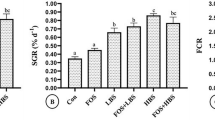
Abstract
Probiotics offer significant health advantages as they enter the digestive system via diet or water intake, playing a crucial role in enhancing immunity, growth, gastrointestinal microbiota, and feed attribute. The main objective of study was to focus on the impact of probiotic functional feed (PFF) on Nile tilapia (Oreochromis niloticus) exposed to challenges from Vibrio harveyi and Vibrio parahaemolyticus. The investigation aims to analyze the genes linked to immunity, hemato-biochemical indices, and the immunological response in tilapia. PFF is a vital component of fish feed production, providing suitable nutrition for various ages and stages to promote healthy growth. The study comprises four treatments: CPF-1 (control group, diet included solely of basal fish feed), the 20% of PFF2 (Rossellomorea marisflavi spp. (DAS-SCF02–1 × 104), PFF3 (Agrococcus spp. (RKDAS1-1 × 106), and PFF4- (DAS-SCF02–1 × 104 + RKDAS1 (1 × 107). A total of 150 Nile tilapia juveniles, weighing 2.56 ± 1.26 g, were administered PFF in triplicates. Significant improvements were observed in hematological indices, encompassing white blood cells (WBC), hemoglobin (Hb), red blood cells (RBC), hematocrit (Htc), and blood performance (BP) in probiotic-treated groups compared to control. Biochemical analysis revealed lower levels of aspartate aminotransferase (AST) and alanine aminotransferase (ALT) in fish fed with probiotics, accompanied by increased total protein, albumin, and globulin levels. Furthermore, probiotic-fed fish exhibited heightened blood glucose, total cholesterol, and triglyceride levels. Immunological assessments demonstrated increased lysozyme activity, intracellular superoxide anion production, reactive nitrogen species synthesis, and myeloperoxidase activity in probiotic-fed groups. Immune gene expression analysis revealed up-regulation of stress response, cytokine signaling, and immune defense-related genes (HSP70, IL-1β, I C3, IFN-α, IFN-γ, GF1, GH, IL-1, and Lyz). In a Vibrio challenge study, probiotic-fed fish exhibited improved survival rates, underscoring the protective effects of probiotics against bacterial infections. Overall, this research underscores the multifaceted benefits of probiotic supplementation in enhancing the health and immunity of tilapia.
Similar content being viewed by others
Introduction
The fish farming sector is experiencing steady growth in response to the world’s increasing population. The adoption of intensive aquaculture strategies aims to meet the rising demand, with the Nile tilapia (Oreochromis niloticus), currently ranking as the third-highest-farmed fish globally in terms of volume (4.5 Mmt) due to continuous advancements in farming practices1. Despite the success of tilapia farming, challenges have emerged, particularly the excessive feed requirements2,3,4. Consequently, there is a growing focus on researching feed formulations and feeding methods to enhance fish gastrointestinal and health while continuing a balanced inhabitation of stomach bacteria. These approaches aim to boosting productivity in the sector of fish farming.
As aquaculture intensifies, the facility’s capacity to address various illnesses and health issues becomes crucial. Ongoing investigations into fish immune responses and physical development seek to develop sustainable and environmentally friendly methods to ensure the well-being of the fish5,6,7,8. Notably, research on immune-stimulating probiotics for fish healthcare shows promising potential in boosting immunity and treating diseases. These immunopotentiators can enhance fish growth, immunological response, intestinal health, and fillet quality5,8,9. Probiotics, active microbes, can thrive and proliferate in the animal’s stomach, contributing to its overall well-being10,11,12,13. They promote fish development by improving gastrointestinal architecture and microbial diversity, influencing the gastrointestinal ecology through the secretion of extracellular digestive enzymes like protease and amylase and the production of compounds such as short-chain fatty acids14.
Researchers have proven the efficacy of functional dietary additives, while also highlighting the detrimental effects of excessive excreta levels on fish and shrimp. In laboratory-cultured common carp (Cyprinus carpio) juveniles, a probiotic functional feed containing Pediococcus acidilactici and formic acid influenced growth, blood biochemical parameters, immune gene upregulation, and survival15. Rossellomorea marisflavi, an aerobic Gram-positive organism, exhibits non-mobile filaments, non-swollen spores, and terminally oval endospores16. When cultured on TSA (tryptic soy agar, M1968-Himedia, Mumbai, India) plates complemented with 2.5% NaCl (w/v) at pH 7.0 and 37 °C for 24 h, this strain demonstrated robust growth conditions, thriving between pH 6.0 and 9.0, at temperatures ranging from 15 to 45 °C, and tolerating NaCl concentrations from 0 to 25% (w/v)17.
The genus Rossellomorea marisflavi, proposed by18, is closely related to the Lactobacillaceae genus of lactic acid-producing bacteria and belongs to the Bacillaceae family within the phylum Firmicutes. Notably, Rossellomorea was part of a bacterial consortium that enhanced the potential of the halophyte Arthrocaulon macrostachyum for phytoremediation applications by improving seed development, growth, and heavy metal preservation in the root system, particularly in heavy metal-polluted soils17,19,20,21. Agrococcus lahaulensis, an aerobic, non-acid-fast, Gram-positive bacterium, forms distinctly margined, opaque, round, lemon-colored colonies on TSA media. The colonies range in size from 0.7 to 3.0 mm, exhibit resilience to up to 7.0% NaCl, and thrive optimally at temperatures ranging from 25 to 37 °C. Growth occurs within the pH range of 6.0 to 10.0, with the ideal pH being 8.022. Agrococcus spp. SD01-s17, a bioactive variant of Agrococcus, possesses diverse pharmacological and biocidal activities. It functions as an antioxidant, demonstrates anticancer, antibiotic, and antifungal properties, and has the potential to inhibit the growth of harmful bacteria, as reported by23. The antibacterial properties of Agrococcus lahaulensis against S. aureus and B. cereus24. Although the combination of Rossellomorea marisflavi and Agrococcus lahaulensis has not been previously explored for improving fish health, this study aims to investigate their potential in enhancing immunity and promoting fish health. The primary objective is to evaluate the impact of Probiotic Functional Feed (PFF) on Nile tilapia challenged by Vibrio harveyi and Vibrio parahaemolyticus. Specifically, the study aims to analyze the genes linked to immunity, assess hemato-biochemical indices, and evaluate the immunological responses in tilapia.
Materials and methods
Preparation of nutrient-enriched probiotic feed
The probiotic functional diet, designed to provide essential nutrients for juvenile tilapia, utilized feed materials sourced from CP Aquaculture (India) Pvt Ltd at 104, GNT Road, Nallur, Red Hills, Chennai, Tamil Nadu, India. The materials underwent crushing and sieving before meticulous blending. Dried ingredients were thoroughly mixed before the addition of liquid components. Employing a feed processing machine, the feed was produced, cut into appropriate sizes, and subjected to a two-hour oven-drying at 60 °C. After cooling, the feed was sealed in plastic bags for storage until required (Table 1).
Probiotics culture preparation
The probiotics were derived from two novel strains of bacteria: Agrococcus spp. RKDAS1 and Rossellomorea marisflavi spp. DAS-SCF02, which were isolated from Indian Snakehead fish (Channa straiata) in freshwater lakes and the Tamiraparani River at Tirunelveli, and sludge from the Muttukadu boat house in Kanchipuram, Tamil Nadu. Isolates of Agrococcus spp. RKDAS1 and Rossellomorea marisflavi spp. DAS-SCF02 were cultivated for a full day in NB (nutrient broth) to produce the probiotic cultures. The probiotic cells were centrifuged, cleaned, and suspended in 0.85% saline solution. Using a spectrophotometer, their concentrations were then adjusted to an absorbance OD of 600. Then, the washed probiotic strain suspensions were added to the basal feeds in a ratio of 20: 100 (w/w) between probiotic strain suspensions and basal supplementations for sustaining probiotics viability25,26.
Experimental grouping and preparation of probiotic functional feeds enriched
The study involved the production of four distinct feed types: PFF2: Containing Agrococcus spp. RKDAS1 (single probiotic treatment); PFF3: Containing Rossellomorea marisflavi spp. DAS-SCF02 (single probiotic treatment); PFF4: Containing a mixture of both Agrococcus spp. RKDAS1 and Rossellomorea marisflavi spp. DAS-SCF02 (dual probiotic treatment); CPF1: Control feed without any probiotic additives. Each experimental group consisted of three replicates, with each tank initially stocked with 10 tilapia fish. Blood samples were collected from 5 fish per tank at three time points, totalling 15 fish sampled over the course of the experiment. The remaining 5 fish per tank were used for the challenge phase of the experiment.
The base feed underwent autoclaving, followed by blending with the prepared probiotic culture. Serial dilution with a 0.85% sterile saline solution was performed to quantify the probiotic concentration in the feed. To enumerate viable probiotics, 100 µL samples from specified dilutions were cultured on Nutrient Agar (NA) or MRS Agar plates and incubated at 37 °C for 24 h. Colony counts were then performed to quantify the probiotic concentration in the feed. After preparation, all feeds were stored under refrigeration to maintain the probiotic dosage integrity27. After preparing the probiotic-enriched feed, it was air-dried to reduce excess moisture and stored in airtight containers under refrigeration (4 °C) to maintain probiotic viability. The feed preparation procedure was repeated weekly to ensure consistent probiotic dosage for the trial.
Media preparation and probiotic enumeration
For the enumeration of viable probiotics, NA and MRS Agar were used as growth media, depending on the specific bacterial strains. Nutrient Agar, a general-purpose medium that supports the growth of a wide range of bacteria, was used for counting Agrococcus spp. RKDAS1. The medium was prepared by dissolving 5 g of peptone, 3 g of yeast extract, 8 g of sodium chloride (NaCl), and 15 g of agar in 1 L of distilled water, which was then autoclaved at 121 °C for 15–20 min. Plates were poured after cooling to 50 °C and were incubated at 37 °C for 24 h.
For Rossellomorea marisflavi spp. DAS-SCF02, a lactic acid bacterium, MRS Agar was used to provide the optimal environment for its growth. MRS Agar was prepared by dissolving 10 g of peptone, 10 g of meat extract, 4 g of yeast extract, 20 g of glucose, and 5 g of sodium acetate in 1 L of distilled water, adjusting the pH to 6.2. The medium was autoclaved and poured into Petri dishes for colony enumeration. The plates for both media were incubated at 37 °C for 24 h. After incubation, colony counts were performed to determine the probiotic concentration in the feed. All media were sourced from reputable suppliers, including Sigma-Aldrich (Nutrient Agar, Cat. #70187; MRS Agar, Cat. #M6435) and Thermo Fisher Scientific (Nutrient Agar, Cat. #211829; MRS Agar, Cat. #R0079)27.
Experimental design
Fish collection
Juvenile Oreochromis niloticus was obtained from the Freshwater Aquaculture Sector of the University of Fisheries, Thanjavur, Tamil Nadu, weighing 2.56 ± 1.26 g. Before the experiment, the general health of each animal was evaluated based on its swimming ability in the tank, regular feeding patterns, the absence of blemishes, the presence of undamaged and vibrant scales, the absence of protruding eyes, and the integrity of fins without tears or ragged edges.
Feeding practices and maintenance protocols in a juvenile fish study
As part of Probiotic functional feed (PFF) trial, the fish were hand-fed a basal diet twice a day after being placed into ten twenty/100-L plastic tanks. They underwent a seven-day acclimatization phase to adapt to the experimental environment. The experiment divided into four groups, CPF-1 (control group, diet included solely of basal fish feed), the 20% of PFF2 (Rossellomorea marisflavi spp. (DAS-SCF02–1 × 104), PFF3 (Agrococcus spp. (RKDAS1-1 × 106), and PFF4- (DAS-SCF02–1 × 104 + RKDAS1 (1 × 107) was conducted in triplicate (three independent tanks per group). Each tank housed twenty juvenile fish in static water, with a routine refresh of approximately half the water volume through a flow-through aquarium system and clearing of accumulated excrement was conducted. The probiotics functional feeds (PFF2, PFF3, and PFF4) were administered for eight weeks, providing the animals with 3% of their daily feed allocation in three meals at 9:00, 15:00, and 21:0028.
Water quality indices monitoring
Water samples were collected biweekly throughout the experimental period. Temperature and dissolved oxygen (DO) were measured using a digital oxygen meter equipped with both oxygen and temperature sensors. pH was recorded using a calibrated pH meter. Salinity was determined using a handheld refractometer (Erma, Japan). Total ammonia nitrogen (TAN) levels in the pond water were analyzed using a commercial test kit (Advance Pharma, Thailand), while unionized ammonia (NH₃) concentrations were calculated following the method described by29,30.
Comprehensive assessment of hematological, biochemical, and environmental parameters in Tilapia during Long-Term PFF supplementation
At 20-days intervals, various hematological and biochemical indices were assayed, including total albumin, globulin, hemoglobin (Hb), albumin–globulin ratio, total serum protein, total leukocyte count (TLC), total erythrocyte count (TEC), acetylcholine esterase (AChE), alanine aminotransferase (ALT), lactate dehydrogenase (LDH), aspartate aminotransferase (AST), and total adenosine triphosphatase (ATPase).
Hemato-biochemical indices assessment
In order to collect plasma and perform enzyme assays, the fish were anesthetized with 100 mg/L of 3-aminobenzoic acid ethyl ester (MS-222, Sigma-Aldrich, St. Louis, MO, USA) on days 20, 40, and 60 of the experiment. The first portion of the blood sample was obtained using an anticoagulant 10% ethylene diamine tetra acetate (EDTA) to estimate the hematocrit (Htc) and white blood cells (WBCs). To harvest plasma samples, five fish from each treatment group including CPF-1 (Control), PFF2, PFF3, and PFF4 was conducted in triplicate (three independent tanks per group). Tissue homogenization was carried out in a cold sucrose buffer (0.25 M) using a Teflon-coated mechanical tissue homogenizer (Remi, India), the samples were centrifugation for 10 min at 5000 g at 4 °C followed by lysis. The enzyme activity was assessed using the supernatant obtained as the enzyme resource. All steps of the enzyme synthesis process were conducted in a refrigerated environment, with sample dilution performed where necessary.
For blood extraction, a sterilized 2-milliliter BD syringe cleaned with EDTA buffer (2.7%) was utilized. Blood was drawn into small glass vials containing 20 µl of EDTA buffer (2.7%) as an anticoagulant.
The TEC and TLC were determined according to the method outlined by31. A hemocytometer (Feinoptik, Germany) was employed for cell counting, with results reported as follows:
Here, Nr represents the total number of RBC measured in each square of the hemocytometer, and Nw indicates the total amount of WBC determined in each square. The factor acquired after accounting for the initial dilution factors is 10,000.
The blood Hb content was determined using Darbkins fluid and the Cyanmethemoglobin technique with a commercial kit (Qualigens, Mumbai, India).
Hematic indices
The blood collection procedure involved using a 2 mL BD sterilized syringe with 0.2 mL of anticoagulant, drawing blood from the caudal part of the fingerling fish, and transferring it to a fresh 1.5 mL Eppendorf centrifuge tube. The blood was permitted to clot for 45 min at room temperature with the tube tilted, followed by a 30-minute incubation at 4 °C. Subsequently, the tube was centrifuged for 10 min at 3000 g at 4 °C. Blood plasma was then collected in sterile Eppendorf centrifuge tubes and analyzed using Qualigens diagnostic kits in a semi-automated analyzer (AR 601, Qualigens, Mumbai, India) for various serum indices. The examined serum indices included LDL, HDL, triglyceride, albumin (using the bromocresol green binding method) (ALB), cholesterol (CHO), and total serum protein (using the biuret method employing biuret reagent and buffered dye reagent). Globulin - albumin ratios were calculated by dividing albumin concentration by globulin content, and globulin content by albumin concentration.
Immunological evaluates
Lysozyme activity
Lysozyme performance was evaluated with slight modifications following the procedure outlined by Parry et al., 196532. In brief, a 96-well microplate was filled in triplicate with 25 µL of plasma. Subsequently, each well received 0.2 mg mL− 1 of Micrococcus lysodeikticus solution in a buffered sodium phosphate solution (pH 6.2), totaling 175 µL. The reaction was monitored using a spectrophotometer set to measure reactions at 540 nm, with readings taken and recorded every minute for ten minutes. The blood activity of lysozyme was quantified as a 0.001 min− 1 reduction at 540 nm and reported in units of mL− 1.
Intracellular superoxide anion (SOA)
The assessment of intracellular superoxide anion (SOA) and respiratory burst activity involved conducting NBT (nitroblue tetrazolium) reduction reactions, adapted from the Secombes method (Secombes, 1990). In brief, microplates (96-well) were utilized to contain three batches of WBCs (6 × 10^6 cells). To each well, 25µL of NBT was added, and the plates were protected at room temperature for two hours. After the incubation, 150µL of absolute methanol was introduced to each well, and the residual fluid was discarded. Subsequently, the wells underwent repeated cleaning with a 70% methanol solution. Finally, 100µL of DMSO and 150µL of 2 M KOH were added to each well. Following thorough mixing, the absorbance value, indicative of the reaction, was measured at 540 nm using a UV-Vis spectrophotometer (201/220, Thermo Scientific).
Production of reactive nitrogen species (RNS) analysis
The Griess reagent technique, which is related to the change of nitrite from nitric oxide, was used to assess the nitric oxide (NO) in the plasma of tilapia33. Using a conventional curve representing the level of nitrate in the serum of tilapia, the amount of nitrite present was determined.
MPO (myeloperoxidase)
The MPO activity in plasma was assessed using34. An absorption change was considered to be one unit, and the activity was represented as U mg plasma− 1. The 90 µL of HBSS solution was used to dilute a 10 µL serum sample. This mixture was then mixed with a solution that contained hydrogen peroxide and 3, 3′, 5, 5′-tetramethyl benzidine dihydro chloride. They were stopped with 35 µL sulfuric acid after two minutes, and measurements were taken at 450 nm at 24 ◦C using a multiscan microplate reader.
Immune-related gene expression
Isolation of RNA and cDNA construction
For total RNA isolations, liver samples were dissected from three animals per treatment group. A concentration of 20 ng µL− 1 was targeted for liver RNA. Total RNA was isolated using a commercial Kit (RNA mini kit, Cat No. 74,104, Qiagen, Germany) according to the manufacturer’s guidelines. The purity of the RNA was assessed through gel electrophoresis (1.2% agarose gel) and NanoDrop spectrophotometry (NanoDrop 2000, Thermo Scientific). Subsequently, cDNA synthesis was performed using a cDNA RT Kit (Applied Biosystems, Cat# no. 4368813, USA), adhering to the manufacturer’s instructions.
Quantitative (qPCR) RT-PCR examination
Quantitative RT-PCR analysis was conducted using the Applied Biosystems 96 Real-time qPCR System, USA, to assess the expression of genes, including β-actin household genes, hsp70, IL-1β, IC3, TNF-α, IFN-γ, GF1, GH, IL-1, and Lyz. The DNA primer sequences used for amplification are presented in Table 2. The SYBR green technique with the SensiFast SYBR Lo-Rox kit (Bioline) was employed for RT-PCR. Amplification conditions consisted of 45 cycles: 10 s at 95 °C, 30 s at 63 °C, and 30 s at 72 °C. Subsequently, the 2−∆∆CT method was applied to determine the relative expression levels of the target genes.
Vibrio challenge
The Vibrio strains of V. harveyi and V. parahaemolyticus were isolated from the infected Tilapia fish at Ramayanpatti, in the Tamil Nadu district of Tirunelveli, India. The conventional morphological, biochemical, and pathogenicity assays identified the Vibrio bacterial isolates. The isolates were pre-enriched with an alkaline peptone solution (APS- M618, Himedia) before being diluted in conventional saline (0.85% NaCl w/v). Each isolate was surface dispersed on three agar media: TCBS (thiosulphate citrate bile salt sucrose agar-M870S, Himedia), SWC (seawater complex agar- M592, Himedia), and Vibrio specific agar medium (VSAM- M820, Himedia). A dark room at 30 °C was the perfect temperature for finding bio-luminous colonies on SWC agar. The Vibrio isolates were compared to strains of V. harveyi (MTCC 3438) and V. parahaemolyticus (MTCC 443) as positive and negative controls, respectively, and then further confirmed by PCR. The PCR confirmed Vibrio isolates were used in this study. To prepare separately fresh V. harveyi and V. parahaemolyticus, a single colony of Vibrio was inoculated into Nutrient Broth with 2% of NaCl and cultured for 24 h at 30 °C. Cell harvesting was performed by centrifuge at 5,000 rpm and 4 °C for 10 min, after that three washing and re-suspending of the cells in a 0.85% saline buffer. The suspension of V. harveyi and V. parahaemolyticus was modified to 106 CFU/ml with 0.85% saline buffer before injection. After the feeding trial, ten fish from each group were randomly selected and intraperitoneally injected with 0.1 ml of V. harveyi and V. parahaemolyticus (106 CFU/ml) based on the procedure outlined by Fatima et al.35.
Statistical analyses
Statistical analyses were conducted using a two-way ANOVA for evaluating the effects of probiotic treatments and time points, followed by the Duncan multiple range test for post-hoc comparisons. A mixed model regression analysis was also performed to account for the repeated measurements over time. Data analysis was performed using SPSS software (version X).
Results and discussion
Water quality indices
The study found that tilapia culture systems treated with Agrococcus spp (RKDAS1) and Rossellomorea marisflavi (DAS-SCF02) significantly improved dissolved oxygen levels, indicating enhanced oxygen availability within the system. The enhanced DO levels are likely due to the combined metabolic activity of inoculated strains, including RKDAS1 and DAS-SCF02 which improve nitrification, denitrification, and organic matter degradation pathways36,37. The co-application of RKDAS1 and DAS-SCF02, to tilapia rearing water reduced microbial oxygen competition, promoting efficient carbon and nitrogen cycling, lowering biochemical oxygen demand, and improving metabolic performance and immunity38,39. Improved DO availability is directly linked to better tilapia health, as sufficient oxygen supports efficient energy metabolism, enhances immune responses, and reduces stress-related mortality30. In our study, tilapia exposed to the mixed treatment group (RKDAS1 + DAS-SCF02) also exhibited higher survival rates and more stable water quality parameters, further supporting the beneficial role of these RKDAS1 and DAS-SCF02 inoculants in Tilapia culture (Table 3).
Hematological indices
The Fig. 1 depicts each of the four treatment groups and their respective hematological indices findings. The evaluated hematological indices exhibited significantly increased levels (P < 0.05) in the comparison of the fish treated with all three types of probiotics to those treated with an untreated diet. Fish raised in the Rossellomorea marisflavi spp. DAS-SCF02 and Agrococcus spp. RKDAS1 (PFF3) treatment demonstrated much greater levels (P < 0.05) of Hb, WBC, RBC, Htc, and BP relation to the control group (CF). Likewise, after the trial, adding PFF4 to the feed led to a substantial increase (P < 0.05) in the numbers of neutrophils, lymphocytes, and monocytes compared to the control group. The present study results are agreed well with the reports of40,41,42.
Shows the alterations in hematological parameters among Nile tilapia juveniles following a 60-day dietary supplementation with PFF. The means and standard errors are depicted for each of the three replications, with (*, **) asterisk denoting significant differences (P < 0.05) between the treatments.
Hematological biochemical indices
In tilapia supplemented with probiotics, specifically Rossellomorea marisflavi spp. DAS-SCF02 and Agrococcus spp. RKDAS1 (PFF3) and Rossellomorea marisflavi spp. DAS-SCF02 and Agrococcus spp. RKDAS1 (PFF4), serum levels of ALT, and AST were lower (Fig. 2a). However, the CF3 diet resulted in the least significant (P < 0.05) values of AST and ALT. Fish-fed PFF2, PFF3, and PFF4 exhibited higher serum concentrations of total protein, albumin, and globulin compared to other diets (P < 0.05). The CF4 diet, containing Rossellomorea marisflavi spp. DAS-SCF02 and Agrococcus spp. RKDAS1 showed the highest levels of total globulin, albumin, and protein.
Depict alterations in hematological biochemical indices among Nile tilapia juveniles following a 60-day dietary supplementation with PFF. (A) LDL, HDL, globulin, total protein and albumin. (B) Glucose, ALT, AST, cholesterol and triglyceride. For each of the three replications, the means and standard errors are provided, with (*, **) asterisk indicating significant differences (P < 0.05) between the treatments.
Furthermore, tilapia fish treated with three types of probiotics functional feed (PFF) additive mixed probiotics had expressively (P < 0.05) greater glucose, total cholesterol, and triglyceride levels than the control group. In comparison to fish fed the CP diet, all groups administered dietary PFF showed significantly lower plasma TG levels (P < 0.05), and all PFF-fed fish exhibited little variation in CHO or TG (P > 0.05). Additionally, with dietary supplementation of PFF, plasma LDL content was lowered, although no significant difference was observed (P > 0.05). The dual probiotics mix added to the feed dramatically raised albumin, globulin, and total protein levels in the serum, with PFF2 and PFF3 showing superiority over the CPF1 control group (Fig. 2b). The present study results are well agreed with43,44 studies. Furthermore, one plausible explanation for this lowering effect on plasma lipid profiles could be the fermentation of indigestible carbohydrates derived from intestinal food to produce short-chain fatty acids, inhibiting the synthesis of cholesterol in the liver and/or returning cholesterol to the liver45.
Immunological indices
Lysozyme activity
Lysozyme, a bactericidal peptide crucial for the fish’s innate immune response, plays a significant role in inhibiting biofilm formation by promoting phagocytes and the complement system46. It also prevents microbial adhesion and colonization47. In this study, during the 20th, 40th, and 60th days of dietary probiotic feeding, tilapia serum exhibited a substantial enhance (p < 0.05) in lysozyme related to the untreated experiment (Fig. 3a). The PFF4 group, in particular, demonstrated a significant (p < 0.05) boost in serum lysozyme activity after the 60th day of dietary probiotic feeding. Notably, plasma exhibited the highest lysozyme activity after the 60th day when compared to the 40th day.
*Changes in key immunological indices of Nile tilapia (Oreochromis niloticus) juveniles after 60 days of dietary supplementation with PFF. Parameters include: (A) lysozyme activity, (B) reactive oxygen species (ROS) production, (C) reactive nitrogen species (RNS) production, and (D) myeloperoxidase (MPO) activity. Data are presented as mean ± standard error (SE) from three independent replicates per treatment group. Asterisks indicate statistically significant differences between treatments: *P < 0.05, *P < 0.01.
Intracellular superoxide anion (SOA)
Following the 20th, 40th, and 60th day of dietary probiotic feeding, the SOA production in serum improved significantly (p < 0.05) in the dietary probiotic experiments in assessment with the untreated fish (Fig. 3b). In comparison with serum, SOA generation had been boosted in all dietary probiotic experimental feeds. In both the dietary probiotic experiments, the highest SOA generation occurred following the 60th day of feeding instead of the 20th day. The PFF4 group had the greatest enhancement, whereas the CP1 showed minimal enhancement48,49.
Production of reactive nitrogen species (RNS) analysis
On the 20th, 40th, and 60th days of the dietary probiotic feeding experiment, there was a significant improvement in RNS synthesis in the serum of tilapia (p < 0.05), as compared to the untreated fish (Fig. 3c). Aligned with the formation of Reactive Oxygen Species (ROS), all dietary probiotic experiments exhibited stronger plasma RNS production on the 60th day than on the 40th day. The PFF4 feed demonstrated the highest activity of RNS generation, while the untreated fish showed a limited quantity. Similar results were observed by50.
MPO (Myeloperoxidase)
After the 20th, 40th, and 60th days of dietary probiotic experiment feeding, dramatic improvement in MPO activity in plasma was observed (p < 0.05), contrasting with the untreated group (Fig. 3d). The MPO activity in serum increased for dietary PFF2, PFF3, and PFF4. Across all PFF2, PFF3, and PFF4 groups, the peak MPO activity was observed after the 60th day of feeding compared to the 40th day. These findings align well with the study conducted by40.
Immune gene expression
Probiotics have a well-established ability to non-specifically alter the immune system51,52. In the present study, supplementation with Rossellomorea marisflavi spp. DAS-SCF02, Agrococcus spp. RKDAS1, Rossellomorea marisflavi spp. DAS-SCF02, and Agrococcus spp. RKDAS1 increased the expression of HSP70, IL-1β, IC3, IFN-α, IFN-γ, GF1, GH, IL-1, and Lyz genes, with the highest expression observed in fish fed a PFF4 diet. Figure 4(a-i) presents the transcript of immune-related gene expression experiments conducted on tilapia liver. In the liver of tilapia fed CF4 and those fed Rossellomorea marisflavi spp. DAS-SCF02, Agrococcus spp. RKDAS1 additive diets (CF2, CF3, and CF4), HSP70, IL-1β, IC3, IFN-α, IFN-γ, GF1, GH, IL-1, and Lyz genes were significantly up-regulated (P < 0.05) (Fig. 4a-i). Compared to CP1 tilapia fingerlings, those fed with PFF3 and PFF4 showed up-regulated expression of the immune gene SOD (P < 0.05). The levels of TNF-α gene expression in fish-fed probiotic feed and all Rossellomorea marisflavi spp. DAS-SCF02, Agrococcus spp. RKDAS1 treatment groups were considerably higher (P < 0.05) when compared to the other treatment groups and control group (CF2 and G3). Additionally, fish-fed CF4 exhibited increased TFN-γ gene expression compared to the control group and all treatment groups (P < 0.05).
The changes in significant immune-related genes in Nile tilapia juveniles following a 60-day dietary supplementation with PFF. The genes examined include HSP70 (a), IL-1β (b), IC3 (c), IFN-α (d), IFN-γ (e), GF1 (f), GH (g), Lyz (h), and IL-1 (i). Means and standard errors are provided for each of the three replications, with (*, **) asterisk representing significant differences (P < 0.05) between the treatments.
Vibrio challenge study
Twenty-five days after PFF2, PFF3, and PFF4 were exposed by intraperitoneal administration of fingerling tilapia fish with respectively V. harveyi and V. parahaemolyticus, the Vibrio challenge test was carried out, as illustrated in Fig. 5a & b. Seven days’ post-challenge, the relative percentage survival (RPS) and cumulative mortality were recorded. Fish fed with dietary probiotics PFF2, PFF3, and PFF4 for 24 days exhibited a significantly reduced cumulative mortality rate compared to those fed with CPF1. Cumulative mortality rates for fish fed CPF1, PFF2, PFF3, and PFF4 were 92.44%, 13.65%, 11.18%, and 10.77%, respectively, at the end of the challenge test. A comparison of Tilapia fish that received functional probiotics versus those that did not show no significant difference in survival rates (P > 0.05). Fingerling fish with the infection displayed abnormal diving, darker pigmentation, and a lack of appetite. Moreover, it was observed that the hemorrhages on their bodies, both in flesh and livers, were more pronounced than those observed in typical fish. The results of the Vibrio challenge test indicate that the dietary probiotics PFF2, PFF3, and PFF4 can significantly reduce the cumulative mortality rate in fingerling tilapia fish infected with V. harveyi and V. parahaemolyticus. This was consistent with previous studies that have shown the beneficial effects of probiotics in enhancing shellfish health and disease resistance53. According to a recent study by54, intraperitoneal exposure to kill V. harveyi enhanced the resistance and antibody response of marine red hybrid tilapia to Vibriosis. However, it is worth noting that there were no significant differences in survival rates between fish that received probiotics and those that did not, suggesting that other factors may also influence fish survival in the presence of these pathogens. Further research is needed to explore the potential mechanisms underlying the observed effects of probiotics on fish health and to optimize their application in aquaculture practices.
Depicts the post-challenge cumulative mortality (A) and post-challenge survival rate (B) of Nile tilapia juveniles fed with PFF after 24 days from V. harveyi and V. parahaemolyticus challenge. Means and standard errors are provided for each of the three replications, with (*, **) asterisk representing significant differences (P < 0.0001) between the treatments.
Survival rate after Vibrio challenge (SR)
On days five and six following the V. harveyi and V. parahaemolyticus challenge, the tilapia fish began to perish. The affected fish exhibited increased mucus discharges, scale detachment, and hemorrhages on numerous areas of their external body surface. Autopsy investigation revealed a pale, swollen liver with colorless nodules dispersed across its surface and a bloated gallbladder. The internal organs of the diseased fish were used to re-isolate Vibrio spp. In a dose-dependent manner, tilapia fish raised with any of the probiotic feed treatments-PFF2, PFF3, and PFF4-showed greater survival levels than those raised in the un-treatment fish group. Significant differences in survival rates were observed between the PFF administrated groups and the control after two weeks of the Vibrio spp. challenge55,56. Notably, a dual probiotic mixed feed (PFF4) led to an increased survival rate among tilapia fish exposed to Vibrio spp. compared to the control fish. Fish fed with PFF4 exhibited the highest survival rate (89.23%), followed by those fed with PFF3 (88.82%), PFF2 (86.35%), and CPF1 (7.56%), as shown in Fig. 5b.
Conclusion
In conclusion, the application of Rossellomorea marisflavi spp. (DAS-SCF02) and Agrococcus spp. (RKDAS1) significantly improved water quality in tilapia culture systems, enhancing metabolic function, hematological, biochemical, immune response, and survival rates. These findings highlight their potential for sustainable aquaculture. The observed increases in hematological indices, including Hb, WBC, RBC, Htc, and BP, suggest enhanced overall fish health. Biochemical analysis indicated improved liver function, as reflected by lower ALT and AST levels and increased total protein, globulin, and albumin concentrations. Furthermore, the immunological responses, as evidenced by increased lysozyme activity, superoxide anion production, reactive nitrogen species synthesis, and myeloperoxidase activity, point towards enhanced immune defenses in probiotic-fed fish. The gene expression analysis revealed the up-regulation of genes associated with stress response, cytokine signaling, and immune defense, indicating the activation of key pathways (hsp70, IL-1β, I C3, IFN-α, IFN-γ, GF1, GH, IL-1, and Lyz) in response to probiotic supplementation. Importantly, the probiotic-fed fish exhibited improved survival rates in a Vibrio challenge study, demonstrating the practical relevance of these findings in disease resistance. Overall, this study provides valuable insights into the comprehensive benefits of probiotic supplementation in tilapia aquaculture, emphasizing its potential to enhance fish health, immunity, and resilience against bacterial challenges. These findings contribute to the growing body of knowledge supporting the sustainable and effective use of probiotics in aquaculture practices. This groundbreaking discovery indicates that the production of sufficient quantities of antagonistic bioactive properties against pathogenic bacteria and their infections can enhance the aquatic environment, promoting fish health by boosting immunity.
Data availability
The results presented are adequate to support the conclusion of this study. However, the lead author (B. P.) can provide extra data upon request.
References
FAO. The state of world fisheries and aquaculture. Food and Agriculture Organization of the United Nations, Rome, Italy. (2020).
Ali, M. M. et al. Dietary alphamonolaurin for Nile tilapia (Oreochromis niloticus): stimulatory effects on growth, immunohematological indices, and immune-related gene expressions. Aquac. Res. (2023).
Li, M. Y. et al. Effects of dietary Allium Mongolicum Regel polysaccharide on growth, lipopolysaccharide-induced antioxidant responses and immune responses in Channa argus. Mol. Biol. Rep. 46, 2221–2230 (2019).
Soltan, N. M., Soaudy, M. R., Abdella, M. M. & Hassaan, M. S. Partial dietary fishmeal replacement with mixture of plant protein sources supplemented with exogenous enzymes modify growth performance, digestibility, intestinal morphology, haemato-biochemical and immune responses for nile tilapia, Oreochromis niloticus. Anim. Feed Sci. Technol. 299, 115642 (2023).
Mohammady, E. Y. et al. Response of nile tilapia under Biofloc system to floating or sinking feed and feeding rates: water quality, plankton community, growth, intestinal enzymes, serum biochemical and antioxidant status. Aquac Rep. 29, 101489 (2023).
Saleh, R. S., Mohammady, E. Y., El-Haroun, E. & Hassaan, M. S. Dietary dried periphyton can improve growth, digestive enzyme, serum biochemical, antioxidant response and intestinal morphometric of nile tilapia. Aquac Res. 53 (18), 6463–6477 (2022).
Kesarcodi-Watson, A., Kaspar, H., Lategan, M. J. & Gibson, L. Probiotics in aquaculture: the need, principles and mechanisms of action and screening processes. Aquac 274 (1), 14 (2008).
Yanbo, W. & Zirong, X. Effect of probiotics for common carp (Cyprinus carpio) based on growth performance and digestive enzyme activities. Anim. Feed Sci. Technol. 299 127 (3–4), 283–292 (2006).
Shams, S. M., Dastar, B., Zerehdaran, S., Khomeiri, M. & Moradi, A. Effects of using plant extracts and a probiotic on performance, intestinal morphology, and microflora population in broilers. J. Appl. Poult. Res. 21 (2), 201–208 (2012).
Abou-El-Atta, M. E., Abdel-Tawwab, M., Abdel-Razek, N. & Abdelhakim, T. M. Effects of dietary probiotic Lactobacillus plantarum and Whey protein concentrate on the productive parameters, immunity response and susceptibility of nile tilapia, Oreochromis niloticus (L.), to Aeromonas sobria infection. Aquac Nutr. 25, 1367–1377. https://doi.org/10.1111/anu.12957 (2019).
Martínez Cruz, P., Ib´a˜nez, A. L. & Monroy Hermosillo, O. A. H. C. Ramírez Saad, use of probiotics in aquaculture. ISRN Microbiol. 1–13 (2012).
Hassaan, M. S., Soltan, M. A., Jarmołowicz, S. & Abdo, H. S. Combined effects of dietary malic acid and Bacillus subtilis on growth, gut microbiota and blood parameters of nile tilapia (Oreochromis niloticus). Aquac Nutr. 24 (1), 83–93 (2018).
Dawood, M. A. et al. Physiological response, blood chemistry profile and mucus secretion of red sea Bream (Pagrus major) fed diets supplemented with Lactobacillus rhamnosus under low salinity stress. Fish. Physiol. Biochem. 43, 179–192 (2017).
Abdo, H. S., Mohammady, E. Y., Tonsy, H. D., Ibrahim, A. & Hassaan, M. S. The potential synergistic action of Quercetin and/or Pediococcus acidilactici on nile tilapia, Oreochromis niloticus performance. Aquac 581, 740353 (2024).
Heshmatfar, F. et al. The effects of combined or singular administration of formic acid and Pediococcus acidilactici on stress resistance, growth performance, immune responses and related genes expression in common carp, Cyprinus carpio. Aquac. Rep. 29, 101474 (2023).
Navarro-Torre, S., Carro, L., Igual, J. M. & Montero-Calasanz, M. D. C. Rossellomorea arthrocnemi spp. Nov., a novel plant growth-promoting bacterium used in heavy metal polluted soils as a phytoremediation tool. Int. J. Syst. Evol. Microbiol. 71 (10), 005015 (2021).
Manzoor, A., AZIZ, I., Mujeeb, A., Abideen, Z. & Yong, J. W. H. Resistance in arthrocaulon macrostachyum to Pb and salt stress is associated with water balance, ion fluxes and antioxidant feedback. Ion Fluxes Antioxid. Feedback 4668654 (2023).
Gupta, R. S., Patel, S., Saini, N. & Chen, S. Robust demarcation of 17 distinct Bacillus species clades, proposed as Nov.l Bacillaceae genera, by phylogenomics And comparative genomic analyses: description of Robertmurraya kyonggiensis spp. Nov. And proposal for an emended genus Bacillus limiting it only to the members of the Subtilis And Cereus clades of species. Int. Int. J. Syst. Evol. Microbiol. 70 (11), 5753–5798 (2020).
Navarro-Torre, S., Mateos-Naranjo, E., Caviedes, M. A., Pajuelo, E. & Rodríguez-Llorente, I. D. Isolation of plant-growth-promoting and metal-resistant cultivable bacteria from Arthrocnemum macrostachyum in the Odiel marshes with potential use in phytoremediation. Mar. Pollut Bull. 110 (1), 133–142 (2016).
Navarro-Torre, S. et al. Mateos-Naranjo, bioaugmentation with bacteria selected from the Microbiome enhances Arthrocnemum macrostachyum metal accumulation and tolerance. Mar. Pollut Bull. 117 (1–2), 340–347 (2017).
Navarro-Torre, S. et al. Sustainable agricultural management of saline soils in arid and semi-arid mediterranean regions through halophytes, microbial and soil-based technologies. Environ. Exp. Bot. 105397 (2023).
Mayilraj, S., Suresh, K., Schumann, P., KroppenstedtR.M. & SainiH.S. Agrococcus lahaulensis spp. Nov., isolated from a cold desert of the Indian Himalayas. Int. J. Syst. Evol. MicroBiol. 56 (8), 1807–1810 (2006).
Gatinho, P. et al. From cultural and natural heritage to biomedicine: prospection of bioactive compounds produced by bacterial isolates from caves. Int. Biodeterior. Biodegrad. 190, 105773 (2024).
Tavarideh, F., Pourahmad, F. & Nemati, M. Diversity and antibacterial activity of endophytic bacteria associated with medicinal plant, Scrophularia striata. Vet. Res. Forum. 13, 409–415 (2022).
Selim, K. M., El-Sayed, M. H., El-Hady, M. A. & Reda, R. R. In-vitro evaluation of the probiotic candidtes isolated from the gut of Clarias Gariepinus with special reference to the in vivo assessment of live and heat-inactivated Leuconostoc mesenteroides and Edwardsiella spp. Aquac Int. 27, 33–51 (2019).
Salam, A. M. et al. Gut probiotic bacteria of Barbonymus gonionotus improve growth, hematological parameters and reproductive performances of the host. Sci. Rep. 11, 10692 (2021).
Aly, S. M., Ahmed, Y. A. G., Ghareeb, A. A. A. & Mohamed, M. F. Studies on Bacillus subtilis and Lactobacillus acidophilus, as potential probiotics, on the immune response and resistance of Tilapia nilotica (Oreochromis niloticus) to challenge infections. Fish Shellfish Immunol. 25 (1–2), 128–136 (2008).
Silva, T. A. et al. Effects of the probiotic Bacillus amyloliquefaciens on growth performance, hematology and intestinal morphometry in Cagereared nile tilapia. Lat Am. J. Aquat. Res. 43 (5), 963–971 (2015).
Boyd, C. E. Water Quality (Springer International Publishing AG, 2019).
Boyd, C. E. Water Quality for Pond Aquaculture (Auburn University, 1998).
Schaperclaus, W., Kulow, H. & Schreckenbach, K. Hematological and serological technique. Fish. Dis. 1 (56), 71–108 (1991).
Parry, R. M., Chandan, R. C. & Shahani, K. M. A rapid and sensitive assay of muramidase. Proc. Soc. Exp. Biol. Med. 119, 384–386 (1965).
Green, L. C. et al. Analysis of nitrate, nitrite, and [15 N] nitrate in biological fluids. Anal. Biochem. 126 (1), 131–138 (1982).
Kumari, J. & Sahoo, P. K. Dietary β-1, 3 glucan potentiates innate immunity and disease resistance of Asian catfish, Clarias batrachus (L). J. Fish. Dis. 29, 95–101 (2006).
Fatima, R. et al. Enhancement of immune response and resistance to Vibrio parahaemolyticus in Tilapia fish (Oreochromis mossambicus) by dietary supplementation of Portieria hornemannii. Aquac 547, 737448 (2022).
Arun, M. et al. Bacterial diversity and functional roles in sustainable aquaculture systems. Aquaculture Rep. 25, 101–113 (2023).
Kim, S. Y. et al. Microbial management in aquaculture: A review of microbial consortia and water quality improvement. Microorganisms 9 (12), 2548 (2021).
Wang, Y. et al. Application of probiotics and microbial community modulation in aquaculture: A review. FEMS Microbiol. Ecol. 96 (8), fiaa134 (2020).
Redhwan, A. et al. Effects of water additive mixed probiotics on water quality, growth performance, feed utilization, biochemical analyses and disease resistance against Aeromonas sobria of nile tilapia. Desalination Water Treat. 319, 100480 (2024).
Dawood, M. A., Magouz, F. I., Salem, M. F. & Abdel-Daim, H. A. Modulation of digestive enzyme activity, blood health, oxidative responses and growth-related gene expression in GIFT by heat-killed Lactobacillus plantarum (L-137). Aquac 505, 127–136 (2019).
Hassaan, M. S. et al. Partial dietary fish meal replacement with cotton seed meal and supplementation with exogenous protease alters growth, feed performance, hematological indices and associated gene expression markers (GH, IGF-I) for nile tilapia, Oreochromis niloticus. Aquac 503, 282–292 (2019).
Hassaan, M. S., Mohammady, E. Y., Soaudy, M. R. & Abdel Rahman, A. A. Exogenous Xylanase improves growth, protein digestibility and digestive enzymes activities in nile tilapia, Oreochromis niloticus, fed different ratios of fish meal to sunflower meal. Aquac Nutr. 25 (4), 841–853 (2019).
Jia, R. et al. Effects of dietary Baicalin supplementation on growth performance, antioxidative status and protection against oxidative stress-induced liver injury in GIFT tilapia (Oreochromis niloticus), comp. Biochem. Physiol. C Toxicol. Pharmacol. 240, 108914 (2021).
Dawood, M. A., Abo-Al-Ela, H. G. & Hasan, M. T. Modulation of transcriptomic profile in aquatic animals: probiotics, prebiotics and synbiotics scenarios. Fish. Shellfish Immunol. 97, 268–282 (2020).
Shen, C. et al. Fecal short chain fatty acids modify therapeutic effects of sleeve gastrectomy. Front. Endocrinol. 14, 1277035 (2023).
Abirami, G. et al. Pyrogallol downregulates the expression of virulence-associated proteins in Acinetobacter baumannii and showing anti-infection activity by improving non-specific immune response in zebrafish model. Int. J. Biol. Macromol. 226, 853–869 (2023).
Ferraboschi, P., Ciceri, S. & Grisenti, P. Applications of lysozyme, an innate immune defense factor, as an alternative antibiotic. Antibiotics 10 (12), 1534 (2021).
Lim, K. C., Yusoff, F. M., Shariff, M. & Kamarudin, M. S. Dietary Astaxanthin augments disease resistance of Asian Seabass, Lates calcarifer (Bloch, 1790), against Vibrio alginolyticus infection. Fish. Shellfish Immunol. 114, 90–101 (2021).
Lee, P. T., Tran, H. T. Q., Huang, H. T., Nan, F. H. & Lee, M. C. Sargassum Horneri extracts stimulate innate immunity, enhance growth performance, and upregulate immune genes in the white shrimp Litopenaeus vannamei. Fish. Shellfish Immunol. 102, 276–285 (2020).
Panase, A., Thirabunyanon, M., Promya, J. & Chitmanat, C. Influences of Bacillus subtilis and fructooligosaccharide on growth performances, immune responses, and disease resistance of nile tilapia, Oreochromis niloticus. Front. Veterinary Sci. 9, 1094681 (2023).
Khan, M. I. R. Biotechnological interventions in Coldwater aquaculture health management. In Coldwater Fisheries and Aquaculture Management. 147–177. (Apple Academic, 2024).
Cave, N., Delaney, S. J. & Larsen, J. A. Nutritional management of Gastrointestinal diseases. Appl. Vet. Clin. Nutr. 235–298 (2023).
Harrison, J. et al. The increased prevalence of Vibrio species and the first reporting of Vibrio jasicida and Vibrio rotiferianus at UK shellfish sites. Water Res. 211, 117942 (2022).
Abu Nor, N. et al. Efficacy of whole cell inactivated Vibrio harveyi vaccine against vibriosis in a marine red hybrid tilapia (Oreochromis niloticus× O. mossambicus) model. Vaccines 8 (4), 734 (2020).
Addo, S. et al. Effects of Bacillus subtilis strains on growth, immune parameters, and Streptococcus iniae susceptibility in nile tilapia, Oreochromis niloticus. J. World Aquaculture Soc. 48 (2), 257–267 (2017).
Abarike, E. D. et al. Effects of a commercial probiotic BS containing Bacillus subtilis and Bacillus licheniformis on growth, immune response and disease resistance in nile tilapia, Oreochromis niloticus. Fish. Shellfish Immunol. 82, 229–238 (2018).
Acknowledgements
The authors acknowledge the Manonmaniam Sundaranar University, Tirunelveli, Tamil Nadu, India for their constant support and the grant support from UGC DR. D. S. Kothari Postdoctoral Fellowship (Normal Fellowship): No. F. 4-2/2006 (BSR)/BL/19-20/0298 New Delhi. The authors express their sincere appreciation to the Researchers supporting Project Number (RSP2025R48), King Saud University, Riyadh, Saudi Arabia.
Author information
Authors and Affiliations
Contributions
B.P.: Formal and experimental analysis, Documentation & Formal analysis, Writing—original draft. R. T.: Investigation, Writing, review & editing—original draft, Conceptualization, Investigation, Visualization, Project administration. C. V.: Data curation & Review, K. S.: Supervision, Project administration, editing & review. K. A. A.G. & C. K.: Formal analysis, writing, review, & editing. All authors have read and agreed to the published version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
The experiment was conducted following the protocol involving animal use approved by the experiment was ethically reviewed and approved by the Department of Animal Science and Animal Ethical Committee, through the Animal and Welfare Ethical Review Body by the Manonmaniam Sundaranar University Animal Care and Use Committee MSU-ACUC (BP, PhD, Reg. No. 17214012272124). All fish handling procedures and regulations followed the ARRIVE guidelines for Animal Care and Use. Furthermore, all relevant organizational and government rules and regulations governing the ethical use of the experimental animals were followed. Written informed consent was obtained from the owners of all animals involved in the study.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Paramashivan, B., Thamarai, R., Subramaniam, K. et al. Synergistic effect of Agrococcus and Rossellomorea Marisflavi species assisted probiotic functional feed on Vibrio affected Nile tilapia fish. Sci Rep 15, 21866 (2025). https://doi.org/10.1038/s41598-025-03715-z
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-03715-z