Abstract
Recent studies demonstrated the role of antisense long noncoding RNAs (AS lncRNAs) in regulating gene expression at the transcriptional or translational level. In this study, we assessed the effects of sirtuin 1 (SIRT1) AS lncRNA overexpression and inhibition, along with overexpression of miR-34a, on interleukin (IL)-1β-induced gene expression alterations in human chondrocytes, aiming to understand its role in human chondrocytes. We analyzed gene expression alterations using real-time PCR and assessed SIRT1 protein level alterations through western blotting. IL-1β stimulation significantly upregulated A disintegrin and metalloproteinase with thrombospondin motif 5 (ADAMTS-5) and matrix metalloproteinase 13 (MMP-13). SIRT1 AS lncRNA overexpression significantly inhibited IL-1β-induced upregulation of these genes, whereas SIRT1 AS lncRNA inhibition further increased their expression. Moreover, overexpression of miR-34a significantly increased IL-1β-induced upregulation of ADAMTS-5 and MMP-13 which were rescued by over expression of SIRT1 AS lncRNA. SIRT1 protein levels were significantly increased by SIRT1 AS lncRNA overexpression and significantly reduced by its inhibition. Ribonuclease protection assay indicated the complete binding of SIRT1 AS lncRNA to SIRT1 mRNA. In the osteoarthritis (OA) cartilage, SIRT1 AS lncRNA expression was significantly reduced compared with that in normal cartilage. Our observations indicate that the binding of SIRT1 AS lncRNA to SIRT1 mRNA may suppress IL-1β-induced expression of cartilage-degrading enzymes. Therefore, SIRT1 AS lncRNA may be a novel therapeutic target for OA treatment.
Similar content being viewed by others
Introduction
Osteoarthritis (OA) is a degenerative disease that affects human articular cartilage and is often observed in older populations. It causes joint pain and deformities, thereby impairing daily activities. During the development of OA, various pathological changes occur, including an imbalance between the anabolic and catabolic activities of chondrocytes, increased production of cartilage-degrading enzymes, including matrix metalloprotease (MMP) and A disintegrin and metalloproteinase with thrombospondin motifs (ADAMTS), and increased apoptosis1. Additionally, the pro-inflammatory cytokine, interleukin 1β (IL-1β), has been implicated in OA progression by upregulating proteolytic enzymes2.
Sirtuins are members of the class III histone deacetylase family that regulate diverse cellular activities associated with aging3. Silent information regulator 2 type 1 (SIRT1) is a sirtuin homolog that regulates various signaling pathways, such as DNA repair, myogenic and adipogenic differentiation, mitochondrial biogenesis, and glucose and insulin homeostasis4. Previous studies have demonstrated the role of SIRT1 in OA progression. In human chondrocytes, SIRT1 overexpression inhibits apoptosis and facilitates cartilage-specific gene expression, whereas SIRT1 inhibition regulates the expression of genes related to OA progression1,2,5. Additionally, Sirt1-knockout mouse models exhibited accelerated OA progression, whereas Sirt1-knockin mouse exhibited delayed OA progression1,6. Moreover, Takayama et al. reported that SIRT1 is involved in the apoptosis of human chondrocytes7. These studies demonstrate that SIRT1 may play a significant role in preventing OA progression.
Long noncoding RNAs (lncRNAs) are a diverse group of noncoding transcripts that exceed 200 nt and lack protein-coding capacity8. Recently, numerous lncRNAs have been identified that affect chondrocyte degradation and OA progression9,10,11,12. Among these, antisense lncRNAs (AS lncRNAs) represent a subgroup that is transcribed from the complementary DNA strand compared with sense transcripts, partially overlapping with sense RNA13. Complemental sequence of AS lncRNA to their sense RNA enables duplex formation to alter their stability and its transcription. AS lncRNAs regulate the expression of sense mRNA through several mechanisms, including transcription, RNA-DNA interactions, and nuclear and cytoplasmic RNA-RNA interactions14. SIRT1 AS lncRNA completely overlaps with the SIRT1 mRNA 3’ untranslated region (3’-UTR) through the base complementary paring principle15. In the previous studies, SIRT1 AS lncRNA increased the stability of SIRT1 mRNA by forming lncRNA-mRNA duplex and facilitated C2C12 cell proliferation, and interaction with micro RNAs is also reported16,17. Additionally, SIRT1 AS lncRNA is involved in various diseases by increasing SIRT1 abundance16,18,19,20,21. However, the association between SIRT1 AS lncRNA and chondrocyte degradation remains unclear. Therefore, this study aimed to assess the effects of SIRT1 AS lncRNAs on human chondrocytes.
Results
SIRT1 AS lncRNA was detected in NHAC-kn
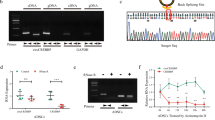
Initially, we assessed the presence of SIRT1 AS lncRNA in normal human articular chondrocytes with knee (NHAC-kn)-extracted RNA through real-time (RT)-PCR, using a strand-specific primer. Electrophoresis of the PCR products demonstrated distinct band formation, indicating the presence of SIRT1 AS lncRNA in NHAC-kn (Fig. 1, supplementary Fig. 1).
The overexpression of SIRT1 AS lncRNA inhibited IL-1β-induced MMP-13 and ADAMTS-5
We assessed the expression of SIRT1 AS lncRNAs using real-time qPCR (Fig. 2A) and confirmed its successful overexpression using an expression plasmid. The protein level of SIRT1 significantly increased, which was confirmed through western blotting (Fig. 2B, supplementary Fig. 2). Subsequently, we assessed the effect of SIRT1 AS lncRNA overexpression under IL-1β. IL-1β stimulation significantly upregulated MMP-13 and ADAMTS-5 mRNA levels. However, SIRT1 AS lncRNA overexpression significantly inhibited the upregulation of MMP-13 and ADATS-5 mRNA. In contrast, the overexpression of SIRT1 AS lncRNA had minimal effects on SRY-related HMG box-containing protein 9 (SOX9) and aggrecan expression. Similarly, the stimulation of IL-1β did not significantly affect SOX9 and aggrecan expression (Fig. 2C). The expression of SIRT1 mRNA was not affected by SIRT1AS RNA overexpression.
The effects of SIRT1 AS lncRNA overexpression. (A) Real-time PCR for sirtuin 1 antisense long noncoding RNA(SIRT1 AS lncRNA). The value is relative to the control expression level (n=3). (B) Western blotting for SIRT1. Representative data are demonstrated from repeated experiments. Relative expression of SIRT1 is demonstrated in the graph (n=5). (C) Real-time PCR for SIRT1,SRY-related HMG box-containing protein 9(SOX9), aggrecan,matrix metalloproteinase 13 (MMP-13), and A disintegrin and metalloproteinase with thrombospondin motif 5 (ADAMTS-5) mRNA (n=5). The values are relative to the control expression level. Data are represented as the dot plot of the individual data and median (n = 5); *p < 0.05,***p<0.001. Full Western blotting is shown in Supplementary Fig. 2.
The inhibition of SIRT1 AS lncRNA increased IL-1β-induced MMP-13 and ADAMTS-5 expression
To further confirm the role of SIRT1 AS lncRNA in regulating MMP-13 and ADAMTS-5 expression, we assessed the effects of SIRT1 AS lncRNA inhibition on the IL-1β-induced expression of MMP-13 and ADAMTS-5 mRNA. Small interfering RNA (siRNA)-induced inhibition of SIRT1 AS lncRNA was confirmed through real-time qPCR (Fig. 3A), and the protein level of SIRT1 was reduced (Fig. 3B, supplementary Fig. 3). The inhibition of SIRT1 AS lncRNA significantly upregulated IL-1β-induced MMP-13 and ADAMTS-5 expression and further downregulated SOX9 expression. The inhibition of SIRT1 AS lncRNA had a significant effect of aggrecan expression, and there was no significant effect on SIRT1 mRNA expression (Fig. 3C).
The effects of SIRT1 AS lncRNA inhibition. (A) Real-time PCR for sirtuin 1 antisense long noncoding RNA (SIRT1 AS lncRNA). The values are relative to the control expression level (n = 3). (B) Western blotting for SIRT1. Representative data are demonstrated from repeated experiments. The relative expression of SIRT1 is demonstrated in the graph (n = 5). (C) Real-time PCR for SIRT1, SRY-related HMG box-containing protein 9 (SOX9), aggrecan, matrix metalloproteinase 13 (MMP-13), and a disintegrin and metalloproteinase with thrombospondin motif 5 (ADAMTS-5) mRNA (n = 5). The values are relative to the control expression level. Data are represented as the dot plot of the individual data and median (n = 5); *p < 0.05, **p < 0.01. Full Western blotting is shown in Supplementary Fig. 3.
Overexpression of miR-34a increased IL-1β-induced MMP-13 and ADAMTS-5 expression, while the upregulation was rescued by overexpression of SIRT1 AS lncRNA
In order to further elucidate the mechanism of the regulation of OA-related gene expressions by SIRT1 AS lncRNA, we investigated the involvement of miR-34a in the regulation since miR-34a targets SIRT122 and has been suggested as one of the micro RNAs associated with progression of OA23,24,25. MiR-34a was overexpressed by transfection and the overexpression of miR-34a was confirmed by real-time qPCR (Fig. 4A). The overexpression of miR-34a significantly upregulated IL-1β-induced MMP-13 and ADAMTS-5 expression, while the upregulations were rescued by co-transfection with SIRT1 AS lncRNA. No significant effect of overexpression of miRNA-34a on SIRT1 mRNA expression was observed (Fig. 4B).
The effects of miR-34a and SIRT1 AS lncRNA overexpression. (A) Real-time PCR for miRNA-34a. The values are relative to the control expression level (n = 3). (B) Real-time PCR for SIRT1, SRY-related HMG box-containing protein 9 (SOX9), aggrecan, matrix metalloproteinase 13 (MMP-13), and a disintegrin and metalloproteinase with thrombospondin motif 5 (ADAMTS-5) mRNA (n = 5). The values are relative to the control expression level. Data are represented as the dot plot of the individual data and median (n = 5); *p < 0.05, **p < 0.01, ****p < 0.0001. mi, miR-34a; AS, SIRT1 AS lncRNA.
SIRT1 AS lncRNA increased the stability of SIRT1 mRNA
We assessed the possibility of RNA duplex formation using an RNase protection assay (RPA) on the RNA extracted from NHAC-kn cells. After applying RNase A that digests only single-stranded RNAs without affecting double-stranded RNAs, single-stranded RNAs are digested and only double-stranded RNAs are expected to be detected. Real-time PCR data of the present study have demonstrated that the overlapping regions of both transcripts were protected from degradation (Fig. 5A, B), indicating that SIRT1 AS lncRNA and SIRT1 mRNA forms an RNA duplex. NHAC-kn cells were then treated with actinomycin D to suppress transcription, and the loss of SIRT1 mRNA was analyzed. The overexpression of SIRT1 AS lncRNA significantly attenuated the decrease of SIRT1 mRNA at three and six hours after applying actinomycin D, suggesting that SIRT1 AS lncRNA plays an important role in regulating SIRT1 mRNA stability (Fig. 5C).
Ribonuclease protection assay (RPA) and stability assay for samples from normal human articular chondrocytes with knee (NHAC-kn). (A) Non-overlapping (P1) and overlapping (P2) primer positions for RPA. (B) Real-time PCR for the products from P1 and P2 primers with and without RNAse. The values are relative to the control expression level. Data are represented as the dot plot of the individual data and median (n = 5); **p < 0.01, ***p < 0.001. (C) RNA stability assay of SIRT1 mRNA expression using samples transfected with control plasmid or SIRT1 AS lncRNA. The values are relative to the control expression level. Data are represented as the mean ± standard deviation (SD) (n = 4); *p < 0.05.
SIRT1 AS lncRNA expression was reduced in OA chondrocytes
We assessed the relationship between SIRT1 AS lncRNA expression and OA chondrocytes. The expression of SIRT1 AS lncRNA was significantly reduced in chondrocytes obtained from patients with OA, compared to those obtained from patients without OA (Fig. 6).
Discussion
In this study, we observed that the overexpression of SIRT1 AS lncRNA inhibited the IL-1β stimulation-induced upregulation of MMP-13 and ADAMTS-5. In contrast, the suppression of SIRT1 AS lncRNA increased the IL-1β-induced expression of these genes. Moreover, overexpression of miR-34a significantly increased IL-1β-induced upregulation of these genes, which were rescued by overexpression of SIRT1 AS lncRNA. The expression of SIRT1 protein was increased by overexpressing SIRT1 AS lncRNA and reduced by suppressing SIRT1 AS lncRNA.
The loss of extracellular matrix caused by cartilage-degrading enzymes is a primary pathological feature of OA, and MMP-13 plays a crucial role in OA26. Additionally, ADAMTS-5 plays a major role in cartilage degradation in OA27. Therefore, the overexpression of SIRT1 protein resulting from SIRT1 AS lncRNA overexpression may contribute to a decline in the IL-1β-induced cartilage degradation. Additionally, our in vivo study revealed that the expression of SIRT1 AS lncRNA was reduced in OA chondrocytes compared with that in non-OA samples. This indicates an association between SIRT1 AS lncRNA and OA progression, highlighting the potential of SIRT1 lncRNA as a diagnostic marker for OA. Recent studies have demonstrated that lncRNAs, including AS lncRNAs, play a crucial role in the development of numerous diseases, including OA. Zhi et al. reported that Opa-interacting protein 5 (OIP5) AS lncRNA regulates chondrocyte apoptosis and inflammatory response by regulating microRNA (miR)−29b-3p/progranulin axis28. Lysyl oxidase-like 1 antisense RNA (LOXL1-AS) contributes to OA progression by targeting the miR-423-5p/lysine-specific demethylase 5 C (KDM5C) axis29. Additionally, X-inactive specific transcript (XIST), a lncRNA that regulates X-linked chromosomal inactivation, has been observed to be involved in the pathogenesis of OA by influencing macrophage polarization and certain miRNAs30,31. Metastasis associated in lung adenocarcinoma transcript-1 (MALATA1) has been shown to regulate key inflammatory signaling pathways, such as the NF-κB and MAPK pathways. Its expression is significantly upregulated in OA chondrocytes, and has been shown to promote OA progression32. Other numerous lncRNAs have been found to be related to OA pathology32,33,34. However, this is the first study to assess the association between SIRT1 AS lncRNAs and gene expression in human chondrocytes.
Our data further demonstrated that SIRT1 AS lncRNA directly binds to SIRT1 mRNA and upregulates SIRT1 protein expression without altering SIRT1 mRNA expression. AS lncRNAs regulate the expression of sense mRNA through several mechanisms, including transcription, RNA-DNA interactions, and nuclear and cytoplasmic RNA-RNA interactions14. Previous studies on AS lncRNAs, such as beta-site amyloid precursor protein cleaving enzyme 1 (BACE1)-antisense, type II mesothelial keratin k7 (KRT7)-antisense, and fibroblast growth factor receptor 3 (FGFR3)-antisense, have demonstrated interactions with their sense target genes (BACE1, KRT7, and FGFR3, respectively) to exert their functions35,36,37. Wang et al. reported that the overexpression of SIRT1 AS lncRNA in C2C12 cells upregulated the SIRT1 protein level without affecting the level of Sirt1 mRNA. However, a study has reported that the overexpression of SIRT1 AS lncRNA has upregulated both SIRT1 mRNA and protein levels simultaneously in RLE-6TN cells21. Although the reason for the contradictory results is unclear, it is possible that the regulatory mechanism for SIRT1 AS lncRNA on SIRT1 mRNA and SIRT1 protein levels is different depending on the cell type. The results of the present study demonstrated that overexpression of SIRT1 AS lncRNA influenced mainly the translational output of SIRT1 protein without significantly influenced on the level of SIRT1 mRNA. Therefore, the main regulatory mechanism of SIRT1 AS lncRNA in chondrocytes might be regulation of its SIRT1 protein stability by forming duplex with SIRT1 mRNA. Additionally, Wang et al. demonstrated that SIRT1 AS lncRNA interacted with Sirt1 mRNA forming an RNA duplex and increased its stability to facilitate SIRT1 translation by competing with miR-34a21 since the target sequence of miR-34a to SIRT1 mRNA was in the overlapping region with SIRT1 AS lncRNA. Endisha et al. reported that the expression of miR-34a-5p, a mature type of miR-34a was found to be increased in the cartilage from patients with an advanced OA and demonstrated its pivotal role in progression of OA using mouse loss- and gain-of-function models24. In the present study, overexpression of miR-34a increased the IL-1β-induced expression of MMP-13 and ADAM TS5, which were rescued by the overexpression of SIRT1 AS lncRNA. These findings showed that SIRT1 AS lncRNA plays a protective role on OA progression partially through competing against miR-34a, preventing SIRT1 mRNA degradation.
The high target specificity of AS lncRNAs, which normally regulate one or a small group of related genes, makes them suitable targets for therapeutic intervention. Recently, a growing number of disease-relevant AS lncRNAs have been discovered, specifically in the field of oncology and degenerative disease, including OA. AS lncRNAs are garnering attention in cancer research because of their diagnostic and therapeutic implications. For example, a minimum of 24 solid tumors are associated with aberrant HOX transcript antisense intergenic RNA (HOTAIR) expression and function, and an increased HOTAIR expression may act as a biomarker for cancer diagnosis, metastasis, therapeutic resistance, and unfavorable survival38,39. Animal models studies have demonstrated the efficacy of using siRNA against HOTAIR in ceasing cancer cell growth and invasion. Ozes et al. developed a peptide-nucleic acid hybrid that blocks HOTAIR’s binding with enhancer of zeste homolog 2 (EZH2), which enhanced the survival of mice with platinum-resistant ovarian tumor40,41. Integrating AS lncRNAs into clinical practice, whether through RNAi-based therapy or CRISPR Cas9 gene editing, there are significant hurdles, including the challenges of drug delivery, cytotoxicity, and off-target effects42.
This study has some limitations. First, this study was primarily conducted in vitro using NHAC-kn. The effect of SIRT1 AS lncRNAs was not tested in OA chondrocytes. Therefore, further in vitro and in vivo studies are necessary to elucidate the effect of SIRT1 AS lncRNAs on OA progression. Histological changes of cartilages should be investigated to truly clarify its effect on OA pathology. Second, although we have identified that SIRT1 AS lncRNA increases the gene expression of cartilage-degrading enzymes by upregulating SIRT1 protein levels, other factors, such as transcription or splicing factors remain unclear. These questions should be addressed in future studies in order to develop a new treatment. Third, the sample size for in vivo comparison of OA and non-OA was relatively small. More robust investigation was ideal. Fourth, the RNA expression changes induced by IL-1β were not consistent throughout the study, although there were some trends toward increasing catabolic markers. Small sample size could have affected the results. Our study demonstrates the possibility of SIRT1 AS lncRNA as a diagnostic marker or therapeutic target in OA pathology. Further studies are required for its clinical application.
Materials and methods
Culture of human chondrocytes
NHAC-kn cells (Lonza Walkersville, Inc., Walkersville, MD, USA), isolated from the knee joints of healthy donors, were maintained following the manufacture’s protocol. Chondrocytes between passages three and five were used for the analysis.
Detection of SIRT1 AS lncRNA in NHAC-kn
After NHAC-kn cells reached 70–80% confluency, total RNA was isolated using a RNeasy Kit (Qiagen, Valencia, CA, USA), following the manufacturer’s protocol. Subsequently, cDNA was reverse transcribed from total RNA (1 µg) using SuperScript IV Reverse Transcriptase (Invitrogen). For the reverse transcription of SIRT1 AS lncRNA, a strand-specific primer was used, whereas random primers were used for glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and SIRT1. PCR amplification was conducted in a PCR apparatus (Bio-Rad) at the cycling conditions of 94 ℃ (45 s), 55 ℃ (30 s), and 72 ℃ (30 s) for 30 cycles, and the PCR products were visualized on a 2% agarose gel. Primers for strand-specific primer and GAPDH and SIRT1 were obtained from Invitrogen (Table 1).
Overexpression of SIRT1
A plasmid DNA (pcDNA) vector that expresses SIRT1 AS lncRNA was synthesized by Invitrogen. Chondrocytes were seeded in 6-well plates (3 × 105 cells/well) and transfected with 2.5 µg of SIRT1 AS lncRNA or pcDNA-empty vector through lipofection, following the manufacturer’s protocol (Lipofectamine 3000; Invitrogen).
Inhibition of SIRT1 AS lncRNA
Chondrocytes were transfected with 25 pmol siRNA or control non-silencing siRNA targeting SIRT1 AS lncRNA. The sequence of guide strand is as follows: 5’-UCUUUAUGCAUAAAACACCCA-3’ (Invitrogen). Cells were seeded in 6-well plates (3 × 105 cells/well) and transfected through lipofection, following the manufacturer’s protocol (Lipofectamine RNAiMAX; Invitrogen).
Overexpression of miR-34a
Chondrocytes were transfected with 25 pmol of miR-34a mimic (mirVana, Invitrogen), pcDNA-empty vector, or both miR-34a mimic and SIRT1 AS lncRNA vector. Cells were seeded in 6-well plates (3 × 105 cells/well) and miR-34a was transfected through lipofection, following the manufacturer’s protocol (Lipofectamine RNAiMAX; Invitrogen).
Stimulation with IL-β
Following a 48-hour transfection, the NHAC-kn cells were treated with 10 ng/mL IL-1β (R&D Systems) for 24 h. Subsequently, RNA and proteins were extracted for further experiments.
Western blotting
Treated NHAC-kn cells were lysed on ice for 30 min in a lysis buffer supplemented with protease and phosphatase inhibitors (Roche Diagnostics, Basel, Switzerland). The separated proteins were electrically transferred onto blotting membranes (Amersham Biosciences, Arlington Heights, IL, USA). The membranes were probed with primary antibodies followed by a horseradish peroxidase (HRP)-conjugated secondary antibody. Proteins were visualized using ECL Plus reagent (Amersham Biosciences) and chemiluminescent analyzer (LAS-3000 mini; Fujifilm, Tokyo, Japan). Band intensities were quantified using the ImageJ software, version 1.53 (NIH; National Institutes of Health, Bethesda, MD, USA). The following antibodies were used: monoclonal anti-β actin (Sigma, St. Louis, MO., USA, product#: A5441, diluted 1:5000), polyclonal anti-human SIRT1 (Bioss, Boston, MA., USA, product#: bs-0921R, diluted 1:2000), and HRP-conjugated anti-rabbit (Cytiva, Tokyo, Japan, product#: NA934VS, diluted 1:5000) and anti-mouse IgG (Cytiva, product#: NA931V, diluted 1:5000).
Real-time PCR
RNA was isolated using a RNeasy Kit (Qiagen, Valencia, CA, USA), following the manufacturer’s protocol. Subsequently, cDNA was reverse transcribed from total RNA (1 µg) using SuperScript IV Reverse Transcriptase (Invitrogen). For the reverse transcription of SIRT1 AS lncRNA, a strand-specific primer was used, whereas random primers were used for other PCR products. Real-time qPCR was performed using Power SYBRTM Green PCR Master Mix (Applied Biosystems, Waltham, MA, USA) and ABI Prism 7700. For the analysis of miR-34a, RNA was isolated using a mirVana miRNA Isolation Kit (Invitrogen), following the manufacturer’s protocol. Subsequently, cDNA was reverse transcribed from total RNA (1 µg) using TaqMan Advanced miRNA cDNA Synthesis Kit (Applied Biosystems). Real-time qPCR was performed using TaqMan Gene Expression Mater Mix (Applied Biosystems). TaqMan MicroRNA Assay for miR-34a and U6 were used (Applied Biosystems). GAPDH served as a housekeeping control to normalize the gene expression using the ΔΔ cycle threshold (ΔΔCt) method. U6 served as a control for miR-34a. Each sample was analyzed in duplicate. In this study, SIRT1 AS lncRNA, SIRT1, SOX9, aggrecan, ADAMTS-5, and MMP-13 (Invitrogen) were assessed. ADAMTS-5 and MMP-13 has been chosen as catabolic cartilage markers, which has been shown to be major catabolic factors and play critical roles in development of OA in the previous studies1,2,6. Aggrecan is a novel anabolic cartilage marker, which has also been used frequently in the previous studies related to OA progression5,6. SOX9 is a major transcription factor during cartilage development, and it is a downstream of NF-κB signaling pathway in OA, and its expression has been shown to be decreased in OA cartilage43. Since previous study has shown that SIRT1 plays important role in NF-κB pathway, by deacetylation of p65, which is a member of NF-κB, SOX9 has also been chosen as an anabolic marker in order to assess the effect of SIRT1 AS lncRNA and SIRT1 mRNA on OA progression. Primer sequences are listed in Table 1.
OA and non-osteoarthritic (non-OA) cartilage sampling and processing
OA cartilage tissues were obtained from the femoral condyle of patients undergoing total knee arthroplasty for primary OA (two males and one female; mean age: 72.4 years). All the patients were Kellgren Lawrence grade three or four. The patients with inflammatory diseases were excluded. Non-OA articular cartilage tissues were obtained from patients undergoing surgery for femoral neck fracture (one male and two females; mean age: 76.4 years). None of the patients exhibited evident progressive OA alterations. Chondrocytes were isolated and cultured in flasks. After reaching 80% confluency, RNA was extracted using a RNeasy Kit (Qiagen). All OA and non-OA cartilage samples were obtained in accordance with the World Medical Association Declaration of Helsinki Ethical Principles for Medical Research Involving Human Subjects. This study was approved by our institutional review board, and all patients provided written informed consent.
Ribonuclease protection assay (RPA)
To detect the sense-antisense RNA duplex, ribonuclease protection assay (RPA) and real-time qPCR analyses were performed on total RNA isolated from untreated NHAC-kn cells, according to previous studies19,44. Isolated RNA was digested with RNase A (Applied Biosystems) to eliminate all single-stranded RNAs. Subsequently, cDNA synthesis and real-time qPCR were performed as described above.
RNA stability assay
In order to investigate the stability of SIRT1 mRNA, NHAC-kn cells were transfected with either control plasmid or SIRT1 AS lncRNA plasmid. Cells were then treated with 2 µg/ml actinomycin D (Invitrogen), which suppresses RNA transcription, as reported in the previous study19. The cells were harvested at 0, 1, 3, 6 h after the treatment. Total RNA was extracted to assess the residual mRNAs by real-time qPCR. GAPDH mRNA was used as the internal control, since its expression is stable for 32 h.
Statistical analysis
All values were expressed as the dot plot of individual experiment and median, unless otherwise described. Comparisons between multiple groups were made using one-way analysis of variance (ANOVA), and Fisher’s protected least significant difference test was used as a post-hoc test; p-values < 0.05 were considered significant. Comparisons between the two groups were performed using the Mann-Whitney U test. Data were analyzed using BellCurve for Excel (Social Survey Research Information Co., Ltd., Tokyo, Japan).
Data availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
References
Matsuzaki, T. et al. Disruption of Sirt1 in chondrocytes causes accelerated progression of osteoarthritis under mechanical stress and during ageing in mice. Ann. Rheum. Dis. 73, 1397–1404 (2014).
Matsushita, T. et al. The overexpression of SIRT1 inhibited Osteoarthritic gene expression changes induced by interleukin-1β in human chondrocytes. J. Orthop. Res. 31, 531–537 (2013).
Deng, Z. et al. The role of Sirtuin 1 and its activator, Resveratrol in osteoarthritis. Biosci Rep 39 (2019).
Hubbard, B. P. & Sinclair, D. A. Small molecule SIRT1 activators for the treatment of aging and age-related diseases. Trends Pharmacol. Sci. 35, 146–154 (2014).
Fujita, N. et al. Potential involvement of SIRT1 in the pathogenesis of osteoarthritis through the modulation of chondrocyte gene expressions. J. Orthop. Res. 29, 511–515 (2011).
Yamamoto, T. et al. Knee osteoarthritis progression is delayed in silent information regulator 2 ortholog 1 knock-in mice. Int J. Mol. Sci 22 (2021).
Takayama, K. et al. SIRT1 regulation of apoptosis of human chondrocytes. Arthritis Rheum. 60, 2731–2740 (2009).
Savary, G. et al. The long noncoding RNA DNM3OS is a reservoir of fibromirs with major functions in lung fibroblast response to TGF-β and pulmonary fibrosis. Am. J. Respir Crit. Care Med. 200, 184–198 (2019).
Pearson, M. J. et al. Long intergenic noncoding RNAs mediate the human chondrocyte inflammatory response and are differentially expressed in osteoarthritis cartilage. Arthritis Rheumatol. 68, 845–856 (2016).
Steck, E. et al. Regulation of H19 and its encoded microRNA-675 in osteoarthritis and under anabolic and catabolic in vitro conditions. J. Mol. Med. 90, 1185–1195 (2012).
Chen, X. et al. Correlation analysis of differentially expressed long non-coding RNA HOTAIR with PTEN/PI3K/AKT pathway and inflammation in patients with osteoarthritis and the effect of Baicalin intervention. J. Orthop. Surg. Res. 18, 34 (2023).
Liu, Q. et al. Long noncoding RNA related to cartilage injury promotes chondrocyte extracellular matrix degradation in osteoarthritis. Arthritis Rheumatol. 66, 969–978 (2014).
Khorkova, O. et al. Natural antisense transcripts as drug targets. Front. Mol. Biosci. 9, 978375 (2022).
Faghihi, M. A. & Wahlestedt, C. Regulatory roles of natural antisense transcripts. Nat. Rev. Mol. Cell. Biol. 10, 637–643 (2009).
Liu, J., Wu, W. & Jin, J. A novel mutation in SIRT1-AS leading to a decreased risk of HCC. Oncol. Rep. 34, 2343–2350 (2015).
Wang, G. Q. et al. Sirt1 AS LncRNA interacts with its mRNA to inhibit muscle formation by attenuating function of miR-34a. Sci. Rep. 6, 21865 (2016).
Wang, Y. et al. Identification, stability and expression of Sirt1 antisense long non-coding RNA. Gene 539, 117–124 (2014).
Mokhberian, N. et al. Sirt1 antisense transcript is down-regulated in human tumors. Mol. Biol. Rep. 46, 2299–2305 (2019).
Li, B. et al. Sirt1 antisense long noncoding RNA promotes cardiomyocyte proliferation by enhancing the stability of Sirt1. J. Am. Heart Assoc. 7, e009700 (2018).
Lou, Z. et al. LncRNA Sirt1-AS upregulates Sirt1 to attenuate aging related deep venous thrombosis. Aging 13, 6918–6935 (2021).
Qian, W., Cai, X. & Qian, Q. Sirt1 antisense long non-coding RNA attenuates pulmonary fibrosis through sirt1-mediated epithelial-mesenchymal transition. Aging 12, 4322–4336 (2020).
Yamakuchi, M., Ferlito, M. & Lowenstein, C. J. miR-34a repression of SIRT1 regulates apoptosis. Proc. Natl. Acad. Sci. U S A. 105, 13421–13426 (2008).
Abouheif, M. M. et al. Silencing microRNA-34a inhibits chondrocyte apoptosis in a rat osteoarthritis model in vitro. Rheumatol. (Oxford). 49, 2054–2060 (2010).
Endisha, H. et al. MicroRNA-34a-5p promotes joint destruction during osteoarthritis. Arthritis Rheumatol. 73, 426–439 (2021).
Liu, H. et al. MicroRNA expression in osteoarthritis: a meta-analysis. Clin. Exp. Med. 23, 3737–3749 (2023).
Mukherjee, A. & Das, B. The role of inflammatory mediators and matrix metalloproteinases (MMPs) in the progression of osteoarthritis. Biomater. Biosyst. 13, 100090 (2024).
Verma, P. & Dalal, K. ADAMTS-4 and ADAMTS-5: key enzymes in osteoarthritis. J. Cell. Biochem. 112, 3507–3514 (2011).
Zhi, L. et al. Downregulation of LncRNA OIP5-AS1 induced by IL-1β aggravates osteoarthritis via regulating miR-29b-3p/PGRN. Cartilage 13, 1345S–1355S (2021).
Chen, K., Fang, H. & Xu, N. LncRNA LOXL1-AS1 is transcriptionally activated by JUND and contributes to osteoarthritis progression via targeting the miR-423-5p/KDM5C axis. Life Sci. 258, 118095 (2020).
Li, L., Lv, G., Wang, B. & Kuang, L. XIST/miR-376c-5p/OPN axis modulates the influence of Proinflammatory M1 macrophages on osteoarthritis chondrocyte apoptosis. J. Cell. Physiol. 235, 281–293 (2020).
Liu, Y. et al. Long non-coding RNA XIST contributes to osteoarthritis progression via miR-149-5p/DNMT3A axis. Biomed. Pharmacother. 128, 110349 (2020).
Hu, K. et al. Deciphering the role of LncRNAs in osteoarthritis: inflammatory pathways unveiled. J. Inflamm. Res. 17, 6563–6581 (2024).
Shakeri, M. et al. Unraveling the molecular landscape of osteoarthritis: a comprehensive review focused on the role of non-coding RNAs. Pathol. Res. Pract. 260, 155446 (2024).
Zhang, X. et al. The emerging role of LncRNAs in osteoarthritis development and potential therapy. Front. Genet. 14, 14:1273933 (2023).
Huang, B. et al. Long non-coding antisense RNA KRT7-AS is activated in gastric cancers and supports cancer cell progression by increasing KRT7 expression. Oncogene 35, 4927–4936 (2016).
Faghihi, M. A. et al. Expression of a noncoding RNA is elevated in alzheimer’s disease and drives rapid feed-forward regulation of beta-secretase. Nat. Med. 14, 723–730 (2008).
Sun, J. et al. Long noncoding RNA FGFR3-AS1 promotes osteosarcoma growth through regulating its natural antisense transcript FGFR3. Mol. Biol. Rep. 43, 427–436 (2016).
Heubach, J. et al. The long noncoding RNA HOTAIR has tissue and cell type-dependent effects on HOX gene expression and phenotype of urothelial cancer cells. Mol. Cancer. 14, 108 (2015).
Gupta, R. A. et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 464, 1071–1076 (2010).
Kishikawa, T. et al. Circulating RNAs as new biomarkers for detecting pancreatic cancer. World J. Gastroenterol. 21, 8527–8540 (2015).
Özeş, A. R. et al. Therapeutic targeting using tumor specific peptides inhibits long non-coding RNA HOTAIR activity in ovarian and breast cancer. Sci. Rep. 7, 894 (2017).
Rehman, S. U. et al. Recent insights into the functions and mechanisms of antisense RNA: emerging applications in cancer therapy and precision medicine. Front. Chem. 11, 1335330 (2023).
Tian, B., Zhang, L., Zheng, J. & Kang, X. The role of NF-κB-SOX9 signalling pathway in osteoarthritis. Heliyon 10, e37191 (2024).
Ogawa, Y., Sun, B. K. & Lee, J. T. Intersection of the RNAi and X-inactivation pathways. Published as: Science.
Acknowledgements
We would like to thank Editage (www.editage.jp) for English language editing.
Author information
Authors and Affiliations
Contributions
T. T. and Ta. M. contributed to the study’s conception, design, acquisition, analysis, data interpretation, and article writing. Ky. N., Ka. N., N. K., Y. H., To. M., and R. K. participated in data acquisition and interpretation. All authors contributed to the critical revisions of the article, ensuring its significant intellectual integrity, and approved the final version.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Tokura, T., Matsushita, T., Nishida, K. et al. SIRT1 antisense long noncoding RNA attenuates interleukin-1β-induced osteoarthritic gene expression in human chondrocytes through its mRNA interaction. Sci Rep 15, 23338 (2025). https://doi.org/10.1038/s41598-025-06565-x
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-06565-x
Keywords
This article is cited by
-
The protective role of dexmedetomidine against anaesthetics-induced neurotoxicity through downregulating miR-34a
Journal of NeuroVirology (2025)