Abstract
Anopheles stephensi, an invasive vector is aggressively expanding its geographic range across Africa, posing significant threat to malaria control. Surveillance of its spread is crucial for mitigating its impact on malaria transmission. Here, we report for the first time incursion of An. stephensi into Gayi, a rural area in southern Niger Republic. A combination of morphological identification, end point PCR and sequencing of fragments of COXI and ITS2 genes confirmed An. stephensi. This species was found resting indoor together with other mosquitoes, including An. coluzzii, An. gambiae s.s., and An. arabiensis. Entomological parameters, including resting densities, human blood index and biting rate, Plasmodium falciparum parasite rate, and entomological inoculation rate were described for the above Anopheles species. The finding of An. stephensi sympatric with the above major malaria vectors in Niger highlights the urgent need for intensified surveillance to develop evidence-based approaches to prevent further spread of this pervasive vector.
Similar content being viewed by others
Introduction
Anopheles stephensi, an invasive malaria vector originally from South Asia and the Arabian Peninsula, is expanding its geographic range across Africa, posing significant threat to malaria control and pre-elimination. Crossing from the Arabian Peninsula1 it was first detected in 2012 in Djibouti2 following an unusual spike in malaria infection. Next, it was reported in Ethiopia in 20163, Sudan in 20164,5,6, Somalia and Somaliland from 20196,7, possibly Nigeria in 20206, Ghana in 20228 and Kenya in 20229. The encroachment of this pervasive species into African continent is a significant cog to the progress made in malaria control, as it is highly plastic - breeding in artificial containers, construction site waters, etc., in the cities, while in rural areas it breeds in streams, catch basins, wells, domestic water-storage containers, etc10,11. The unpredictability of its seasonality based on rainfall1,12 which complicates timing of control measures at peak transmission windows; propensity to blood feed on both humans and animals11; its Plasmodium falciparum and P. vivax transmission capabilities6; as well as its multiple resistance towards the public health insecticides (e.g., the pyrethroids, carbamates, organophosphate and organochloride)13,14,15,16 makes it a formidable challenge to control efforts.
Niger Republic grapples with malaria, ranking third highest globally in malaria mortality (6%), behind Nigeria and Democratic Republic of the Congo17; with highly resistant, native African malaria vectors from An. gambiae s.l. Complex and An. funestus Group documented18,19,20 and evidence of clinical infections with P. falciparum, P. vivax, P. malariae and P. ovale in some sites21. Here, we report the first identification of An. stephensi, in a rural area in Niger Republic, in sympatry with other mosquitoes including An. coluzzii, An. gambiae s.s., An. arabiensis, Culex quinquefasciatus and Aedes aegypti – all caught resting indoor. We also describe entomological and parasitological parameters of transmission from the above Anopheles species, and identified the An. stephensi individuals to species level using PCR amplification, sequencing and analysis of the fragments of cytochrome oxidase I (COXI) and internal transcribed spacer region 2 (ITS2) genes.
Results
Mosquito genus composition
A total of 220 mosquitoes were collected indoors. Among these, 184 (83.64%; 95% CI: 78.75–88.52%) belonged to the Anopheles genus morphologically, 35 (15.91%; 95% CI: 11.08–20.74%) to Culex, and 1 (0.45%; 95% CI: 0.00–1.34%) to Aedes genera (Supplementary Table S1). Within the Anopheles group, 171 (92.93%; 95% CI: 89.23–96.64%) were females (Supplementary Table S1), with 122 blood-fed (71.35%; 95% CI: 64.57–78.12%) and 49 unfed (28.65%; 95% CI: 21.88–35.43%).
Morphological identification of mosquito species
Morphological keys were utilised to differentiate Anopheles mosquitoes from the non-Anopheles, and An. stephensi from An. gambiae s.l. mosquitoes. Eight (8) specimens were morphologically identified as An. stephensi, exhibiting distinct physical characteristics, including the presence of three light bands on the maxillary palps, as well as two characteristic light interruptions in the second main dark area of wing vein 1 (Fig. 1a, b and c). Typical environmental conditions and habitat structures of mosquito collection sites in Gayi village are illustrated in Fig. 1d to f.
Morphological identification of An. stephensi in Gayi, Niger Republic and sampling site characteristics. (a). sample of An. stephensi suspect showing characteristic maxillary palpus with the two broad apical bands, indicated by the white arrows; (b). An. stephensi suspect showing the second major dark area on wing vein 1, characterized by two pales interruptions, indicated by the white arrows; (c). comparison of body sizes between An. gambiae and An. stephensi from Gayi; (d)., (e). and (f). examples of domestic animal pen, mud housing structures and mud bricks making site near a pond, highlighting the environmental features of Gayi.
Identification of Anopheles mosquitoes to molecular level and analysis of genetic diversity of An. stephensi
SINE200 PCR of 155 F₀ females An. gambiae s.l. confirmed 129 as An. coluzzii (79% of all Anopheles, including An. stephensi samples), and 13 each as An. arabiensis and An. gambiae s.s. (8% each) (Supplementary Fig. 1).
Amplification of COXI from 8 An. stephensi suspects provided a characteristic band of about 710 bp fragments (Fig. 2a), from which 7 were sequenced successfully and utilised for NCBI BLASTn identification. The top hit was found to be 614 bp SM147 COXI sequences (Accession OM801708, see Table 1), from An. stephensi described in Ethiopia22 with 99% query cover and sharing 98.15% identity with six Gayi samples.
Agarose gel electrophoretic image showing the results of amplification of fragments of COXI and ITS2 from Anopheles species. (a). COXI gel showing a diagnostic band at 710 bp. Lanes 1 and 17: Hyperladder 100 DNA ladder [Bioline 100–1013 bp]; Lane 2 and 16: Negative controls; Lane 3–10: An. stephensi; Lane 11, 12, 13, 14 and 145 An. coluzzii, An. arabiensis, and An. gambiae s.s., Culex, and Ae. aegypti, respectively. (b). ITS2 gel showing An. stephensi diagnostic band at 438 bp (lanes 2–9) from simplex PCR with primers st-F and UD2-R only; Lane 1: Hyperladder 100 DNA ladder [Bioline 100–1013 bp]; lanes 11 and 25: Hyperladder 1 kb [Bioline 200–10037]; lane 10: Negative control; lanes 12–24: gel showing variable control bands (> 600 bp) using st-F, U5.8 S-F, and UD2-R primers which targets a fragment common to arthropods; lanes 12–19: An. stephensi amplicons; lanes 20–24: An. coluzzii, An. arabiensis, and An. gambiae s.s., Culex and Ae. aegypti.
Phylogenetic analysis was conducted by comparing COXI sequences from Niger to other An. stephensi sequences (from Ethiopia, Kenya, Sudan, Somaliland, Yemen, India, United Arab Emirates and India) and sequences from other 11 Anopheles species (Fig. 3a). Analysis of the polymorphism patterns of COXI fragments (709 bp) revealed relative homogeneity, with haplotypes forming a separate cluster in the maximum likelihood phylogenetic tree. The sequences exhibit a close relationship to An. stephensi sequences from African countries, particularly from Ethiopia and Somaliland, with significant bootstrap suggesting possible common origin. The COXI fragments are polymorphic, with 3 haplotypes from 7 sequences and 21 polymorphic sites (Fig. 3b and c). Haplotype diversity is relatively high (Hd = 0.66) and nucleotide diversity (π) were computed as 0.01. A neutrality test of all sequences revealed Tajima’s D (−0.937) and Li and Fu’s D* (−0.814) as negative, but not statistically significant.
Amplification of ITS2 from 8 An. stephensi suspects provided a characteristic ~ 438 bp fragments (Fig. 2b), from which 7 were sequenced successfully and utilised for NCBI BLASTn identification. The topmost hit, 100% identical to 5 sequences were haplotypes 5–8 of ITS2 (Accession: MW676295, see Table 1) from India23. The ITS2 sequences were compared to other sequences of An. stephensi from 13 countries, and five sequences from other Anopheles species (Fig. 3d). Niger ITS2 sequences were more homogenous than COXI sequences from the same mosquitoes, with their haplotypes forming a cluster in the maximum likelihood phylogenetic tree. They exhibit close relationship to An. stephensi sequences from Myanmar, India, followed by African countries. Also, the ITS2 fragments are less polymorphic, with 3 haplotypes from 7 sequences and only 4 polymorphic sites (Fig. 3e and f). Haplotype diversity (Hd) and nucleotide diversity (π) were computed as 0.52 and 0.002, respectively. A neutrality test of all sequences revealed Tajima’s D (−1.43) and Li and Fu’s D* (−1.50) as negative, but not statistically significant.
Pattern of the genetic variability and polymorphism of Niger An. stephensi COXI and ITS2 fragments. (a). maximum likelihood phylogenetic tree of COXI sequences; (b). COXI sequences showing haplotypes and their respective frequencies, as well as (c). polymorphic sites with respect to the major haplotype (Hap_1). (d). A maximum likelihood phylogenetic tree of ITS2 sequences; (e). ITS2 sequences showing haplotypes and their respective frequencies, as well as (f). polymorphic sites with respect to the major haplotype (Hap_1).
Estimation of entomological indicators of malaria transmission
A total of 44 out of 66 female Anopheles have fed on humans (66%), translating into human blood index of 0.66 (Table 2). These include 38 An. coluzzii, 5 An. arabiensis, 2 An. stephensi, and 1 An. gambiae s.s. The rest of the mosquitoes have fed on cows (8 An. coluzzii and 1 An. arabiensis) and goats (7 An. coluzzii and 4 An. arabiensis). Indoor resting density (IRD) was calculated as 9 female Anopheles per house, 1.10 female Culex per house and 0.05 female Ae. aegypti per house. Human biting rate (HBR) was estimated to be 2.21 bites/person/night. Six (6) An. coluzzii mosquitoes out of 122 were found infected with P. falciparum, which translate to a parasite rate of 0.0492 (~ 4.92%). All the other Anopheles were negative for Plasmodia species. From a single day of collection, entomological inoculation rate (EIR) was estimated to be 10.87 bites/person/night.
Discussion
Anopheles stephensi is becoming more invasive, clear and present danger with the ability to disrupt the trajectory of malaria epidemiology and control efforts in Africa. Understanding its spread, bionomics and response to the frontline major vector control tools, such as insecticide treated bed nets, is crucial to mitigating its additive impact on malaria transmission in Africa. Indeed, modelling study has suggested that 126 million people in cities in Africa could be at risk from this new malaria vector if it were to spread unchecked1,6,24 with the WHO’s Global Malaria Programme urging member states, especially those in and around affected countries, to take immediate action against the threat posed by this invasive species24. The WHO has launched an initiative in 2022 to stop the spread of An. stephensi in Africa, with a five-pronged approach, including strengthening entomological and epidemiological surveillance and improving information exchange6.
Our findings of An. stephensi in a rural area is in agreement with the previous observations that these species can be found in both rural and urban settings13,14,25 though the prevailing expectation is that it thrives in urban settings1,13,14,25. Anopheles stephensi is known to exhibit high resistance to multiple classes of public health insecticides commonly used in vector control15,16,26. The finding of this species in Niger is particularly concerning because vector control strategies rely heavily on pyrethroid-based control tools, against An. gambiae complex and An. funestus species, and the incursion of An. stephensi will complicate control efforts. In contrast to previous studies which describe higher zoophilic tendencies in An. stephensi27 we caught human-blood fed females resting indoor, indicating anthropophilic tendencies. We found 8 An. stephensi females resting indoor, two of them fed on humans, a finding which is consistent with the previous observation of An. stephensi (type form) being anthropophilic (preferring to take blood meals from humans), endophilic (preferring to rest indoor) and endophagic (preferring to feed indoors)28 even though it will bite outdoors during the warmer summer months due to greater outdoor activity of humans and domestic animals11. However, our above observations are limited by the low number of the captured An. stephensi mosquitoes. This low number could possibly be due to its recent introduction into Gayi. Also, it is possible that there were An. stephensi among the larvae collected but which died before reaching adulthood, possibly due to our inexperience in raising this species in the insectarium.
Anopheles stephensi larvae were not found in domestic water storage containers, pots, earthen ware containers, and outdoor breeding sites, mud block making sites and the pond in Gayi. This is in contrast to several, recent studies across Africa, in which larvae of this species were found, e.g., in Kenya9 Ethiopia3 Somaliland7 Sudan29 and Ghana8. The absence of larvae can be explained by low density, cryptic tendencies, and possibly by our inability to raise to adulthood larvae of this species which might have been caught in the sampled houses.
Analysis of genetic diversity using COXI barcode gene fragments suggests that the Niger population might have originated from Semera, Eastern Ethiopia22 a region not far from Djibouti and Eritrea. Haplotype analysis revealed striking resemblance to the parameters obtained from a previous multi-site study in Sudan29. For example, our COXI haplotype diversity of 0.66 is close to the 0.609 described in the above Sudanese populations. Likewise, we found a total of 21 polymorphic sites, which is comparable to the 20 polymorphic sites obtained in the above Sudanese population. It is possible that the Gayi An. stephensi have arrived from Ethiopia via Sudan and Chad. This is not farfetched, because since 2021 presence of An. stephensi has been established in Sudan along the borders of six countries previously assumed as free, including Chad, Egypt, Eritrea, Libya, Republic of Central Africa, and South Sudan29. The negative Tajima’s D, is probably due to founding event following recent invasion, and the low nucleotide diversity supports the idea of a founder effect (a few individuals seeding the Gayi An. stephensi population), following a recent invasion from a small genetic pool. However, this needs to be confirmed with larger sample number and a more refined genomics approaches such as whole genome sequencing.
Conclusion
The invasive malaria mosquito An. stephensi has encroached into Niger Republic, a high burden country suffering from extreme malaria-related mortalities, and may spread across the Sahel if efforts are not invested to contain it. In Gayi, it exists in sympatry with other native malaria vectors, including An. gambiae s.s., An. coluzzii and An. arabiensis, leading to high biting and entomological inoculation rates, consistent with the high infection documented in the year 2024. There is an urgent need for enhanced surveillance, targeted vector control strategies, and cross-border collaboration to mitigate the potential spread of this malaria mosquito and its impact on malaria transmission in the Sahel and other regions neighbouring Niger and other parts of Africa.
Methods
Study site and sampling
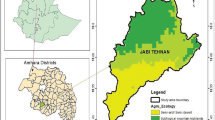
Our team from the Ecology, Health, and Environment (ECOSEN) group, Université André Salifou, Zinder, embarked on a study of biodiversity of mosquitoes (Diptera: Culicidae) in Gayi (13.2402° N, 9.3499 °E), Wacha commune, Magaria Department, Zinder Region (Fig. 4) on 18 of August 2024. Gayi, a Sahelian settlement, characterised with seasonal malaria transmission is located 82 km southeast of Zinder, about 100 km from the border of Nigeria.
The project was approved by the Scientific Council of Université André Salifou, Zinder-Niger (Ref: N°000792 June 13, 2023).
The total population of Gayi was 32,709 inhabitants in 2024. Agriculture, livestock, and local trade constitute the primary livelihood activities, with flux of humans and animals between the locality and border towns in Nigeria. Female blood fed, unfed, and male, adult mosquitoes, resting indoor were collected from 19 houses, early in the morning (6:00–8:00 am) in Gayi using Improved Prokopack battery-operated aspirators (John. W. Hock, Florida, USA). During this month, Gayi had experienced a total of 1,927 reported malaria cases and an average monthly rainfall of 146.9 mm. Environmental conditions on the day of collection were: temperature of 29 °C, wind speeds of 2.14 km/h, and relative humidity of 64.06%. The mosquitoes were transported in temperature-regulated boxes and maintained on 10% sugar at 25 °C ± 2 and 75% relative humidity, before being sorted.
Morphological identification of mosquitoes genus
Mosquitoes were individually identified using morphological keys: for Anopheles gambiae complex30; for An. stephensi31, for Culex genera32 and for Aedes genera33.
Anopheles species identification to molecular levels
SINE 200 PCR identification of an. gambiae s.l. Complex mosquitoes
Genomic DNA (gDNA) was extracted from single legs each from 155 female adult An. gambiae s.l. mosquitoes using the LIVAK protocol34. For mosquitoes belonging to the Anopheles gambiae complex, species identification was performed using SINE200 PCR35.
COXI and ITS2 PCR identification of An. stephensi
Legs from eight (8) female mosquitoes morphologically identified as An. stephensi were also gDNA-extracted. Universal primers LCO1490 and HC0219836 were utilised to amplify a fragment of the mitochondrial COXI gene using KAPA Taq DNA polymerase (KAPA Biosystems, Wilmington, MA, USA). The PCR reaction mix (15 µL) included 1 µL of gDNA, 1.5 µL of 10× Taq Buffer A, 0.4 mM (0.5 µL) each of forward and reverse primers, 1.25 mM (0.75 µL) MgCl2, 0.84 mM (0.5 µL) dNTP mix, and 0.2 µL KAPA Taq DNA polymerase (KAPA Biosystems, Wilmington, MA, USA), with the final volume adjusted using ddH2O. The thermal cycling conditions included an initial denaturation step of 2 min at 94 °C, followed by 35 cycles each of 30 s at 94 °C (denaturation), 30 s at 55 °C (primer annealing), and 30 s at 72 °C (extension). A final extension step of 10 min at 72 °C concluded the reactions. PCR products were resolved on a 1.5% agarose gel stained with peqGREEN, and visualised.
The same gDNA samples were also used for amplification of fragments of ITS2 of ribosomal DNA (rDNA) using size diagnostic PCR primers st-F and UD2-R, which amplify 438 bp An. stephensi–specific diagnostic amplicon, and primers 5.8 S-F and UD2-R which amplify a universal amplicon of varying sizes (> 600 bp) depending upon the length of ITS2 in a particular species37. Details of PCR mix and thermocycling conditions were as above. In addition, for both COXI and ITS2 amplifications gDNA from An. coluzzii, An. arabiensis, and An. gambiae s.s., Culex, and Aedes aegypti were include in the PCRs for comparison.
The PCR products were purified using the QIAquick® Gel Extraction Kit (QIAGEN, Hilden, Germany), ligated into the pJET1.2/blunt cloning vector (ThermoFisher Scientific, Waltham, MA, USA) and transformed into E. coli DH5α cells. Minipreps were prepared using the QIAprep® Spin Miniprep Kit (QIAGEN, Hilden, Germany) and sequenced using pJET1.2/blunt forward and reverse sequencing primers. Sequence trace files were manually examined using BioEdit version 7.7.138. Genetic diversity parameters, including the number of haplotypes (h), haplotype diversity (Hd), the number of polymorphic sites (S), and nucleotide diversity (π), were computed using DnaSP version 6.12.0339. Final sequences were used for BLASTn searches, in NCBI to retrieve previously published sequences from An. stephensi and other Anopheles mosquitoes sharing high similarities from GenBank. Multiple sequence alignments were carried out using CLC Sequence Viewer version 6.9 (http://www.clcbio.com/), and different haplotypes were compared by constructing maximum likelihood phylogenetic trees with MEGA XI40 using Kimura 3 parameter model, with 500 replicates bootstrapping. For outgroup, An. implexus (GQ165788) was used for COXI and An. sawadwongporni (JQ446420) for ITS2, respectively.
Estimation of entomological parameters of transmission
Entomological parameters were calculated, following the procedures outlined by the WHO41. Indoor resting density (IRD) was calculated from the number of Anopheles caught relative to the number of houses sampled. Human biting rate (HBR) was estimated for sampled rooms by dividing the number of mosquitoes captured by the number of occupants sleeping in the room.
To establish anthropophilic index, a set of blood fed females were dissected, heads/thoraces and abdomens used for gDNA extraction using DNeasy Blood and Tissue Kit (QIAGEN, Hilden, Germany) according to manufacturer’s protocol. The gDNA from 66 abdomens were utilised for PCR to detect source of blood. Human blood index was established from the proportion of Anopheles that have fed on humans relative to the total number of blood fed female Anopheles caught, using a cocktail PCR of Kent and Norris42.
A total of 122 gDNA from head/thoraces were tested for Plasmodium infection using nested PCR protocol targeting 18 S rRNA43. Parasite rate was calculated as percentage of mosquitoes with Plasmodium relative to the total number of the females tested. Entomological Inoculation Rate (EIR), which estimates the number of infectious bites per person per night, was calculated by multiplying the mean HBR by the parasite rate41.
Data availability
All sequences generated in this study have been deposited in NCBI GenBank under accession numbers: PV200295-PV200301 for COXI and PV208146-PV208152 for ITS2. The folders for raw sequences of the above gene fragments have been deposited in Zenodo for access and re-use (https://zenodo.org/records/15013332).
References
Sinka, M. E. et al. A new malaria vector in Africa: Predicting the expansion range of Anopheles stephensi and identifying the urban populations at risk. Proc. Natl. Acad. Sci. 117, 24900–24908 (2020).
Faulde, M. K., Rueda, L. M. & Khaireh, B. A. First record of the Asian malaria vector Anopheles stephensi and its possible role in the resurgence of malaria in djibouti, Horn of Africa. Acta Trop. 139, 39–43 (2014).
Carter, T. E. et al. First detection of Anopheles stephensi liston, 1901 (Diptera: culicidae) in Ethiopia using molecular and morphological approaches. Acta Trop. 188, 180–186 (2018).
Ahmed, A., Khogali, R., Elnour, M. A. B., Nakao, R. & Salim, B. Correction to: Emergence of the invasive malaria vector Anopheles stephensi in Khartoum state, central Sudan. Parasit. Vectors. 14, 562 (2021).
Ahmed, A., Khogali, R., Elnour, M. A. B., Nakao, R. & Salim, B. Emergence of the invasive malaria vector Anopheles stephensi in Khartoum state, central Sudan. Parasit. Vectors. 14, 511 (2021).
WHO WHO initiative to stop the spread of Anopheles stephensi. In Africa, 2023 Update Vol. 4 (World Health Organization, 2023).
Ali, S., Samake, J. N., Spear, J. & Carter, T. E. Morphological identification and genetic characterization of Anopheles stephensi in Somaliland. Parasit. Vectors. 15, 247 (2022).
Afrane, Y. A. et al. First detection of Anopheles stephensi in Ghana using molecular surveillance. bioRxiv 2023.12.01.569589. https://doi.org/10.1101/2023.12.01.569589 (2023).
Ochomo, E. et al. Detection of Anopheles stephensi mosquitoes by molecular surveillance, Kenya. Emerg. Infect. Dis. J. 29, 2498 (2023).
Sinka, M. E. et al. Correction: The dominant Anopheles vectors of human malaria in the americas: occurrence data, distribution maps and bionomic précis. Parasit. Vectors. 4, 210 (2011).
Sinka, M. E. et al. The dominant Anopheles vectors of human malaria in the americas: occurrence data, distribution maps and bionomic précis. Parasit. Vectors. 3, 1–26 (2010).
Whittaker, C. et al. Seasonal dynamics of Anopheles stephensi and its implications for mosquito detection and emergent malaria control in the Horn of Africa. Proc. Natl. Acad. Sci. 120, e2216142120 (2023).
Balkew, M. et al. An update on the distribution, bionomics, and insecticide susceptibility of Anopheles stephensi in ethiopia, 2018–2020. Malar. J. 20, 263 (2021).
Balkew, M. et al. Correction to: An update on the distribution, bionomics, and insecticide susceptibility of Anopheles stephensi in ethiopia, 2018–2020. Malar. J. 20, 331 (2021).
Samake, J. N., Yared, S., Hassen, M. A., Zohdy, S. & Carter, T. E. Insecticide resistance and population structure of the invasive malaria vector, Anopheles stephensi, from fiq, Ethiopia. Sci. Rep. 14, 27516 (2024).
Yared, S. et al. Insecticide resistance in Anopheles stephensi in Somali region, Eastern Ethiopia. Malar. J. 19, 180 (2020).
WHO. World Malaria Report 2023 (World Health Organization, 2023).
Ibrahim, S. S. et al. High Plasmodium infection and multiple insecticide resistance in a major malaria vector Anopheles coluzzii from Sahel of Niger Republic. Malar. J. 18, 181 (2019).
Labbo, R. et al. Longitudinal follow-up of malaria transmission dynamics in two villages in a Sahelian area of Niger during a nationwide insecticide-treated Bednet distribution programme. Med. Vet. Entomol. 26, 386–395 (2012).
Moustapha, L. M. et al. First identification of Microsporidia MB in Anopheles coluzzii from Zinder city, Niger. Parasit. Vectors. 17, 39 (2024).
Garba, M. N. et al. Circulation of Non-falciparum species in niger: implications for malaria diagnosis. Open. Forum Infect. Dis. ofae474. https://doi.org/10.1093/ofid/ofae474 (2024).
Waymire, E. et al. Wolbachia 16S rRNA haplotypes detected in wild Anopheles stephensi in Eastern Ethiopia. Parasit. Vectors. 15, 178 (2022).
Mishra, S., Sharma, G., Das, M. K., Pande, V. & Singh, O. P. Intragenomic sequence variations in the second internal transcribed spacer (ITS2) ribosomal DNA of the malaria vector Anopheles stephensi. PLOS ONE. 16, e0253173 (2021).
Samarasekera, U. A missed opportunity? Anopheles stephensi in Africa. Lancet 400, 1914–1915 (2022).
de Santi, V. P. et al. Role of Anopheles stephensi mosquitoes in malaria outbreak, djibouti, 2019. Emerg. Infect. Dis. 27, 1697–1700 (2021).
Acford-Palmer, H. et al. Genome wide population genetics and molecular surveillance of insecticide resistance in Anopheles stephensi mosquitoes from Awash Sebat Kilo in Ethiopia. Sci. Rep. 15, 16443 (2025).
Thomas, S. et al. Resting and feeding preferences of Anopheles stephensi in an urban setting, perennial for malaria. Malar. J. 16, 111 (2017).
Alomar, A. A. & Alto, B. W. Anopheles stephensi Liston, 1901 (Insecta: Diptera: Culicidae). EDIS 2022 (2023).
Abubakr, M. et al. The phylodynamic and spread of the invasive Asian malaria vectors, Anopheles stephensi, in Sudan. Biology 11, 409 (2022).
Coetzee, M. Key to the females of Afrotropical Anopheles mosquitoes (Diptera: Culicidae). Malar. J. 19, 70 (2020).
Das, B. P., Rajagopal, R. & Akiyama, J. Pictorial key to the species of Indian anophline mosquitoes. Zoology 2, 131–162 (1990).
Reuben, R., Tewari, S. C., Hiriyan, J. & Akiyama, J. Illustrated keys to species of Culex (Culex) associated with Japanese encephalitis in Southeast Asia. Mosq. Syst. 26, 075–096 (1994).
Rueda, L. M. Pictorial keys for the identification of mosquitoes (Diptera: Culicidae) associated with Dengue Virus Transmission. Zootaxa 589 (2004).
Livak, K. J. Organization and mapping of a sequence on the Drosophila melanogaster X and Y chromosomes that is transcribed during spermatogenesis. Genetics 107, 611–634 (1984).
Santolamazza, F. et al. Insertion polymorphisms of SINE200 retrotransposons within speciation Islands of Anopheles gambiae molecular forms. Malar. J. 7, 163 (2008).
Folmer, O., Hoeh, W., Lutz, R. & Vrijenhoek, R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar. Biol. Biotechnol 294–299 (1994).
Singh, O. et al. Molecular tools for early detection of invasive malaria vector Anopheles stephensi mosquitoes. Emerg. Infect. Dis. J. 29, 36 (2023).
Hall, T. A. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. in vol. 41, pp 95–98 (Oxford, 1999).
Rozas, J. et al. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 34, 3299–3302 (2017).
Tamura, K., Stecher, G. & Kumar, S. MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38, 3022–3027 (2021).
WHO. A Toolkit for Integrated Vector Management in Sub-Saharan Africa (World Health Organization, 2016).
Kent, R. J., Thuma, P. E., Mharakurwa, S., Norris, D. E. & Seasonality Blood feeding behavior, and transmission of Plasmodium falciparum by Anopheles arabiensis after an extended drought in Southern Zambia. Am. J. Trop. Med. Hyg. 76, 267–274 (2007).
Snounou, G. et al. High sensitivity of detection of human malaria parasites by the use of nested polymerase chain reaction. Mol. Biochem. Parasitol. 61, 315–320 (1993).
Funding
The field work associated with the paper was sponsored by UAS Zinder and other expenses, including molecular analyses, were privately sponsored by S.S.I.
Author information
Authors and Affiliations
Contributions
L. M. M., and S. S. I., conceived and designed this study. N. H., T. M. M., I. Y. L., and A. D. M., carried out the field collection. L. M. M., N.H., T. M. M., I. Y. L., and A. D. M., conducted morphological identification. N. H., M. M. M., A. M., H. K. E., Y. Y. A., and S. A., carried out molecular analyses. L. M. M., and S. S. I., carried out data analyses and wrote the manuscript with contributions from H. S., E. A., M. L. I., and M. D. All authors read and approved the last version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval
This study involves the collection and analysis of mosquito samples only. The project was approved by the Scientific Council of Université André Salifou, Zinder, Niger Republic (Ref: N°000792 June 13, 2023).
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Moustapha, L.M., Sanda, AN.H., Mukhtar, M.M. et al. First detection and entomological characterisation of invasive malaria vector Anopheles stephensi in sympatry with other vectors in Gayi Southern Niger Republic. Sci Rep 15, 22647 (2025). https://doi.org/10.1038/s41598-025-07389-5
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-07389-5