Abstract
Sarcopenia is defined as a progressive decrease in skeletal muscle mass and strength due to ageing, and is associated with adverse outcomes such as falls, fractures, and physical disability, leading to increased healthcare expenditures and mortality risks. Although sarcopenia has recently become a major challenge, there are currently no effective interventions or drugs marketed for sarcopenia. Therefore, new biomarkers for early diagnosis and targeted drugs for the treatment of sarcopenia are urgently needed. In the present research, transcriptomics and proteomics analyses were combined and experiments at the transcriptional and protein levels were conducted to identify key genes and molecular mechanisms underlying sarcopenia in senescence-accelerated mouse prone 8 (SAM-P8) mice, followed by molecular docking to predict targeted drugs. The combined omics analysis identified 8 key genes, while the experiments verified that only CD9 significantly decreased in sarcopenia. The gene set enrichment analysis (GSEA) results suggested that CD9 involved in ATP biosynthesis, mitochondrial biogenesis, and oxidative phosphorylation. Besides, dapoxetine, levomilnacipran, and milnacipran were predicted to target CD9 through molecular docking. Our study reported for the first time that CD9 is a novel potential biomarker of sarcopenia, and targeting CD9 may be a new idea for the development of therapeutic drugs for sarcopenia.
Similar content being viewed by others
Introduction
Sarcopenia was first proposed in 1989 by Irwin Rosenberg1,2. And now, the Global Leadership Initiative in Sarcopenia (GLIS) defines sarcopenia as the age-related loss in muscle mass and strength/function3. It is conservatively estimated that 10%-16% of elderly individuals worldwide suffer from sarcopenia4. In China, the prevalence of sarcopenia among the elderly population is even higher (20.7%), and varies by sex and age; that is, the prevalence among women (21.6%) is greater than that among men (19.2%), and the prevalence among people aged ≥ 80 years (45.4%) is the highest, followed by people aged 70–79 years (27.2%) and 60–69 years (15.7%)5. In addition, the muscle mass decreases at an annual rate of 1–2% approximately after the age of 50, and the muscle strength decreases by 1.5% between the ages of 50 and 60 years and by 3% thereafter6. The occurrence of sarcopenia leads to instability, an increased risk of falls and consequently an increased demand for medical and social care. Indeed, sarcopenia was found to be closely associated with all-cause mortality7,8.
However, the pathogenesis mechanism of sarcopenia remains unclear, and studies suggest that it is associated with ageing, lifestyle changes, physical activity, nutritional levels, and neurological factors9,10,11. These factors contribute to skeletal muscle inflammation, oxidative stress, mitochondrial dysfunction, and disorders of protein metabolism12,13,14, eventually leading to sarcopenia. Given the enormous socioeconomic burden that sarcopenia can impose, preventive measures and effective treatments are urgently needed. At present, interventions for sarcopenia are limited, including exercise therapy and nutritional support15,16,17,18,19, and patients benefit little, especially for patients who are unsuitable for exercise or are bedridden. Although several medications have been shown to be potentially effective in treating sarcopenia, such as formoterol20, allopurinol21, and myostatin-specific antibodies22, no drugs have been marketed to date23. As a result, new methods for the rapid and efficient early diagnosis and treatment of sarcopenia are urgently needed, including novel biomarkers, underlying mechanisms, and targeted drugs.
Naturally, ageing mice are the preferred choice for sarcopenia research, but their application has been hampered by their long rearing time and high cost. Fortunately, animal models of accelerated ageing shorten the time required to wait for ageing. The senescence-accelerated mouse (SAM) is one of the most common ageing models developed by Takeda et al.24, and is considered a "fast-forward" option for ageing studies. The line consists of 18 different strains, 11 of which are SAM-prone (SAM-P, susceptible to ageing) and 7 of which are SAM-resistant (SAM-R, resistant to ageing)25. Compared with SAM-R1 mice, SAM-P8 mice have significantly lower skeletal muscle weight and contractility, and more obvious atrophy of type II muscle fibres, indicating the significant characteristics of accelerated skeletal muscle ageing26, thus becoming a common animal model for the study of sarcopenia27,28.
The main process of gene expression includes transcription and protein synthesis, where mRNA is the intermediate of gene expression, and protein is the executive of gene function. Transcriptomics and proteomics elucidate the changes in related genes and molecular mechanisms involved in the occurrence, development and prognosis of diseases from the perspectives of gene transcription and protein synthesis, respectively29. As a result, the combined analysis of transcriptomics and proteomics has become an important means to discover and study the biomarkers of diseases from the the source30,31.
In this study, a combined transcriptomics and proteomics analysis strategy was employed to screen key genes and possible signalling pathways that exhibit significant changes throughout the pathological process of sarcopenia in SAM-P8 mice, aiming to identify novel biomarkers and possible regulatory mechanisms for the early recognition of and interventions for sarcopenia. Furthermore, biomarkers for targeted drugs are predicted to provide new directions for the development of targeted drugs for sarcopenia.
Results
Twenty-eight-week-old SAM-P8 mice develop sarcopenia characterized by the significant weakening of skeletal muscles
Initially, the treadmill test and forelimb grip strength test were performed, and the results revealed that muscular endurance (Fig. 1A) and explosive power (Fig. 1B) were dramatically lower in 28-week-old SAM-P8 mice (model group) than in SAM-R1 mice (NC group) (### p value < 0.001). Furthermore, the gastrocnemius (gastro), extensor digitorum longus (EDL) and soleus were excised and photographed. Obvious changes in the phenotypes of the skeletal muscles in the model group were observed (Fig. 1C and D). Each type of skeletal muscle was subsequently weighed, and muscle tissue indices (muscle weight/body weight) were calculated. We found notably lower muscle weights in the model group than in the NC group (Fig. 1E, ### p value < 0.001, # p value < 0.05), and the same was true for muscle tissue indices (Fig. 1F, ### p value < 0.001, # p value < 0.05). In addition, haematoxylin‒eosin (HE) staining was performed to examine the changes in muscle fibre size in the model group (Fig. 1G), and the cross-sectional areas (CSAs) of the gastrocnemius, EDL and soleus clearly decreased (Fig. 1H, ### p value < 0.001). The results of the present study indicated that 28-week-old SAM-P8 mice developed sarcopenia.
Identification of sarcopenia in 28-week-old SAM-P8 mice. (A) Treadmill test, n = 15–25 mice/group. (B) Forelimb grip strength test, n = 12–21 mice/group. (C, D) Representative images of muscle morphology in hindlimb of mice. (E) Muscle weights of gastro, EDL, soleus, n = 10 mice/group. (F) Muscle indexes of gastro, EDL, soleus, n = 10 mice/group. (G) Representative images of H&E staining of gastro, EDL and soleus (200 × magnification). (H) Cross-sectional areas of gastro, EDL, and soleus, n = 5 mice/group, 3 views/sample. Data described are means ± SEMs. Unpaired two-tailed Student’ s t test was applied for statistical comparisons. # p < 0.05, ### p < 0.001 vs. the NC group. EDL, extensor digitorum longus; gastro, gastrocnemius; NC, normal control; SAM-R1, senescence-accelerated mouse resistant 1; SAM-P8, senescence-accelerated mouse prone 8.
Screening of 637 DEGs and 94 DEPs in mice with sarcopenia
The PCA results revealed great consistency and high heterogeneity between the model and NC groups (Fig. 2A and D). Furthermore, in the model group, there were 637 DEGs in the transcriptome, with 521 upregulated genes and 116 downregulated genes (Fig. 2B and C), and 94 DEPs in the proteome, with 46 increased proteins and 48 decreased proteins (Fig. 2E and F). These DEGs and DEPs served as the basis for subsequent analyses.
Identification of 37 overlapping genes in the PPI network of transcriptome DEGs
The STRING was applied to construct a PPI network of the above 637 transcriptome DEGs (Fig. 3A), and the genes were sequenced by EPC (Fig. 3B), MNC (Fig. 3C), closeness (Fig. 3D) and MCC (Fig. 3E) algorithms via the cytoHubba plug-in. The top 50 DEGs of the four algorithms were subsequently selected, and a total of 37 overlapping genes were obtained in the model group, including Plg, Alb, Apoa5, Cyp2c70, F2, Apob, Ugt2b1, Cyp3a11, Apoc3, Apoh, Apoa1, Cyp2c50, Cyp2a12, Ahsg, Hp, Cyp2c54, Serpinc1, Cyp3a25, Fga, Apoa2, Fgg, Cyp1a2, Pzp, Serpina1d, Fgb, Ugt2b36, Hpx, Serpina1b, Ambp, Serpina1a, Apoe, Ugt2b5, Serpina1c, Apoa4, Gc, Cyp2c29 and Ugt2a3 (Fig. 3F).
PPI network of transcriptome DEGs and intersection of TOP50 genes. (A) PPI network of 637 DEGs in transcriptome. (B–E), The PPI network of TOP50 genes analyzed by 4 algorithms: (B) Closeness, (C) EPC, (D) MCC, and (E) MNC. (F) Intersection of TOP50 genes obtained from results of 4 algorithms. DEGs, differentially expressed genes; EPC, edge percolated component; MCC, maximal clique centrality; MNC, maximum neighborhood component.
Functional enrichment analysis of the 37 overlapping genes in the transcriptome
The enrichment of the 37 overlapping genes was assessed via GO and KEGG pathway analyses using the “ClusterProfiler” R package. A total of 515 GO terms and 30 KEGG signalling pathways were enriched. The top 5 GO pathways were triglyceride rich lipoprotein particle remodelling, steroid metabolic process, protein containing complex remodelling, phospholipid efflux and coagulation regulation; the top 5 KEGG pathways were retinol metabolism, chemical carcinogenesis-DNA adducts, steroid hormone biosynthesis, complement and coagulation cascades and cholesterol metabolism, as shown in Fig. 4A. In addition, the specific biological processes of the DEGs involved in the top 5 pathways identified above are shown in Fig. 4B.
Functional enrichment analysis of 37 overlapping genes. (A) Enrichment bubble diagram of TOP5 results of 37 overlapping genes in BP, CC, MF and KEGG. (B) Differential expression of genes in TOP5 items of BP and KEGG enrichment results. Abbreviations: BP, biological process; CC, cell component; MF, molecular function; KEGG, Kyoto encyclopedia of genes and genomes.
Identification of 50 overlapping proteins in PPI network of proteome DEPs and functional enrichment analysis
A protein–protein interaction (PPI) network of 94 DEPs in the proteome of the model group was constructed (Fig. 5A), and 50 overlapping proteins were obtained for functional enrichment analysis. As shown in Fig. 5B, GO analysis indicated that 281 pathways were enriched: the top 5 pathways were translation at synapses, nitrobenzene metabolic processes, the detoxification of nitrogen compounds, glutathione metabolic processes and the regulation of the force of heart contractions. In addition, a total of 25 KEGG signalling pathways were enriched, and the top 5 were ribosomes, glutathione metabolism, coronavirus disease-COVID-19, cardiac muscle contraction and motor proteins. The specific biological processes associated with the DEPs involved in the top 5 pathways identified above are shown in Fig. 5C.
PPI network of proteome DEPs and functional enrichment analysis. (A) PPI network of 94 DEPs in proteome. (B) Enrichment bubble diagram of TOP5 results of 50 overlapping proteins in BP, CC, MF and KEGG. (C) Differential expression of genes in TOP5 items of BP and KEGG enrichment results. BP, biological process; CC, cell component; DEGs, differentially expressed genes; MF, molecular function; KEGG, Kyoto encyclopedia of genes and genomes; PPI, protein–protein interaction.
Identification of the 8 key genes in mice with sarcopenia by integrated transcriptomics and proteomics analysis
The GO and KEGG analyses revealed pathways enriched simultaneously in the transcriptome and proteome, including 22 BP pathways, 2 CC pathways, 3 MF pathways and 7 KEGG pathways (Fig. 6A). Furthermore, 29 genes and 33 proteins involved in the above pathways were used for subsequent correlation analysis. Nine quadrants were used to show the differential expression of genes in the transcriptome and proteins in the proteome and the correlations between them. The 1st and 5th quadrants represented the genes and proteins that were differentially expressed and positively correlated; the 3rd and 7th quadrants represented the genes and proteins that were differentially expressed but negatively correlated; genes in quadrants 2 and 6 were differentially expressed, but there was no difference in protein expression; the genes in quadrants 4 and 8 were not differentially expressed, but the proteins were; and quadrant 9 represented genes no significant difference in gene or protein expression (Fig. 6B). A heatmap was used to analyse the correlation between the two kinds of omics data (Fig. 6C), and key genes related to sarcopenia, including Apoa1, Apoe, Apoa2, Rpl21, Nefm, Cd9, Rpl6 and Cd81 (Fig. 6D) were obtained.
Determining key genes in sarcopenia by integrated transcriptomics and proteomics analysis. (A) Enrichment bubble diagram of the TOP5 items enriched by transcriptomics and proteomics simultaneously. (B) The nine-quadrant diagram, the horizontal and vertical coordinates represent the differential expression multiples of genes in the transcriptome and proteome, respectively. (C) Heatmap of correlation between transcriptome gene (vertically) and proteome gene (horizontally). (D) Correlation network diagram of genes and proteins.
CD9 may be a novel biomarker of sarcopenia
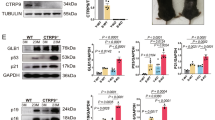
The key genes associated with sarcopenia identified above were subjected to qRT-PCR validation. As shown in Fig. 7A, only CD9 significantly reduced in the model group (## p < 0.01). Furthermore, the immunofluorescence (IF) assay was performed to assess the expression of CD9 at the protein level. The results indicated that CD9 was obviously decreased in mice with sarcopenia (Fig. 7B and C,### p < 0.001), suggesting that CD9 may be a novel biomarker of sarcopenia.The gene set enrichment analysis (GSEA) of CD9 was subsequently performed using the “clusterProfiler” package, and the screening criterion was p ≤ 0.05. The results suggested that CD9 regulated aerobic respiration and ATP biosynthesis and participated in amyotrophic lateral sclerosis (ALS), diabetic cardiomyopathy, Huntington disease, mitochondrial biogenesis, and oxidative phosphorylation, which are possibly the mechanisms by which CD9 is involved in the occurrence and development of sarcopenia (Fig. 7D).
CD9 expression and single gene GSEA analysis. (A) CD9 expression at transcriptional level. Data described are means ± SEMs, n = 6 mice/group. (B) Representative images of immunofluorescence staining of nuclei and CD9 (40 × magnification). (C) Relative fluorescence intensity normalized with nuclei. Data described are means ± SEMs, n = 5 mice/group, 3 views/sample. (D) Single-gene GSEA analysis of CD9. Left: GO enrichment; Right: KEGG enrichment. A, C: Unpaired two-tailed Student’ s t test was applied for statistical comparisons. ## p < 0.01, ### p < 0.001 vs. the NC group. GO, gene ontology; GSEA, gene set enrichment analysis; KEGG, Kyoto encyclopedia of genes and genomes.
Prediction of drugs that target CD9 and molecular docking
To further discover possible targeted drugs for sarcopenia, we used the DGIdb database to predict targeted drugs for the biomarker CD9 and obtained a total of 28 drugs, as shown in Fig. 8A. Subsequently, molecular docking with CD9 was performed for each drug; the greater the degree of binding is, the lower the molecular binding energy. Table 1 shows the top 3 drugs with the lowest binding energy, namely, dapoxetine, levomilnacipran, and milnacipran, and the binding sites are shown in Fig. 8B–D.
Discussion
In this study, we revealed that compared with SAM-R1 mice, SAM-P8 mice developed ageing-induced sarcopenia at 28 weeks of age, as determined by dramatically reduced muscle strength, muscle weight, and muscle fibre size. We subsequently performed a combined transcriptomic and proteomic analysis of the gastrocnemius muscle and identified 8 key genes that were significantly altered in the mice with sarcopenia. Through further experimental verification at the transcription and protein levels, CD9 was identified as a novel biomarker of sarcopenia that may participate in the occurrence and development of sarcopenia by regulating ATP biosynthesis and mitochondrial biogenesis. In addition, potential drugs that target CD9 were predicted, and dapoxetine, levomilnacipran, and milnacipran were found to bind stably to CD9, the role of which in sarcopenia deserves further investigation (Fig. S1).
Compared with SAM-R1 mice, 22-week-old SAM-P8 mice exhibited sarcopenia, as determined by decreases in muscle weight and fibre CSA32, suggesting that 22-week-old SAM-P8 mice are a sufficient model for premature ageing and are a good choice for sarcopenia studies. In our study, the occurrence of sarcopenia in SAM-P8 mice was detected at 28 weeks of age. In addition, 8 key genes associated with sarcopenia were screened by the combined analysis of transcriptomics and proteomics. The relationships of Apoa133, Apoe34, and Cd8135 with sarcopenia have been preliminarily reported, providing evidence of the reliability of the results of our study. However, the relationships of Apoa2, Rpl21, Rpl6, Nefm, and Cd9 with sarcopenia hava not been reported, thus serving as novel findings of this study.
Originally found in lymphohaematopoietic progenitor and acute lymphoblastic leukaemia cells36, cluster of differentiation 9 (CD9) is a membrane protein that belongs to the tetraspanin family, also known as tetraspanin-29 (TSPAN29) and motility-related protein-1 (MRP-1). CD9 is involved in many cellular processes, such as intercellular signal transduction, cell–extracellular matrix interactions, membrane fusion, integrin-dependent cell migration, proliferation and differentiation, apoptosis, and inflammation37. There are a number of studies on CD9 in various diseases, including cardiac hypertrophy38, inflammation39, and cancers40, but no studies on CD9 in sarcopenia have been reported. Surprisingly, our study identified CD9 as a novel potential biomarker for sarcopenia through a combination of dual omics analysis and dual level experimental verification. This marker is undoubtedly a beneficial supplement to the existing regular markers of sarcopenia and also provides favorable evidence for future research on the pathophysiological mechanism of sarcopenia and the development of targeted drugs. It is expected to provide a basis for the clinical prevention and treatment of sarcopenia, which will bring huge social and economic benefits. Besides, in our previous study30, we found that CD9 was a potential biomarker of skeletal muscle atrophy induced by dexamethasone, which is consistent with the results of our present study, suggesting that CD9 may be an important marker for multiple skeletal muscle diseases.
The CD9-targeted drug identified in this study, dapoxetine, is a potent selective serotonin reuptake inhibitor (SSRI) that is primarily used clinically to treat premature ejaculation41 in men; levomilnacipran and milnacipran42 are the selective serotonin norepinephrine reuptake inhibitors (SNRIs) that are clinically used to treat depression. However, the effects of these drugs on sarcopenia have not been reported, providing a research direction for future studies, and new therapeutic effects and molecular mechanisms from the “new use of old drugs” strategy are expected.
The limitations of this study are that the exact molecular mechanism by which CD9 is involved in sarcopenia was not explored and that the effects of CD9-targeting drugs on sarcopenia were not further explored. Therefore, in future studies, the mechanism of CD9 in sarcopenia will be confirmed, and the potential of drugs targeting CD9 for the treatment of sarcopenia will be studied. In addition, the sequence of occurrence of the significant changes in CD9 and sarcopenia is an important issue for our future in-depth research.
In conclusion, this is the first report that CD9 is a novel biomarker for sarcopenia, not only offering an experimental basis for the early diagnosis and treatment of sarcopenia but also providing new inspiration for the development of targeted drugs for sarcopenia, that is, targeting CD9 to treat sarcopenia.
Materials and methods
Animal care
Male senescence-accelerated mouse prone 8 (SAM-P8, 30–35 g, 27 weeks) were obtained from Beijing HuaFuKang Bioscience Co., Ltd. (Beijing, China), which were set as the sarcopenia group (model group). Male senescence-accelerated mouse resistant 1 (SAM-R1, 30–35 g, 27 weeks) were obtained from Department of Experimental Animal Science, Peking University Health Science Center (Beijing, China), which were set as the normal control group (NC group). The mice were housed at a temperature of 22 ± 3 °C and a humidity of 55 ± 5% with a 12-h light/dark cycle and standard mice chow diet and water were provided. All animal procedures involved in this study were performed in strict accordance with ARRIVE guidelines and research guidelines for the care and use of laboratory animals. We got the approval from the Experimental Animal Welfare and Ethics Committee of Shanxi Bethune Hospital (approval number: SBQLYLL-2024-008).
Skeletal muscle strength detection
The treadmill test was introduced to evaluate muscle endurance. Firstly, the mice were initially acclimatized to the treadmill (Beijing Zhishuduobao Biological Technology, Beijing, China) by running 10 min/day at the speed of 15 M/min for 3 days 43. Setting the stimulation current to 0.6 mA, the stimulus tolerance time to 10 s, and the initial speed to 15 M/min, after which the speed increased by 5 M/min every 5 min. Then, the mice were placed at the beginning of each track and the switches of power and electric shock were turned on at the same time. The mice stayed at the area of electric shock site for more than 10 s (stimulus tolerance time set previously) were considered exhausted and the running displacement (m) was recorded.
The forelimb grip strength test was employed to determine muscle explosive force using a grip strength meter provided by Beijing Zhishuduobao Biological Technology (Beijing, China) as previously described44. In brief, the mice were positioned to grasp the metal bar with forelimbs only and then pulled horizontally until letting go. Pulls were repeated thrice with three rounds in sequence in mice. For repetitive pulls, mice were given a 30-s pause in between the pulls; between rounds, mice were given a 30-min rest period. The maximum grip strength (grams, g) of the nine pulls was used for analysis.
Tissue processing and omics data collection
At the end of the experiment, mice were weighed and placed in an airtight container with 4% (v/v) isoflurane until completely anesthetized, followed by cervical dislocation immediately. After the confirmation of humanely sacrifice, skeletal muscles containing mixed type skeletal muscle (gastrocnemius, gastro), fast type skeletal muscle (extensor digitorum longus, EDL), and slow type skeletal muscle (soleus) were excised, and the bilateral muscles were weighed to calculate muscle tissue indexes (muscle weight/body weight). The gastro were then frozen in liquid nitrogen and stored at – 80 ℃ for qRT-PCR detection and transcriptome and proteome sequencing, or fixed in 4% paraformaldehyde for hematoxylin–eosin (HE) staining and immunofluorescence (IF) staining.
The specific operation process of transcriptome and proteome sequencing was referred to the published literature30 (n = 3 mice/group).
Screening of DEGs and DEPs and construction of PPI network
Trimmomatic software was used to filter low-quality data so that removing contamination and splicing sequences in RNA sequencing data. Then, FastOC software (using default parameters) was employed to evaluate the quality of sequencing data, and the filtered clean reads were compared with the reference genome via the Hisat2 software (using default parameters). After obtaining the position information on the reference genome or gene and the specific sequence characteristics of the sequenced samples, the comparison rate between each sample and the reference genome was calculated, and gene expression quantity was then extracted by FeatureCount software with the default parameters. Meanwhile, Proteomic sequencing data processing also included quality control, protein coverage distribution, peptide length distribution, and protein molecular weight distribution. Subsequently, the consistency and heterogeneity of transcriptome and proteome sequencing data between the NC group and the model group was analyzed by PCA map. Then, R package “DEseq2” was used to screen DEGs and DEPs between the NC group and the model group, and the threshold was set as |log2FC|> 0.5, p < 0.05. Further, the STRING database and Cytoscape software (v3.9.1) were applied to the construction of PPI network of transcriptome DEGs and proteome DEPs, respectively. The construction parameters were the maximum number of interactors = 0 and the confidence score ≥ 0.7 in transcriptome, while the maximum number of interactors = 0 and the confidence score ≥ 0.4 in the proteome. Next, genes were sorted by EPC, MNC, closeness and MCC algorithms, and the Top 50 DEGs of each algorithm were selected, followed by intersecting for core genes obtaining.
Functional enrichment analysis of core genes and integrated analysis of transcriptome and proteome
GO and KEGG pathway enrichment analysis were performed for transcriptome DEGs and proteome DEPs using “ClusterProfiler” R package. The significance threshold was set as p value < 0.05. Then, the pathways that were enriched both in transcriptome and proteome were found, where the genes and proteins involved were collected for correlation analysis by nine quadrants. The key genes were identified based on the absolute value of the correlation between transcriptome and proteome was greater than or equal to 0.9 and p value < 0.05.
Prediction of targeted drugs of key genes and molecular docking
Using DGIdb database to predict targeted drugs of key genes, and Cytoscape software was applied for visualization. Subsequently, corresponding protein structures of key genes were downloaded from AlphaFold Protein Structure Database, while 3D structure of targeted drugs were obtained from PubChem database. Then, Autodock Vina system was used to perform molecular docking between the protein structure of the key genes and the corresponding targeted drugs. The binding energy between them was calculated. Generally, the more stable the binding conformation of the two, the lower the required binding energy. The molecular binding energy ≤ − 5.0 kcal/mol represents good binding activity.
Quantitative real-time PCR
Total RNA was isolated according to the TRIzol isolation reagent protocol (Invitrogen, USA) and then Direct-zol RNA kits (ZYMO Research, USA) were used to purify RNA. 1 μg of total RNA with SuperScript III reverse transcriptase (Invitrogen, USA) and Oligo-dT17 as primers were used to synthesize cDNA. Gene expression was detected by SsoFast™ EvaGreen® Supermix (Bio-Rad, USA) and a CFX-96 real-time PCR System (Bio-Rad, USA). The results were normalized with Tbp45 before being quantified. And gene-specific primer pairs can be found in Table S1.
Histological and immunofluorescent staining
The gastro fixed with 4% paraformaldehyde were processed to hematoxylin–eosin (HE) staining. The high-content screening machine (Pannoramic DESK, P-MIDI, P250, 3D HISTECH, Hungary) was used to capture images. The CSA of muscle fibers were determined by ImageJ software (NIH, USA). Then, the sections were permeabilized, blocked and incubated with the primary antibody against CD9 (98327, 1:50, Cell Signaling Technology, MA, USA). After overnight culture at 4 °C, a mixture of appropriate secondary antibodies was added for 2 h at 37 °C. Subsequently, DAPI (G1012, Servicebio, Wuhan, China) was used to counterstain the nucleus and images were captured by aforementioned machine. The fluorescence intensity was determined by ImageJ software (NIH, USA), which was normalized with fluorescence intensity of nuclei before analysis.
Statistical analysis
Data are showed as the means ± SEM. The metric and the algorithm were used to reduce the noise and eliminate the faults in the measurements46,47,48. Statistical analysis was carried out using GraphPad Prism 7.0 (GraphPad Software, Inc. CA, USA). For statistical comparisons, Wilcoxon rank sum test or unpaired two-tailed Student’s t test was used as appropriate. Differences were considered significant at P ≤ 0.05.
Data availability
All data generated or analysed during this study are available within the article and/or its supplementary materials.
Abbreviations
- Apoa1:
-
Apolipoprotein A1
- Apoa2:
-
Apolipoprotein A2
- Apoe:
-
Apolipoprotein E
- BP:
-
Biological process
- CC:
-
Cell component
- CSA:
-
Cross-sectional area
- DEGs:
-
Differentially expressed genes
- DEPs:
-
Differentially expressed proteins
- EDL:
-
Extensor digitorum longus
- EPC:
-
Edge percolated component
- gastro:
-
Gastrocnemius
- GO:
-
Gene ontology
- GSEA:
-
Gene set enrichment analysis
- HE:
-
Hematoxylin–eosin
- IF:
-
Immunofluorescence
- IHC:
-
Immunohistochemistry
- KEGG:
-
Kyoto encyclopedia of genes and genomes
- MCC:
-
Maximal clique centrality
- MF:
-
Molecular function
- MNC:
-
Maximum neighborhood component
- MRP-1:
-
Motility-related protein-1
- NC:
-
Normal control
- Nefm:
-
Neurofilament medium chain
- PPI:
-
Protein–protein interaction
- Rpl6:
-
Ribosomal protein L6
- Rpl21:
-
Ribosomal protein L21
- SAM-R1:
-
Senescence-accelerated mouse resistant 1
- SAM-P8:
-
Senescence-accelerated mouse prone 8
- SNRI:
-
Serotonin norepinephrine reuptake inhibitor
- SSRI:
-
Selective serotonin reuptake inhibitor
- TSPAN29:
-
Tetraspanin-29
References
Epidemiologic and methodologic problems in determining nutritional status of older persons. Proceedings of a conference Albuquerque, New Mexico, October 19–21, 1988. Am. J. Clin. Nutr. 50, 1121–1235 (1989).
Rosenberg, I. H. Sarcopenia: origins and clinical relevance. J. Nutr. 127, 990s–991s (1997).
Kirk, B. et al. The conceptual definition of sarcopenia: Delphi Consensus from the Global Leadership Initiative in Sarcopenia (GLIS). Age Ageing 53, 563 (2024).
Yuan, S. & Larsson, S. C. Epidemiology of sarcopenia: prevalence, risk factors, and consequences. Metabolism 144, 155533 (2023).
Meng, S. et al. The prevalence of sarcopenia and risk factors in the older adult in China: a systematic review and meta-analysis. Front. Public Health 12, 1415398 (2024).
Hao, Q. et al. Prevalence of sarcopenia and associated factors in hospitalised older patients: a cross-sectional study. Austral. J. Ageing 37, 62–67 (2018).
Wu, G. et al. Sarcopenia and mild kidney dysfunction and risk of all-cause and cause-specific mortality in older adults. Nephrol. Dial. Transplant 39, 989–999 (2024).
Benz, E. et al. Sarcopenia and Sarcopenic obesity and mortality among older people. JAMA Netw. Open 7, e243604 (2024).
Cai, Z. et al. The role and therapeutic potential of stem cells in skeletal muscle in sarcopenia. Stem Cell Res. Ther. 13, 28 (2022).
Cailleaux, P. E., Déchelotte, P. & Coëffier, M. Novel dietary strategies to manage sarcopenia. Curr. Opin. Clin. Nutr. Metab. Care 27, 234–243 (2024).
Arosio, B. et al. Sarcopenia and cognitive decline in older adults: targeting the muscle-brain axis. Nutrients 15, 1583 (2023).
Nishikawa, H., Asai, A., Fukunishi, S., Nishiguchi, S. & Higuchi, K. Metabolic syndrome and sarcopenia. Nutrients 13, 3519 (2021).
Zhang, H. et al. Oxidative stress: roles in skeletal muscle atrophy. Biochem. Pharmacol. 214, 115664 (2023).
Tan, K. T., Ang, S. J. & Tsai, S. Y. Sarcopenia: tilting the Balance of Protein Homeostasis. Proteomics 20, e1800411 (2020).
Hurst, C. et al. Resistance exercise as a treatment for sarcopenia: prescription and delivery. Age Ageing 51, 2 (2022).
Lu, L. et al. Effects of different exercise training modes on muscle strength and physical performance in older people with sarcopenia: a systematic review and meta-analysis. BMC Geriatr. 21, 708 (2021).
Giacosa, A., Barrile, G. C., Mansueto, F. & Rondanelli, M. The nutritional support to prevent sarcopenia in the elderly. Front. Nutr. 11, 1379814 (2024).
McKendry, J. et al. Nutritional supplements to support resistance exercise in countering the sarcopenia of aging. Nutrients 12, 2057 (2020).
Robinson, S., Granic, A., Cruz-Jentoft, A. J. & Sayer, A. A. The role of nutrition in the prevention of sarcopenia. Am. J. Clin. Nutr. 118, 852–864 (2023).
Ryall, J. G., Schertzer, J. D. & Lynch, G. S. Attenuation of age-related muscle wasting and weakness in rats after formoterol treatment: therapeutic implications for sarcopenia. J. Gerontol. A Biol. Sci. Med. Sci. 62, 813–823 (2007).
Ferrando, B. et al. Allopurinol and its role in the treatment of sarcopenia. Rev. Esp. Geriatr. Gerontol. 49, 292–298 (2014).
Hong, Y., Lee, J. H., Jeong, K. W. & Choi, C. S. Amelioration of muscle wasting by glucagon-like peptide-1 receptor agonist in muscle atrophy. J. Cachexia Sarcopenia Muscle 10, 903–918 (2019).
Rolland, Y., Dray, C., Vellas, B. & Barreto, P. S. Current and investigational medications for the treatment of sarcopenia. Metabolism 149, 155597 (2023).
Takeda, T. et al. A new murine model of accelerated senescence. Mech. Ageing Dev. 17, 183–194 (1981).
Huang, Y. et al. Ferroptosis in a sarcopenia model of senescence accelerated mouse prone 8 (SAMP8). Int. J. Biol. Sci. 17, 151–162 (2021).
Derave, W., Eijnde, B. O., Ramaekers, M. & Hespel, P. Soleus muscles of SAMP8 mice provide an accelerated model of skeletal muscle senescence. Exp. Gerontol. 40, 562–572 (2005).
Baek, K. W. et al. Two types of mouse models for sarcopenia research: senescence acceleration and genetic modification models. J. Bone Metab. 28, 179–191 (2021).
Xie, W. Q. et al. Mouse models of sarcopenia: classification and evaluation. J. Cachexia Sarcopenia Muscle 12, 538–554 (2021).
Fagerberg, L. et al. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol. Cell Proteom. 13, 397–406 (2014).
Yin, L., Wu, S., Bai, P. & Wang, X. Combination of transcriptomics and proteomics for analyzing potential biomarker and molecular mechanism underlying skeletal muscle atrophy. J. Proteom. 309, 105283 (2024).
Sivakumar, P. et al. Integrated plasma proteomics and lung transcriptomics reveal novel biomarkers in idiopathic pulmonary fibrosis. Respir. Res. 22, 273 (2021).
Zhao, W. et al. The regulation of MFG-E8 on the mitophagy in diabetic sarcopenia via the HSPA1L-Parkin pathway and the effect of D-pinitol. J. Cachexia Sarcopenia Muscle 15, 934–948 (2024).
Sun, Y., Liu, B. W., Zhang, X. & Yin, F. Z. Relationships between the apolipoprotein levels and sarcopenia in inpatients with type 2 diabetes mellitus: a cross-sectional study. Diabetes Metab. Syndr. Obes. 17, 2085–2094 (2024).
Lee, H. J. et al. Sex differences in the association between sarcopenia and mild cognitive impairment in the older Korean population. BMC Geriatr. 23, 332 (2023).
Dai, H. et al. Inhibiting uptake of extracellular vesicles derived from senescent bone marrow mesenchymal stem cells by muscle satellite cells attenuates sarcopenia. J. Orthop. Transl. 35, 23–36 (2022).
Kersey, J. H. et al. P-24: a human leukemia-associated and lymphohemopoietic progenitor cell surface structure identified with monoclonal antibody. J. Exp. Med. 153, 726–731 (1981).
Lorico, A., Lorico-Rappa, M., Karbanová, J., Corbeil, D. & Pizzorno, G. CD9, a tetraspanin target for cancer therapy?. Exp. Biol. Med. (Maywood) 246, 1121–1138 (2021).
Li, Y. et al. CD9 exacerbates pathological cardiac hypertrophy through regulating GP130/STAT3 signaling pathway. iScience 26, 108070 (2023).
Brosseau, C., Colas, L., Magnan, A. & Brouard, S. CD9 tetraspanin: a new pathway for the regulation of inflammation?. Front. Immunol. 9, 2316 (2018).
Ondruššek, R. et al. Prognostic value and multifaceted roles of tetraspanin CD9 in cancer. Front. Oncol. 13, 1140738 (2023).
Vieiralves, R. R. & Favorito, L. A. Dapoxetine and premature ejaculation. Int. Braz. J. Urol. 49, 511–514 (2023).
Wu, Y. et al. Levomilnacipran improves lipopolysaccharide-induced dysregulation of synaptic plasticity and depression-like behaviors via activating BDNF/TrkB mediated PI3K/Akt/mTOR signaling pathway. Mol. Neurobiol. 61, 4102–4115 (2024).
Dungan, C. M., Gordon, B. S. & Williamson, D. L. Acute treadmill exercise discriminately improves the skeletal muscle insulin-stimulated growth signaling responses in mice lacking REDD1. Physiol. Rep. 7, e14011 (2019).
Yin, L. et al. Puerarin ameliorates skeletal muscle wasting and fiber type transformation in STZ-induced type 1 diabetic rats. Biomed. Pharmacother. 133, 110977 (2021).
Radonić, A. et al. Guideline to reference gene selection for quantitative real-time PCR. Biochem. Biophys. Res. Commun. 313, 856–862 (2004).
Mercorelli, P. Recent advances in intelligent algorithms for fault detection and diagnosis. Sens. Basel 24, 8 (2024).
Mercorelli, P. A fault detection and data reconciliation algorithm in technical processes with the help of Haar Wavelets packets. Algorithms 10, 13 (2017).
Mercorelli, P. Denoising and harmonic detection using nonorthogonal wavelet packets in industrial applications. J. Syst. Sci. Complex. 20, 325–343 (2007).
Acknowledgements
This work was financially supported by National Natural Science Foundation of China (82401854), Fundamental Research Program of Shanxi Province (202303021222315), The central government guides local funds for science and technology development (YDZJSX2024D069), Fund Program for the Scientific Activities of Selected Returned Overseas Professionals in Shanxi Province (20240054), Research Project Supported by Shanxi Scholarship Council of China (2024-144), 2024 Annual “Promising Candidates” Cultivation Project for National Natural Science Foundation at Shanxi Bethune Hospital (2024GZRZ22), and Research and Innovation Team Project for Scientific Breakthroughs at Shanxi Bethune Hospital (2024ZHANCHI09).
Author information
Authors and Affiliations
Contributions
L.Y. conceived and designed the experiments, performed the experiments, prepared figures, authored the paper, and approved the final draft; N.L., P.B., S.W. and X.W. performed the experiments, analyzed the data, and reviewed drafts of the paper; B.L. conceived and designed the experiments, reviewed and approved the final draft. All authors reviewed the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Yin, L., Li, N., Bai, P. et al. CD9, a novel potential biomarker of sarcopenia. Sci Rep 15, 19099 (2025). https://doi.org/10.1038/s41598-025-04331-7
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-04331-7