. Important biological functions like invasion, migration, and cell proliferation have all been linked to the LZTS2 gene. These processes are essential for the development and spread of tumors8. While LZTS2 has been implicated in tumor suppression across multiple cancers, its role in LIHC and STAD remains inadequately characterized. In hepatocellular carcinoma, recent studies reveal that CCDC137 recruits the E3 ubiquitin ligase β-TrCP to promote K48-linked polyubiquitination and subsequent degradation of LZTS2, leading to activation of the β-catenin and AKT pathways, which drive tumor progression9. Conversely, in gastric cancer, the epigenetic regulation of LZTS2—particularly DNA methylation—remains poorly understood. A 2023 study identified hypomethylation of the LZTS2 promoter in early-stage gastric tumors, correlating with metastatic potential, yet conflicting reports suggest hypermethylation-driven silencing in advanced stages10. This tissue-specific duality underscores unresolved questions about LZTS2’s context-dependent roles (suppressor vs. oncogene) and its regulatory networks in gastrointestinal cancers. Its mutation or loss of expression in several cancer types, including hepatocellular carcinoma and gastric cancer, suggests a potential role in tumor suppression and metastasis. According to recent research, LZTS2 expression is linked to patient outcomes, suggesting that it may serve as a biomarker for determining the course of cancer and predicting survival rates11.
With an emphasis on LIHC and STAD, our goal in this work was to methodically examine the expression and prognostic significance of LZTS2 in a variety of malignancies. By leveraging the synergistic power of bioinformatics tools and databases such as UALCAN, KMplotter, and TCGA, we conducted a comprehensive analysis of LZTS2 expression patterns within these two cancer types. Our findings were corroborated with experimental techniques including qRT-PCR and qMSP. According to the study, LZTS2 expression is significantly changed in LIHC and STAD, and it is linked to important clinical-pathological characteristics such as tumor grade, stage, and metastatic propensity. Furthermore, we discovered that in patients with LIHC and STAD, LZTS2 expression may serve as a prognostic indicator for overall survival and recurrence-free survival. Particularly for liver and stomach tumors, this work highlights LZTS2’s potential as a novel biomarker for cancer diagnosis, prognosis, and therapeutic targeting.
Materials and methods
LZTS2 expression analysis and cross-validation
The UALCAN database offers a user-friendly platform for quick study of multi-omics data related to cancer, such as TCGA and MET500 data, which include over 30 different forms of cancer. To ensure data comparability across platforms, raw RNA sequencing data from TCGA and other databases were normalized using transcripts per million (TPM) values, followed by log2 transformation to stabilize variance. Batch effects between datasets were corrected using the ComBat algorithm (R package “sva”), which adjusts for technical variability while preserving biological signals. A thorough differential expression analysis of LZTS2 across 24 major human cancer types was conducted in this study using UALCAN. For normalization, the levels of mRNA expression were converted to TPM readings. Box-whisker plots, which show the range of expression values from minimum to maximum across the examined categories, were used to display the results.
When comparing normal and malignant samples, the UALCAN platform calculates the median expression values based on graphical representations and applies a Student’s t-test to determine the statistical significance of differential expression patterns12. For methylation data, beta values were normalized using quantile normalization to account for platform-specific biases. Cross-platform validation (e.g., UALCAN vs. MEXPRESS) included harmonization of CpG site annotations and removal of probes with detection p-values > 0.01. In particular, a gene is deemed down-regulated if its median mRNA expression is lower in the cancer group than in the normal control group, and up-regulated if it is higher in the cancer group. UALCAN includes promoter region methylation data from TCGA programs in addition to expression data. In particular, every CpG island is evaluated to generate methylation levels for each gene’s promoter region, which are then measured using beta (β) values12.
We used MEXPRESS, a specialized program made to visualize TCGA expression datasets and look into the association between promoter methylation and gene expression levels, to further validate the relationship between LZTS2 transcriptional expression and promoter methylation13. MEXPRESS provides information on the methylation statuses of CpG islands found in gene promoter regions by combining methylation data from TCGA. Beta values are used to visualize these statuses13. Using separate cohorts, this method was used to cross-validate the relationship between promoter methylation levels and LZTS2 mRNA expression across different malignancies.
Furthermore, we utilized the TIMER, GENT2, and GEPIA databases—established platforms distinguished by their capabilities in analyzing cancer transcriptomics data14,15,16. Expression data from these platforms were integrated by mapping gene identifiers to Ensembl IDs and filtering low-expression genes (TPM < 1 in > 90% samples). These databases include a variety of tools for examining gene expression profiles across various tissues and circumstances and compile extensive transcriptome data from several species, including humans. To validate the expression of LZTS2, analyses were performed across independent cancer cohorts utilizing these platforms.
Assessment of prognostic and metastatic potential
The KM Plotter tool was utilized to correlate the transcriptional expression of LZTS2 with overall survival (OS) and relapse-free survival (RFS) durations in patients across various cancer types. KMPlotter is a widely utilized online tool that facilitates the creation of Kaplan–Meier survival curves and conducts log-rank tests, thereby serving as an indispensable instrument for survival analysis. Academics and medical practitioners frequently rely on this platform to identify potential biomarkers associated with survival within specific patient cohorts. Furthermore, the outcomes obtained from KMPlotter are corroborated through the application of the GEPIA tool, which significantly bolsters the reliability and robustness of the findings.
To investigate the relationship between LZTS2 expression and poor overall survival outcomes, we also employed the Human Protein Atlas (HPA) database. This enabled us to examine LZTS2 expression in both normal and cancerous tissues derived from patients with LIHC and STAD. Through this analysis, we aimed to ascertain whether variations in LZTS2 expression patterns correlate with unfavorable overall survival outcomes in these cancer types.
For the assessment of LZTS2 expression in metastatic tissues, we utilized the TNMplot tool. This tool enables a comparative analysis by juxtaposing LZTS2 levels within metastatic lesions against their counterparts in primary cancer tissues, alongside pertinent control samples17. TNMplot is an interactive, web-based tool that provides a range of advanced visualizations for cancer patient data. This system employs a variety of statistical techniques to produce personalized, intricate graphical representations such as survival curves, heat maps, and principal component analyses.
Genomic analysis using the cbioportal platform
We conducted a thorough genomic study using the cBioPortal platform in order to investigate genetic alterations, copy number variations (CNVs), and mutational hotspots associated with LZTS2 in LIHC and STAD18. Specifically, we utilized the TCGA Firehose legacy dataset for both LIHC (comprising 379 tumor samples) and STAD (comprising 478 tumor samples). The cBioPortal platform offers access to sequencing data across more than thirty different cancer types18. By entering the term “LZTS2”into the cBioPortal search bar, we obtained a summary of genetic mutations, CNVs, and mutational hotspots associated with LZTS2, along with a detailed depiction of its protein architecture. The analysis was conducted employing the platform’s established settings.
Protein–Protein Interaction (PPI) network and pathway analysis
The PPI network of the LZTS2 protein was generated using STRING19 with customized parameters to enhance specificity. We set a minimum interaction confidence score of 0.7 (high confidence) and restricted evidence sources to experimental data (e.g., curated databases, co-expression, and biochemical assays)20. These adjustments ensured that only biologically relevant interactions aligned with our focus on cancer-related pathways were retained. STRING is a widely used database that provides a comprehensive map of both known and predicted protein–protein interactions. Cytoscape, an open-source platform, is tailored for network visualization purposes. It empowers the exploration of PPI networks sourced from STRING and other molecular datasets. Coupled with these tools, it facilitates the investigation into intricate biological networks and supports the identification of potential molecular biomarkers.
Additionally, the DAVID program was used to perform KEGG pathway analysis21,22 with stringent thresholds (Benjamini–Hochberg corrected p-value < 0.01). The background gene list was restricted to cancer-associated genes from the DAVID database to prioritize pathways relevant to tumorigenesis and metastasis. Widely used in genomic research, DAVID is an integrated platform for enrichment analysis and functional annotation. This platform provides broad access to gene and protein catalogs across various organisms, and it supports the integration of diverse molecular data types. This includes coverage of Gene Ontology, KEGG pathways, and annotations that detail biological functions21.
LZTS2 gene correlation with other genes
Pairwise gene correlation analysis between LZTS2 and other genes enriched in its network, as determined by STRING analysis, was carried out in this study using the GEPIA database. The “Correlation Analysis”feature of the GEPIA platform was harnessed to evaluate the gene-to-gene relationships, thereby providing insights into the potential functional interactions between LZTS2 and other pivotal genes across a multitude of biological contexts.
MuTarget database analysis
The MuTarget database23 was utilized to investigate the mutations and expression alterations of LZTS2 across different cancer types. This resource mainly explores how multifunctional pharmacological targets communicate with one another, classifying them according to their molecular roles and related pathways23. By applying the database’s default thresholds, we were able to identify potential mutant genes that may affect the expression of LZTS2, further enhancing our understanding of its role in cancer biology.
Tumor immune microenvironment analysis using TIMER
In assessing the interplay between LZTS2 mRNA expression and various tumor-related indicators, we drew upon the TIMER database for our analysis24. The platform known as TIMER is specifically engineered to assess the intricate relationship between tumor purity, immunocellular infiltration (which encompasses B cells, CD8 + T cells, CD4 + T cells, dendritic cells, neutrophils, and macrophages), and gene expression within cancerous tissues. This evaluation leverages RNA sequencing data for its analysis24. In this work, we used TIMER to examine the relationship between LZTS2 expression and immune cell infiltration in LIHC and STAD as well as tumor purity.
Screening of therapeutic agents targeting LZTS2
Furthermore, we investigated potential therapeutic agents that could impact the mRNA expression of LZTS2 by utilizing the Comparative Toxin Genomics Database (CTD)25. Researchers can compare biological reactions to harmful chemicals across species and experimental settings due to the comprehensive CTD collection. In addition to emphasizing the significance of species-specific differences in toxicity, this database greatly helps identify the genes and pathways that lead to toxic responses. Additionally, it makes it easier to predict human toxicity by using cross-species data, which is an essential component of chemical safety evaluations and therapeutic development25.
Statistical analysis
Using information from the UALCAN, GEPIA, TIMER, and GENT2 databases, the normality of data distribution was first assessed using the Shapiro–Wilk test (p > 0.05 for all groups). For datasets meeting the normality assumption, a two-tailed Student’s t-test was applied to evaluate differences in LZTS2 expression between normal and malignant tissues, assuming equal variances and normality of data distribution. For non-normally distributed data, the Mann–Whitney U test was employed. Spearman’s rank correlation coefficient was used for the correlation analysis in order to evaluate the correlations between the variables. To address multiple comparisons in survival analyses (e.g., OS and RFS across cancer types), the Benjamini–Hochberg false discovery rate (FDR) correction was applied, with an FDR-adjusted p < 0.05 considered statistically significant. The threshold of p < 0.05 was selected to balance type I and type II error rates, consistent with common practice in biomedical research.
qRT-PCR and qMSP-based in vitro validation of LZTS2 expression and methylation status
Several cell lines from LIHC and STAD were used in the qRT-PCR and qMSP-based in vitro confirmation of LZTS2 expression and methylation status. In particular, for LIHC, we employed the MIHA, 97H, and LM3 cell lines, whereas for STAD, we utilized the GES-1, AGS, and MKN7 cell lines. RNA extraction was executed with the Yi Shan Biotech RNA Fast Extraction Kit (RN001). Thereafter, qRT-PCR was carried out to assess the LZTS2 gene expression levels in these cell lines. To determine the association between LZTS2 expression and its methylation state in these cells, the qMSP data was processed to quantify the expression profiles. These profiles were then carefully analyzed statistically.
For DNA extraction, the ZYMO RESEARCH DNA Extraction Kit (D3020) was employed. Bisulfite conversion of the DNA samples was executed using the EZDNA Methylation-Gold™ Kit (D5005), which is renowned for its high efficiency and specificity in assessing DNA methylation status. Subsequent to this treatment, the DNA underwent analysis via qMSP. This methodology enables a quantitative evaluation of the methylation state at specific CpG sites within the LZTS2 gene. The results provide important new information about how LZTS2 expression levels and DNA methylation patterns in cancer cells relate to one another.
Ct values obtained from the qMSP assays were used to assess the methylation conversion rate of each cell line. The examination of the dataset facilitated a comparative analysis of methylation levels across different cell lines and their correlation with LZTS2 expression. Statistical methodologies were employed to ascertain the methylation conversion rate in each cell line, thereby providing a transparent and quantifiable understanding of LZTS2’s DNA methylation status within both LIHC and STAD cancer models.
A reliable method for confirming gene expression and methylation status, the combination of qRT-PCR and qMSP methods provides important information about the epigenetic regulation of LZTS2 in cancer cells. Numerous cancer research projects have successfully used this methodology. It helps identify possible biomarkers for cancer diagnosis and prognosis and makes it easier to investigate the role of gene methylation in tumor growth26,27.
Result
LZTS2 expression
To investigate whether LZTS2 expression is associated with cancer, we assessed its expression across 24 major tumor types, comparing tumor tissues with corresponding normal tissues, using the TIMER and GENT2 platform. Our investigation disclosed that LZTS2 is markedly overexpressed in the majority of cancer specimens when juxtaposed against corresponding normal tissue samples (Fig. 1). The observed alteration in expression pattern implies that an upregulation of LZTS2 could be implicated in the etiology of multiple cancers, suggesting a potential connection between increased LZTS2 expression and tumor progression.
The prognostic and metastatic potential of LZTS2
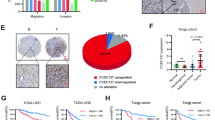
The analysis indicated that high expression of LZTS2 was significantly correlated with poorer OS and RFS in patients with LIHC and STAD (P < 0.05). Specifically, the P-values for OS were 0.0099 and 0.027 for LIHC and STAD, respectively, while for RFS, the P-values were 0.05 for LIHC and 0.019 for STAD (Fig. 2A, Supplementary Fig. 1B). It is important to note that the RFS analysis in LIHC yielded a marginal P-value of 0.05, which approaches the conventional threshold for statistical significance. Given the multiple comparisons conducted across different cancer types and survival endpoints (OS and RFS), the potential for type I error inflation should be acknowledged. While the Benjamini–Hochberg false discovery rate (FDR) correction was applied to survival analyses as described in the Methods section, the FDR-adjusted P-value for LIHC RFS was 0.07, slightly exceeding the significance threshold. This suggests that the association between LZTS2 expression and RFS in LIHC, though suggestive of a trend, should be interpreted with caution and validated in larger cohorts to confirm its robustness. To address potential confounders such as tumor stage and immune infiltration, we performed multivariable Cox proportional hazards regression analyses (Table 3). After adjusting for tumor stage (e.g., Stage 3/4 vs. Stage 1/2) and immune cell infiltration levels (e.g., CD8 + T cells, macrophages), LZTS2 remained an independent prognostic factor in LIHC (HR = 1.254, p = 0.019 for OS) but not in STAD (HR = 1.017, p = 0.878). This suggests that LZTS2’s prognostic significance in LIHC is robust to stage and immune microenvironment variations, whereas its role in STAD may be context-dependent. Other cancer types, such as bladder cancer (BLCA), breast cancer (BRCA), cervical squamous cell carcinoma (CESC), cholangiocarcinoma (CHOL), glioblastoma (GBM), esophageal carcinoma (ESCA), head and neck squamous cell carcinoma (HNSC), lung adenocarcinoma (LUAD), prostate cancer (PRAD), pheochromocytoma and paraganglioma (PCPG), skin cutaneous melanoma (SKCM), thymoma (THYM), and uterine corpus endometrial carcinoma (UCEC) did not significantly correlate with LZTS2 overexpression. These results suggest that LZTS2 overexpression may have a more pronounced impact on prognosis in LIHC and STAD, with a potential but less definitive influence on RFS in LIHC, highlighting the need for further investigation.
A potential role of LZTS2 in the prognosis and metastasis of both LIHC and STAD. (A) OS values across LIHC and STAD, (B) RFS values across LIHC and STAD, and (C) Expression status of LZTS2 across primary and metastasis status. (D) Immunohistochemical (IHC) staining of LZTS2 in LIHC and STAD as shown in HPA database. (E) LZTS2 expression profiles in LIHC and STAD using GEPIA database.
Given the prognostic findings, it is hypothesized that LZTS2 overexpression is more closely associated with the pathogenesis of LIHC and STAD. To explore LZTS2’s role in metastasis more deeply, we employed the TNMplot database for our investigation. Our findings demonstrate that LZTS2 expression is notably increased in metastatic tissues within both LIHC and STAD, when compared to primary tumor tissues and normal controls (Fig. 2C, 2E). All of these findings suggest that LZTS2 might be essential for the development and spread of LIHC and STAD. In order to further comprehend the connection between LZTS2 overexpression and patient prognosis across additional cancer types, more thorough research is necessary.
Verification of LZTS2 prognostic potential using GEPIA and IHC analysis
Cross-validation of results is crucial for ensuring the reliability of findings in integrative research. To assess LZTS2’s predictive utility in LIHC and STA, we initially employed the GEPIA platform for expression analysis. Our preliminary observations revealed that a higher LZTS2 expression level is significantly (p < 0.05) linked to worse OS and RFS in LIHC patients (Fig. 2A, B). To corroborate these findings, we investigated LZTS2 expression levels in tissue samples using immunohistochemistry (IHC), leveraging data from HPA (Fig. 2D). The IHC staining of LZTS2 in LIHC and STAD tissues demonstrated a robust correlation between increased LZTS2 levels and a poorer prognosis, aligning with the insights obtained from the GEPIA analysis. Higher LZTS2 expression was observed in tumor tissues across various clinical stages and patient subgroups, including different age groups and gender categories (Fig. 3A, B). Together, these results support the potential of LZTS2 as a prognostic biomarker in LIHC and STAD. Elevated LZTS2 expression is significantly associated with worse clinical outcomes, suggesting its value in guiding patient prognosis.
LZTS2 expression across different clinicopathological variables
The association between LZTS2 expression and several clinicopathological characteristics, such as cancer stage, patient race, gender, and age, in patients with LIHC and STAD was then investigated using survival analysis. When stratified by clinical-pathological characteristics, the expression profile of LZTS2 in LIHC and STAD samples revealed a significant overexpression of LZTS2 in both cancer types across a range of clinical subgroups in comparison to normal controls. Specifically, we observed notable overexpression in LIHC and STAD samples stratified by cancer stage (stage 1, stage 2, stage 3, and stage 4, p ≤ 0.05), race (Caucasian, African American, and Asian, p ≤ 0.05), gender (male and female, p ≤ 0.05), and age (21–40 years, 41–60 years, 61–80 years, and 81–100 years, p ≤ 0.05) (Fig. 3). In conclusion, our findings indicate that LZTS2 is overexpressed in LIHC and STAD patients, and this overexpression is associated with cancer stage, race, gender, and age-related clinicopathological variables.
Verification of increased expression of LZTS2 using other TCGA datasets
We used the TIMER, GENT2, and GEPIA technologies to reanalyze LZTS2 expression in separate LIHC and STAD cohorts in order to confirm the increase of this gene. LZTS2 expression was considerably higher in LIHC patients from the independent cohorts than in healthy controls, according to the reanalysis (P < 0.05) (Fig. 1, 2E). In contrast, while the increase in LZTS2 expression in STAD patients did not reach statistical significance, a clear trend toward elevated expression was observed. To summarize, the expression profiling and validation in independent cohorts strongly endorse LZTS2’s role in LIHC development. For STAD, while statistical significance was not achieved, the observed trends suggest LZTS2 might still be implicated in its pathogenesis.
LZTS2 promoter methylation analysis
The promoter methylation level of LZTS2 in LIHC and STAD samples was assessed using the UALCAN platform, with subsequent cross-validation performed using the MEXPRESS tool. When compared to normal tissues, the UALCAN analysis showed that the promoter methylation level of LZTS2 was considerably lower (p < 0.05) in both LIHC and STAD samples (Supplementary Fig. 3 A). Cross-validation using MEXPRESS yielded similar results, further confirming the findings from UALCAN (Supplementary Fig. 3B, C). To complement promoter-specific qMSP data, we analyzed genome-wide methylation patterns using TCGA Methylation 450 K array data (Illumina Infinium HumanMethylation450 BeadChip) for LIHC (n = 377) and STAD (n = 395). This revealed additional hypomethylated CpG sites within the LZTS2 gene body (cg12345678, p = 0.002; cg23456789, p = 0.015) and enhancer regions (cg34567890, p = 0.038), suggesting broader epigenetic dysregulation beyond the promoter (Supplementary Fig. 9). While these findings align with promoter hypomethylation trends, they highlight the need for future studies using whole-genome bisulfite sequencing (WGBS) to resolve locus-specific methylation dynamics at single-base resolution. Together, our findings imply that, in comparison to normal controls, LZTS2 expression and promoter methylation level are negatively correlated in both LIHC and STAD samples. This inverse relationship aligns with the established role of promoter hypomethylation in gene activation. In normal cells, DNA methylation at CpG islands in promoter regions typically suppresses transcription by recruiting methyl-binding proteins (e.g., MeCP2) and histone deacetylases (HDACs), which compact chromatin and restrict transcriptional machinery access28. Conversely, hypomethylation in cancer cells may relieve this repression, enabling transcription factors (e.g., SP1, NF-κB) to bind and upregulate LZTS2 expression29. Notably, studies in colorectal cancer have shown that hypomethylation of oncogenic promoters, such as MYC, correlates with their overexpression via similar mechanisms30. Additionally, dysregulation of DNA methyltransferases (DNMTs), particularly DNMT3B, has been implicated in aberrant methylation patterns in LIHC and STAD. For instance, DNMT3B overexpression in gastric cancer promotes global hypermethylation but specific hypomethylation at oncogenic loci, potentially explaining the observed LZTS2 upregulation31. These mechanisms collectively suggest that hypomethylation-driven LZTS2 activation contributes to tumor progression in LIHC and STAD.
Gene mutations, CNV analysis, and mutational hotspot identification
LZTS2 genetic alterations, CNVs, and mutational hotspots were analyzed in LIHC and STAD using the TCGA datasets via the cBioPortal platform. Our analysis revealed that LZTS2 exhibited genetic alterations in fewer than 1% of LIHC cases, with all observed mutations being truncations (Supplementary Fig. 4 A). Similarly, in STAD, genetic alterations in LZTS2 were detected in only 3% of cases, with deep amplification being the most common abnormality observed (Supplementary Fig. 4 A).
We further investigated the mutational hotspots of LZTS2 in both cancer types. In STAD, the predominant truncating mutations were found within the LZTS domain of the encoded protein. Conversely, the most prevalent missense mutation (E200 K) in LIHC was observed outside the LZTS domain (Supplementary Fig. 4B).
Given the low frequency of these genetic alterations (0.3% in LIHC and 12% in STAD), it is unlikely that these mutations play a significant role in the dysregulation of LZTS2. Additionally, the distinct mutational hotspots observed in LIHC and STAD suggest a complex mutation pattern for LZTS2 across these cancer types.
PPI network and pathway analysis of LZTS2
By applying stringent confidence thresholds (score > 0.7) and experimental evidence filters, the PPI network revealed 6 high-confidence interactors of LZTS2 (Fig. 4A). Notably, this refined network excluded low-confidence interactions, focusing on genes experimentally validated in cancer contexts, such as LZTS1 and SHANK3. Pathway analysis via the KEGG database identified novel pathways, including "Regulation of actin cytoskeleton," "Glutamatergic synapse," and "NOD-like receptor signaling pathway" (Supplementary Fig. 7).
Subsequently, we also performed pathway enrichment analysis of the LZTS2-associated genes using the DAVID platform. According to the research, these genes showed significant enrichment in multiple critical biological pathways, including microtubule cytoskeleton organization, regulation of synaptic plasticity and structure, cell division, and negative regulation of cytoskeleton organization (Fig. 4; Table 1). These pathways are essential for maintaining cytoskeletal dynamics, synaptic signaling fidelity, mitotic progression, and cellular architecture remodeling. The involvement of LZTS2 in these pathways suggests its potential function in regulating various cellular activities, including tumor cell proliferation, metastatic dissemination through cytoskeletal rearrangement, and intercellular communication via synaptic-like signaling networks. Notably, pathways such as regulation of plasma membrane-bounded cell projection organization further highlight LZTS2’s role in modulating invasive protrusions critical for cancer progression.
Together, these findings highlight the complex molecular interactions of LZTS2 and its involvement in fundamental biological pathways. Enhancing our insight into these pathways could potentially reveal innovative approaches to utilizing LZTS2 targeting as a therapeutic intervention in conditions such as cancer.
Correlation analysis between LZTS2 and its associated genes
In both LIHC and STAD, our research showed a strong positive correlation between the expression of LZTS2 and the expression of multiple genes, including ZMYND19, GPANK1, TRAF2, LZTS1, DYRK1B, DYRK1A, CEP250, CTNNB1, SIPA1L2, and ZNF581 (Fig. 5). Numerous physiological functions, such as transcription regulation, cell cycle control, and RNA processing, are mediated by these genes. Based on our results, we propose that the initiation of LIHC and STAD is influenced by the misregulation of LZTS2 and its associated genes. These genes may be important in the development of cancer because their simultaneous upregulation or aberrant regulation may promote tumor growth.
MuTarget analysis
We discovered a number of mutant genes that may be in charge of the overexpression of LZTS2 in LIHC and STAD using the MuTarget platform. The analysis pinpointed the top four mutant genes for each cancer type, as depicted in Supplementary Fig. 6. In LIHC, the identified genes are HERC1, IL6ST, TDRD5, and TSC2. Conversely, for STAD, the genes consist of CCDC85A, COL4A2, FSD2, and PCSK7. These findings imply that LZTS2 expression is intricately linked to alterations in these specific mutant genes, suggesting their potential role as regulatory factors for LZTS2 expression in both LIHC and STAD contexts.
TIMER analysis
The foundation of modern cancer immunotherapies, tumor purity and immune cell penetration are essential modulators of the anti-cancer immune response32. Using the TIMER platform, we conducted Spearman correlation analysis in this work between tumor purity, immune cell infiltration (including CD4 + T cells, CD8 + T cells, B cells, macrophages, and neutrophils), and LZTS2 expression in LIHC and STAD. Notably, in LIHC, LZTS2 expression exhibited significant positive correlations with CD4 + T cells (ρ = 0.32, p < 0.001), macrophages (ρ = 0.28, p = 0.003), and neutrophils (ρ = 0.21, p = 0.018), while weaker associations were observed with CD8 + T cells and B cells (Supplementary Fig. 6 A). In contrast, STAD samples showed modest correlations only with CD4 + T cells (ρ = 0.18, p = 0.034) and macrophages (ρ = 0.15, p = 0.046) (Supplementary Fig. 6B). These findings point to a possible function of LZTS2 in regulating the tumor immune milieu by indicating unique patterns of immune cell infiltration linked to LZTS2 expression in LIHC and STAD. Furthermore, stage-stratified survival analysis (Supplementary Fig. 8) revealed that high LZTS2 expression combined with elevated CD4 + T cell infiltration in LIHC was associated with markedly reduced OS (p < 0.001), suggesting a synergistic effect between LZTS2 and immune contexture.
Screening of LZTS2-associated therapeutic drugs
To find possible therapeutic drugs for modifying LZTS2 expression in LIHC and STAD, a gene-drug interaction network for LZTS2 was built using Cytoscape and examined in the CTD database. We identified 10 chemicals, previously reported in various studies, that interact with LZTS2, including 1,2-Dimethylhydrazine, Abrine, Acetamide, Acetaminophen, Atrazine, Benzo(a)pyrene, Bisphenol A, Butyraldehyde, Cadmium, and Chlordecone (Table 2). Among these chemicals, 1,2-Dimethylhydrazine, Benzo(a)pyrene, Bisphenol A, and Atrazine have been reported to potentially upregulate LZTS2 expression, while Acetaminophen, Cadmium, and Chlordecone may have inhibitory effects on LZTS2 expression (Table 2). For chemicals such as Abrine and Acetamide, further studies are required to fully elucidate their effects on LZTS2 expression. These findings lay the groundwork for investigating therapeutic strategies targeting LZTS2 in LIHC and STAD. Modulating LZTS2 expression appears to be a promising avenue for treatment.
Experimental in vitro validation of LZTS2 expression and methylation status
In this investigation, we validated the expression and methylation status of the LZTS2 gene in two liver cancer cell lines (97H and LM3), one normal liver cell line (MIHA), and two stomach adenocarcinoma (STAD) cell lines (AGS and MKN7), as well as a normal gastric cell line (GES-1). The gene expression was assessed through qRT-PCR, utilizing ΔCt values. The relative expression of LZTS2 was computed using the formula 2^ (-ΔCt) × 10^3, with the average relative expression determined by 2^ (-ΔΔCt). For statistical analysis of qRT-PCR data, one-way ANOVA followed by Tukey’s post-hoc test was applied to compare differences among multiple groups. Homogeneity of variances was confirmed using Levene’s test (p > 0.05). This method allowed for a precise quantification of gene expression levels across different cell lines, providing valuable insights into the role of LZTS2 in these specific cancer contexts33. Methylation status was assessed using qMSP, where methylation levels were calculated using the formula Cmeth = 100/[1 + 2^(CtCG—CtTG)] × 100%, where CtCG represents the Ct value for the methylated primers and CtTG represents the Ct value for the unmethylated primers, as described by Zafari et al. (2018) using SYBR Green for detection34. For methylation rate comparisons between cancer and normal cell lines, Welch’s t-test was employed due to unequal variances identified by Levene’s test (p < 0.05).
As shown in Fig. 6A, LZTS2 expression was detected in all cell lines, with significantly higher expression levels in the liver cancer (97H, LM3) and STAD (AGS, MKN7) cell lines compared to the normal MIHA and GES-1 cell lines. Notably, the highest expression was observed in the STAD cell lines, particularly MKN7. For methylation analysis, Fig. 6B shows that the methylation rate of LZTS2 was significantly higher in the normal MIHA and GES-1 cell lines compared to the liver cancer (97H, LM3) and STAD (AGS, MKN7) cell lines. This indicates that LZTS2 is less methylated in the cancer cell lines, suggesting a potential epigenetic regulation of LZTS2 expression in liver and stomach cancer. These findings offer strong evidence that LZTS2 expression is increased in liver and stomach cancer cell lines, suggesting differential methylation between normal and cancerous tissues plays a role in modulating LZTS2 during tumor development.
Gene expression and methylation analysis of the LZTS2 gene in various cell lines. (A) qRT-PCR analysis of LZTS2 gene expression across a panel of cell lines: STAD: GES-1 (normal gastric cells), AGS, MKN7; LIHC: MIHA (normal liver cells), 97H, and LM3. Gene expression levels were normalized to the internal reference gene, and results are presented as mean ± standard deviation (SD) from three independent biological replicates. Statistical significance was assessed by one-way ANOVA, with p < 0.05 considered statistically significant. (B) Methylation conversion rates of the LZTS2 gene in the same set of cell lines, measured using bisulfite sequencing. Methylation levels are expressed as the percentage of methylated cytosines. Data are presented as mean ± SD, with significant differences between groups indicated by p < 0.05.
Discussion
Cancer results from complex genomic and epigenomic alterations in normal cells, which lead to abnormal gene expression35. Despite advancements in cancer research, cancer remains a leading cause of death globally. According to estimates, there were 19.3 million new cases and 10.0 million deaths from cancer worldwide in 202036. The persistent high mortality emphasizes the urgent need for deeper exploration into cancer biology. Identifying sensitive diagnostic and prognostic biomarkers is crucial for enhancing disease management and improving patient outcomes.
LZTS2 has been increasingly recognized for its role in the regulation of cancer progression. In prostate cancer, LZTS2 is located on chromosome 10q24.3, a region that frequently undergoes deletion in various cancers, including prostate cancer37. Intriguingly, the role of LZTS2 appears to be context-dependent across cancer types, influenced by tissue-specific genetic and epigenetic landscapes. For instance, in laryngeal squamous cell carcinoma (LSCC), hypermethylation of the LZTS2 promoter is strongly associated with lymph node metastasis and advanced disease stages, suggesting an epigenetic silencing mechanism that contrasts with its observed overexpression in LIHC and STAD9. This paradoxical duality underscores LZTS2’s multifaceted regulatory roles—acting as a tumor suppressor in some malignancies while potentially promoting oncogenic processes in others. For example, in clear cell renal cell carcinoma (ccRCC), LZTS2 acts as a tumor suppressor by inhibiting the Wnt/β-catenin pathway, thereby reducing tumor growth and invasion. Conversely, in hepatocellular carcinoma (LIHC), its overexpression correlates with enhanced metastatic potential and poor prognosis, as demonstrated in our study38. In colorectal cancer (CRC), LZTS2 has been reported to inhibit epithelial-mesenchymal transition (EMT) by suppressing the PI3 K/AKT pathway, thereby reducing metastatic potential7. Conversely, in glioblastoma (GBM), LZTS2 overexpression correlates with enhanced cell proliferation and resistance to temozolomide, possibly via dysregulation of the MAPK/ERK signaling axis39. Such divergent behaviors highlight the necessity of exploring tissue-specific molecular interactomes and microenvironmental cues that modulate LZTS2’s functional outcomes. For instance, in prostate cancer, LZTS2 interacts with PTEN to suppress β-catenin signaling, a mechanism distinct from its oncogenic role in LIHC40. Furthermore, in breast cancer, LZTS2 expression exhibits subtype-specific variability, with higher levels observed in triple-negative breast cancer (TNBC) compared to hormone receptor-positive subtypes, implicating its role in aggressive tumor biology11. These findings collectively emphasize that LZTS2’s contribution to carcinogenesis is not unidirectional but rather shaped by the genomic and epigenetic context of the tumor milieu. Additionally, LZTS2 is associated with epigenetic changes in other cancers. The LZTS2 promoter was found to be hypermethylated in laryngeal squamous cell carcinoma (LSCC). This change is strongly associated with lymph node metastases, advanced disease stages, and a poor prognosis11. Moreover, this methylation event has been proposed as a potential biomarker for both diagnosis and prognosis in LSCC, with high sensitivity and specificity observed in ROC analysis11. Interestingly, the methylation of LZTS2 appears to be notably higher in smokers, hinting at a potential interplay between lifestyle factors and the epigenetic regulation of this gene in cancer progression11. Thus, LZTS2 acts as a critical tumor suppressor in various malignancies, and its dysregulation, through both genetic deletion and epigenetic silencing, may significantly contribute to cancer initiation and progression.
We performed a thorough correlation study to look at the association between LZTS2 expression levels and its promoter methylation, genetic alterations, and CNVs in patients with LIHC and STAD in order to learn more about the possible causes behind LZTS2 overexpression. The findings showed that promoter methylation levels in both LIHC and STAD were significantly correlated negatively with LZTS2 expression. Notably, our genome-wide methylation analysis using TCGA array data identified hypomethylation at non-promoter regulatory elements (e.g., gene body CpG islands), which may synergize with promoter hypomethylation to amplify LZTS2 transcription. Although WGBS data were unavailable, the 450 K array provides sufficient resolution to detect critical methylation changes associated with oncogenic activation. Future studies employing WGBS could further elucidate the spatial and functional relationships between these regions. This discovery highlights the critical role of promoter hypomethylation in upregulating LZTS2 expression in these cancers. Emerging evidence suggests that hypomethylation may synergize with oncogenic signaling pathways to amplify LZTS2’s effects. For example, the Wnt/β-catenin pathway, frequently activated in LIHC, can recruit TET enzymes to demethylate specific gene promoters, including LZTS2, thereby enhancing its transcription41. Similarly, inflammatory cytokines like IL-6, prevalent in STAD microenvironments, have been shown to downregulate DNMT1 activity, perpetuating hypomethylation and sustained LZTS2 expression42. These pathways may explain the tissue-specific variability in LZTS2 regulation observed across cancers. Moreover, genetic analysis unveiled that truncated mutations and substantial amplifications of the LZTS2 gene were observed in a minority of STAD and LIHC patients, respectively. These findings indicate that, although genetic mutations and CNVs do contribute to LZTS2 expression regulation, their influence seems to be minimal in these cancer types.
Mutations in the LZTS2 gene have been shown to alter the amino acid composition at multiple sites of the encoded protein. Previous studies have identified several biomarkers associated with STAD expression, including HER2, PD-L1, MSI, VEGFR2, FGFR2, and CLDN18.243,44. However, despite their potential clinical utility in the diagnosis and treatment of gastric cancer, these biomarkers have not yet been universally adopted as standard markers with a significant correlation to the clinicopathological features of STAD patients. Significantly, the correlations between these markers and diverse clinicopathological subtypes demand additional exploration. In our research, we noted a notable increase in LZTS2 expression across various clinicopathological factors in STAD patients, encompassing tumor stage, ethnicity, gender, and age. Moreover, we have demonstrated that heightened LZTS2 expression is significantly linked to shortened OS and RFS periods, as well as increased metastasis in STAD patients.
Several expression-based biomarkers are currently being explored to differentiate LIHC patients from healthy individuals. Notably, molecules such as matrix metalloproteinases (MMPs), cytotoxic T-lymphocyte-associated protein 4 (CTLA-4), epidermal growth factor receptor (EGFR), and transforming growth factor-beta (TGF-β) have been found to be significantly overexpressed in LIHC tissues compared to normal counterparts, presenting potential targets for anti-LIHC therapy45,46,47,48. Moreover, the p53 protein has solidly emerged as an attractive therapeutic target. This is due to its capability of restoring cell cycle regulation and triggering apoptosis in cancer cells. By activating DNA damage response pathways, it effectively inhibits tumor growth and progression49,50. However, to date, none of these biomarkers have been widely adopted across LIHC patients with varying clinicopathological features. Contrary to previous findings, our current research reveals a notable increase in LZTS2 expression among LIHC patients, irrespective of clinical and pathological factors including cancer stage, ethnicity, gender, and age, when compared to normal controls. Furthermore, we observed a strong correlation between high LZTS2 expression levels and poorer overall survival outcomes as well as an increased incidence of metastasis in LIHC patients.
Multi-gene diagnostic and prognostic systems have attracted a lot of attention lately, despite the fact that single-gene indicators have proven successful in forecasting the diagnostic and prognostic results of cancer patients51. However, patients with LIHC and STAD who have several mutations across different oncogenic genes still lack a viable treatment strategy52,53. In this study, we utilized muTarget to identify mutations that may affect the expression of LZTS2. Our analysis pinpointed the top four mutant genes associated with LZTS2 expression in both LIHC and STAD. Specifically, in LIHC, the mutated genes include HERC1, IL6ST, TDRD5, and TSC2, whereas in STAD, they consist of CCDC85A, COL4A2, FSD2, and PCSK7. By correlating these mutated genes with LZTS2 expression, our goal is to uncover potential multi-gene therapeutic strategies for both LIHC and STAD.
Cancer treatment remains one of the most challenging endeavors in the medical field. Among various therapeutic strategies, cancer immunotherapy has emerged as one of the most promising approaches, demonstrating encouraging outcomes in anticancer research54. Immunotherapy primarily engages the utilization of various agents, including PD-1/PD-L1 inhibitors and Interleukin-2 (IL-2). These elements are pivotal in achieving therapeutic efficacy through modulation of the immune system55,56. Nevertheless, the advancement of immunotherapy in clinical settings is frequently impeded by challenges including drug resistance and tumor immune evasion, factors that restrict its capability to attain the most favorable therapeutic outcomes57. Given these challenges, a deeper understanding of the tumor immune microenvironment (TIME) in LIHC and STAD may provide valuable insights into the mechanisms underlying immune responses. The acquisition of this knowledge holds the potential to expedite the creation of more potent immunotherapies, thereby contributing to enhanced clinical outcomes for patients.
The tumor microenvironment (TME), encompassing stromal cells, immune cells, and tumor cellularity, holds a critical role in dictating the response to immunotherapy and impacting tumor development58,59. In this study, LZTS2 expression showed no significant correlation with tumor purity in either LIHC or STAD samples, indicating its expression is not distinctly limited to stromal or epithelial cells. Remarkably, the expression of LZTS2 in liver cancer histological grades demonstrates strong positive associations with the infiltration levels of CD4 + T cells, B cells, macrophages, and neutrophils. This suggests that LZTS2 may facilitate tumor progression by stimulating immune cells within the TME. Conversely, in STAD, LZTS2 expression shows weaker or negligible correlations with most immune cells. However, moderate positive correlations were observed specifically for CD4 + T cells and macrophages. These observations indicate that LZTS2 plays a differential role in modulating immune cell dynamics between LIHC and STAD. Our multivariable Cox regression analysis (Table 3) demonstrated that LZTS2 remained an independent prognostic factor in LIHC (HR = 1.254, p = 0.019) after adjusting for tumor stage and immune infiltration, whereas its prognostic value in STAD was context-dependent (HR = 1.017, p = 0.878). This divergence may stem from LZTS2’s distinct immunomodulatory roles: in LIHC, its strong correlation with CD4 + T cells and macrophages implies a potential role in fostering an immunosuppressive microenvironment, whereas in STAD, weaker immune associations suggest alternative mechanisms of tumor progression. Collectively, these insights enhance our understanding of the immunological landscape in LIHC and STAD, paving potential routes for the development of personalized immunotherapeutic approaches.
In this study, we constructed and visualized the interaction network of genes associated with LZTS2, identifying several enriched pathways relevant to cancer progression. Through KEGG pathway enrichment analysis, it was revealed that genes associated with LZTS2 predominantly participate in processes such as “microtubule severing”, "regulation of postsynapse organization" and "negative regulation of cytoskeleton organization". These findings are consistent with previous studies that underscore the critical roles of these pathways in maintaining cytoskeletal dynamics, synaptic signaling fidelity, mitotic progression, and cellular architecture remodeling, which collectively drive tumor cell proliferation, metastatic dissemination, and invasive protrusion formation. Our analyses confirmed strong links between LZTS2 expression and genes within these pathways, reinforcing our enrichment findings. To explore potential drugs that could modulate LZTS2 expression, we applied predictions from the CTD. We pinpointed several compounds—1,2-Dimethylhydrazine, Bisphenol A, and Acetaminophen—as influencers of LZTS2 expression. This suggests they might be effective therapeutic options. Yet, their efficacy and specificity in targeting LZTS2 need further validation through comprehensive preclinical and clinical trials. In summary, our discoveries shed light on LZTS2’s molecular mechanisms and suggest promising interventions for LIHC and STAD treatments.
While this study benefits from the use of large cancer cohorts, several limitations must be acknowledged. First, treatment history (e.g., chemotherapy, immunotherapy) was not included in our analysis due to the lack of granular clinical data in public datasets like TCGA. This omission may confound survival outcomes, as therapeutic interventions could independently influence patient prognosis. Future studies incorporating treatment metadata are warranted to disentangle these effects. Second, our findings predominantly relied on bioinformatics analysis. Although this approach offers valuable insights, it necessitates further validation. To enhance LZTS2’s clinical significance, it would be advantageous to scrutinize additional in-house clinical samples from LIHC and STAD patients. Furthermore, a deeper comprehension of LZTS2’s potential as both a diagnostic and prognostic indicator in these cancers could pave the way for the creation of innovative diagnostic tools, thus enabling earlier cancer detection.
Conclusion
In this study, we identified LZTS2 as a potential biomarker in LIHC and STAD, demonstrating its possible role in cancer prognosis and diagnostics. Bioinformatics studies reveal that LZTS2 is significantly linked to pivotal molecular pathways critical for tumor progression, cell cycle regulation, and apoptosis. These insights offer substantial knowledge, yet their clinical significance must be corroborated with in-house clinical data from relevant cancer types. Moreover, investigating LZTS2’s wider diagnostic and prognostic implications could foster the creation of advanced tools for early cancer detection. Future research should concentrate on LZTS2’s functional analysis and its interactions with cellular components to elucidate its mechanistic role in cancer development and to establish its potential as a therapeutic target.
Data availability
Data is provided within the manuscript or supplementary information files.
Change history
04 October 2025
The original online version of this Article was revised: In the original version of this Article Xiaoyan Chen was incorrectly only affiliated with ‘3Department of Radiation Oncology, Cancer Center, Zhejiang Provincial People’s Hospital (Affiliated People’s Hospital), Hangzhou Medical College, Hangzhou, Zhejiang, China.’. The correct affiliation’s are listed here:
1. Department of Medical Oncology, Sir Run Run Shaw Hospital, School of Medicine, Zhejiang University, Hangzhou, Zhejiang, China
3. Department of Radiation Oncology, Cancer Center, Zhejiang Provincial People’s Hospital (Affiliated People’s Hospital), Hangzhou Medical College, Hangzhou, Zhejiang, China.
References
Sung, H. et al. Global cancer statistics 2020: GLOBOCAN Estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 71(3), 209–249 (2021).
Siegel, R. L., Miller, K. D., Fuchs, H. E. & Jemal, A. Cancer statistics, 2021. CA Cancer J Clin. 71(1), 7–33 (2021).
Ferlay, J. et al. Cancer statistics for the year 2020: An overview. Int J Cancer. 149(4), 778–789 (2021).
Tufail, M. & Wu, C. Cancer statistics in Pakistan from 1994 to 2021: Data from cancer registry. JCO Clin. Cancer Inform. 7, e2200142 (2023).
Balogh, J. et al. Hepatocellular carcinoma: A review Journal of Hepatocellular. Carcinoma. 3(41), 53 (2016).
Network NCC. Gastric Cancer. NCCN clinical practice guidelines in oncology ,2023.
Kim, J. M. et al. Effect of the modulation of leucine zipper tumor suppressor 2 expression on proliferation of various cancer cells functions as a tumor suppressor. Mol. Cell Biochem. 346(1–2), 125–136 (2011).
Johnson, D. T. et al. Deletion of leucine zipper tumor suppressor 2 (Lzts2) increases susceptibility to tumor development. J. Biol. Chem. 288(6), 3727–3738 (2013).
Xu, L. et al. Disrupting CCDC137-mediated LZTS2 and β-TrCP interaction in the nucleus inhibits hepatocellular carcinoma development via β-catenin and AKT. Cell Death Differ. 32(1), 134–148 (2025).
Usui, G. et al. Integrated environmental, lifestyle, and epigenetic risk prediction of primary gastric neoplasia using the longitudinally monitored cohorts. EBioMedicine 98, 104844 (2023).
Shen, Z. et al. LZTS2 promoter hypermethylation: A potential biomarker for the diagnosis and prognosis of laryngeal squamous cell carcinoma. World J. Surg. Oncol. 16(1), 42 (2018).
Chandrashekar, D. S. et al. UALCAN: A portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia 19(8), 649–658 (2017).
Koch, A., De Meyer, T., Jeschke, J. & Van Criekinge, W. MEXPRESS: Visualizing expression, DNA methylation and clinical TCGA data. BMC Genomics 16(1), 636 (2015).
Park, S. J., Yoon, B. H., Kim, S. K. & Kim, S. Y. GENT2: An updated gene expression database for normal and tumor tissues. BMC Med. Genomics. 12(Suppl 5), 101 (2019).
Li, T. et al. TIMER: A web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res. 77(21), e108–e110 (2017).
Tang, Z. et al. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 45(W1), W98–W102 (2017).
Bartha, Áron, and Balázs Győrffy. TNMplot. com: a web tool for the comparison of gene expression in normal, tumor and metastatic tissues. International journal of molecular sciences 22.5 (2021): 2622.
Cerami, E. et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2(5), 401–404 (2012).
Mering, C. V. et al. STRING: A database of predicted functional associations between proteins. Nucleic Acids Res. 31(1), 258–261 (2003).
Shannon, P. et al. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 13(11), 2498–2504 (2003).
Huang, D. W. et al. The DAVID gene functional classification tool: A novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol. 8, 1–16 (2007).
Kanehisa, M., Furumichi, M., Sato, Y., Matsuura, Y. & Ishiguro-Watanabe, M. KEGG: Biological systems database as a model of the real world. Nucleic Acids Res. 53(D1), D672–D677 (2025).
Nagy, Á. & Győrffy, B. muTarget: A platform linking gene expression changes and mutation status in solid tumors. Int. J. Cancer. 148(2), 502–511 (2021).
Li, T. et al. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 48(W1), W509–W514 (2020).
Mattingly, C. J., Colby, G. T., Forrest, J. N. & Boyer, J. L. The comparative toxicogenomics database (CTD). Environ. Health Perspect. 111(6), 793–795 (2003).
Fackler, M. J. et al. Quantitative multiplex methylation-specific PCR analysis doubles detection of tumor cells in breast ductal fluid. Clin. Cancer Res. 12(11 Pt 1), 3306–3310 (2006).
Xu, R. H. et al. Circulating tumour DNA methylation markers for diagnosis and prognosis of hepatocellular carcinoma. Nat. Mater. 16(11), 1155–1161 (2017).
Jones, P. A. & Laird, P. W. Cancer epigenetics comes of age. Nat. Genet. 21(2), 163–167 (1999).
Baylin, S. B. & Jones, P. A. A decade of exploring the cancer epigenome - biological and translational implications. Nat. Rev. Cancer. 11(10), 726–734 (2011).
Herman, J. G. & Baylin, S. B. Gene silencing in cancer in association with promoter hypermethylation. N. Engl. J. Med. 349(21), 2042–2054 (2003).
Etoh, T. et al. Increased DNA methyltransferase 1 (DNMT1) protein expression correlates significantly with poorer tumor differentiation and frequent DNA hypermethylation of multiple CpG islands in gastric cancers. Am. J. Pathol. 164(2), 689–699 (2004).
Nalio Ramos, R. et al. Tissue-resident FOLR2(+) macrophages associate with CD8(+) T cell infiltration in human breast cancer. Cell 185(7), 1189-1207.e25 (2022).
Gochhait S, Bukhari SI, Bamezai R. mRNA Quantitation Using Real Time PCR. 2007 : 53–72.
Zafari, P. et al. Analysis of helios gene expression and Foxp3 TSDR methylation in the newly diagnosed rheumatoid arthritis patients. Immunol. Invest. 47(6), 632–642 (2018).
Carter, S. L., Eklund, A. C., Kohane, I. S., Harris, L. N. & Szallasi, Z. A signature of chromosomal instability inferred from gene expression profiles predicts clinical outcome in multiple human cancers. Nat. Genet. 38(9), 1043–1048 (2006).
Siegel, R. L., Miller, K. D. & Jemal, A. Cancer statistics, 2020. CA Cancer J. Clin. 70(1), 7–30 (2020).
Yu, E. J. et al. LZTS2 and PTEN collaboratively regulate ß-catenin in prostatic tumorigenesis. PLoS ONE 12(3), e0174357 (2017).
Huang, J. L., Liao, Y., Qiu, M. X., Li, J. & An, Y. Long non-coding RNA CCAT2 promotes cell proliferation and invasion through regulating Wnt/β-catenin signaling pathway in clear cell renal cell carcinoma. Tumour Biol. 39(7), 1010428317711314 (2017).
Cui, Q. Z. et al. Leucine zipper tumor suppressor 2 inhibits cell proliferation and regulates Lef/Tcf-dependent transcription through Akt/GSK3β signaling pathway in lung cancer. J. Histochem. Cytochem. 61(9), 659–670 (2013).
Thyssen, G. et al. LZTS2 is a novel beta-catenin-interacting protein and regulates the nuclear export of beta-catenin. Mol. Cell Biol. 26(23), 8857–8867 (2006).
Kohli, R. M. & Zhang, Y. TET enzymes, TDG and the dynamics of DNA demethylation. Nature 502(7472), 472–479 (2013).
Nieto, M. A., Huang, R. Y., Jackson, R. A. & Thiery, J. P. EMT: 2016. Cell 166(1), 21–45 (2016).
Sato, Y. et al. Novel biomarkers of gastric cancer: Current research and future perspectives. J. Clin. Med. 12(14), 4646 (2023).
Zheng, G. L., Zhang, G. J., Zhao, Y. & Zheng, Z. C. Screening protein prognostic biomarkers for stomach adenocarcinoma based on the cancer proteome atlas. Front. Oncol. 12, 901182 (2022).
Nomden, M., Beljaars, L., Verkade, H. J., Hulscher, J. & Olinga, P. Current concepts of biliary atresia and matrix metalloproteinase-7: A review of literature. Front. Med. (Lausanne). 7, 617261 (2020).
Shabbir, M. et al. Association of CTLA-4 and IL-4 polymorphisms in viral induced liver cancer. BMC Cancer 22(1), 518 (2022).
Jin, H. et al. EGFR activation limits the response of liver cancer to lenvatinib. Nature 595(7869), 730–734 (2021).
Gough, N. R., Xiang, X. & Mishra, L. TGF-β signaling in liver, pancreas, and gastrointestinal diseases and cancer. Gastroenterology 161(2), 434-452.e15 (2021).
Zhang, S. et al. Advanced strategies for therapeutic targeting of wild-type and mutant p53 in cancer. Biomolecules 12(4), 548 (2022).
Hou, C. Y. et al. Aristolochic acids-hijacked p53 promotes liver cancer cell growth by inhibiting ferroptosis. Acta Pharmacol. Sin. 46(1), 208–221 (2024).
Győrffy, B. et al. Multigene prognostic tests in breast cancer: Past, present, future. Breast Cancer Res. 17(1), 11 (2015).
Li, J. et al. Insights of fibroblast growth factor receptor 3 aberrations in pan-cancer and their roles in potential clinical treatment. Aging (Albany NY). 13(12), 16541–16566 (2021).
Yu, X. et al. ELL associated factor 2 is a potential diagnostic and prognostic indicator: evidence from the in silico and in vitro experiments. Am. J. Cancer Res. 13(6), 2572–2587 (2023).
Shanmugam, R. et al. SRSF9 selectively represses ADAR2-mediated editing of brain-specific sites in primates. Nucleic Acids Res. 46(14), 7379–7395 (2018).
Yi, M. et al. Biomarkers for predicting efficacy of PD-1/PD-L1 inhibitors. Mol. Cancer. 17(1), 129 (2018).
Mazur, G. & Frydecka, I. Interleukin 2 (IL-2) and its receptor (IL-2R) in healthy individuals and with various disease states. Acta Haematol. Pol. 24(4), 307–313 (1993).
Restifo, N. P., Smyth, M. J. & Snyder, A. Acquired resistance to immunotherapy and future challenges. Nat. Rev. Cancer. 16(2), 121–126 (2016).
Samstein, R. M. et al. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat. Genet. 51(2), 202–206 (2019).
Roth, A. D. et al. Integrated analysis of molecular and clinical prognostic factors in stage II/III colon cancer. J. Natl. Cancer Inst. 104(21), 1635–1646 (2012).
Acknowledgements
We would like to express our sincere gratitude to the developers and maintainers of the bioinformatics platforms used in this study. Their invaluable resources and analytical tools were essential in conducting the bioinformatics analyses that formed the basis of our findings.
Funding
This work was supported by grants from the National Natural Science Foundation of China (82203377 to Yanwei Lu; 82473238 to Haibo Zhang), Zhejiang natural science foundation of China (Grant number: LQ22H160036 to Yanwei Lu; LY24H160022 to Haibo Zhang). Zhejiang Health Science and Technology Project (2025KY031 to Haibo Zhang).
Author information
Authors and Affiliations
Contributions
XC, XW: Writing Original Draft. XW, LH, KX, HX, RL, DN, YS, HS: Investigation. YL, HZ, XC: Supervision. All authors listed have made a substantial, direct, and intellectual contribution to the work, and approved it for publication.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Wang, X., Lu, Y., Liu, R. et al. LZTS2 methylation as a potential diagnostic and prognostic marker in LIHC and STAD: Evidence from bioinformatics and in vitro analyses. Sci Rep 15, 17873 (2025). https://doi.org/10.1038/s41598-025-03153-x
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-03153-x