Abstract
The mechanism underlying the role of trefoil factor family 3 (TFF3) in intestinal metaplasia remains unclear. This study reveals the molecular mechanism by which TFF3, in the process of gastric mucosal epithelial cell intestinal metaplasia (IM) induced by high salt, activates the JAK2/STAT3/CDX2 pathway, providing a potential target for the occurrence of IM. An in vitro model of high salt-induced intestinal metaplasia was established using bioinformatics to screen the GEO dataset for significantly differentially expressed genes related to intestinal metaplasia. The gastric epithelial cell line GES-1 was cultured in high-salt medium, and changes in cell function and the expression of TFF3, JAK2, STAT3, and CDX2 were examined following TFF3 knockdown or overexpression. Subsequent experiments disrupted the TFF3-JAK2/STAT3-CDX2 pathway to assess its effects on gene expression and cell function. The expression of TFF3 is upregulated during intestinal metaplasia, which promotes cell proliferation and migration. TFF3 regulates the expression of JAK2, STAT3, and CDX2 and activates the JAK2/STAT3 pathway to induce CDX2 expression in gastric epithelial cells, leading to intestinal metaplasia. Functional assays revealed that the TFF3-JAK2/STAT3-CDX2 pathway enhances both cell proliferation and migration. TFF3 induces intestinal metaplasia in gastric epithelial cells through the JAK2/STAT3-CDX2 pathway, providing new insights into the underlying mechanism and therapeutic strategies for intestinal metaplasia.
Similar content being viewed by others
Introduction
Gastric cancer remains one of the most prevalent and deadly malignancies worldwide, ranking fifth in incidence and fourth in mortality as of 20201. The Correa cascade model describes the progressive stages of gastric cancer development and reveals the relationships among gastric mucosal inflammation, atrophy, intestinal metaplasia, and tumor formation, with intestinal metaplasia being a critical factor in the progression to gastric cancer. Research has demonstrated that IM serves as an independent risk factor for gastric cancer. For every 10,000 individuals, the annual occurrence of gastric cancer in those with IM is approximately 12.4 cases2. Consequently, investigating the critical pathways involved in IM holds substantial importance. The precise mechanisms underlying intestinal metaplasia remain elusive, while some studies suggest an elevated risk associated with prolonged high-salt diets3. This paper focuses on the molecular mechanisms of intestinal metaplasia in gastric epithelial cells under high-salt conditions.
Most current models of gastric epithelial cell intestinal metaplasia are induced by bile acids4,5, with no established high-salt-induced models. This study revealed significant upregulation of CDX2 expression in GES-1 gastric epithelial cells cultured in high-salt media (80 mM NaCl) for 24 h. CDX2 is an intestine-specific nuclear transcription factor that is rarely expressed in the gastric mucosa. Ectopic expression of CDX2 is closely associated with intestinal metaplasia. Additionally, the expression of intestinal-specific markers such as KLF4, MUC2, and VIL1 also increased, indicating the successful establishment of a high-salt-induced intestinal metaplasia model.
By utilizing the GEO database to identify genes associated with IM, LASSO and SVM-RFE model analyses were used to identify DEGs, including TFF3, FABP1, CCL25, MUC2, GIP, ITLN1, and MTTP. Among these genes, TFF3, FABP1, and CCL25 were upregulated in the established model, with TFF3 being the most significantly expressed. Given the limited research on the role of TFF3 in gastric epithelial cell intestinal metaplasia, this study investigated TFF3.
IM is a significant precancerous condition associated with gastric cancer6, and its underlying molecular mechanisms remain incompletely understood. In recent years, TFF3 has gained attention as a marker for IM7, while abnormal activation of the JAK2/STAT3 signaling pathway has been linked to the progression of various malignancies8. Nevertheless, there is currently no direct evidence demonstrating that TFF3 contributes to the development of IM by modulating the JAK2/STAT3/CDX2 pathway. Research indicates that the positive expression rate of TFF3 in gastric cancer cases is 42.7%, and it serves as an independent prognostic factor for survival in patients with gastric cancer9. Despite reports on the role of the JAK2/STAT3 pathway in gastric cancer10, it remains unclear whether TFF3 influences the onset and progression of gastric cancer through regulation of this pathway.While TFF3 can act through the JAK2/STAT3 signaling pathway in malignancies8, its relationship with gastric epithelial cell intestinal metaplasia is not yet clear. This study revealed that TFF3, JAK2, STAT3, and CDX2 expression was upregulated in a high-salt diet-induced model, suggesting that TFF3 may induce CDX2 expression through the JAK2/STAT3 pathway, thereby inducing intestinal metaplasia in gastric epithelial cells and offering new perspectives for the treatment and prevention of IM.
Materials and methods
Intestinal metaplasia cell model construction
Cells were cultured in media supplemented with various concentrations of NaCl (20 mM, 40 mM, 80 mM, or 100 mM) for different durations (4 h, 8 h, 12 h, 24 h, or 48 h). Following incubation, cell RNA was extracted, and PCR was conducted to determine the expression levels of intestinal markers.
Cell culture
GES-1 cells were obtained from the Digestive Laboratory of Xuzhou Medical University. TFF3 overexpression plasmids were purchased from Youbao Biology. siRNAs, including siTFF3, siJAK2-1876, siJAK2-1542, siJAK2-2649, siSTAT3-450, siSTAT3-930, siSTAT3-1208, siCDX2-404, siCDX2-548, and siCDX2-738, were acquired from Suzhou GenePharma Biotech Co., Ltd. TFF3, JAK2, STAT3, and CDX2 antibodies were obtained from Abcam, USA.
GES-1 cells were cultured in DMEM, passaged when they reached approximately 90% confluence, and maintained in an incubator at 37 °C with 5% CO2.
Bioinformatics analysis
Three intestinal metaplasia datasets (GSE60662, GSE78523, GSE106656) were obtained from the Gene Expression Omnibus (GEO) database, consisting of 27 IM samples and 33 normal gastric mucosa controls (detailed sample metadata provided in Supplementary Table S1). Raw microarray data were normalized using Robust Multi-array Average (RMA) to minimize inter-dataset batch effects.
Differentially expressed genes (DEGs) between IM tissues and normal controls were identified using stringent thresholds of |log2FC| ≥ 1 and adjusted P-value < 0.05 (Benjamini-Hochberg correction), with low-expression genes filtered out (mean CPM > 1 in ≥ 50% samples). The top 47 DEGs ranked by statistical significance were visualized through volcano plots generated with ggplot2 and heatmaps created using pheatmap.
For feature selection, we employed both LASSO regression implemented via the glmnet R package with 10-fold cross-validation (selecting the optimal λ value to minimize overfitting while retaining predictive features) and SVM-RFE executed using the caret package with a radial basis kernel (iteratively eliminating the lowest-weight 10% of features until convergence to the final gene subset).
qRT‒PCR experiment
Cellular RNA was extracted and reverse transcribed. The expression levels of various genes were detected using specific primers, and the data were analyzed using the 2−ΔΔCT method. The sequences of primers used are listed in Table 1.
Western blot analysis
Total protein was extracted from the cells, and the protein concentration was determined using a BCA protein assay kit. Protein samples were prepared and subjected to SDS-PAGE, followed by membrane transfer, blocking, antibody incubation, and finally detection and imaging. Antibody sources are detailed in Table S2.
Plasmid transformation, amplification, and extraction
The DNA plasmid intended for amplification was introduced into competent DH5α cells. The mixture was kept on ice for 25 min, heat-shocked at 42 °C for 45 s and immediately transferred back to ice for 2 min. Sterile, antibiotic-free LB medium (700 µl) was added, and the culture was shaken for 1 h. After centrifugation, a portion of the bacterial suspension was resuspended and plated on solid LB media. Following sufficient growth, single colonies were picked for amplification, and plasmids were extracted according to the kit instructions.
Transient cell transfection
An appropriate number of cells were seeded in culture dishes. Transfection with siRNA was performed when the cell density reached 30–50%, and transfection with plasmids was performed when coverage was 70–80%, following the instructions of the Lipo2000 transfection reagent. The siRNA sequences used are listed in Table 2.
Immunohistochemistry
Pathological sections of normal gastric tissue and intestinal metaplasia were obtained from the Department of Pathology, Affiliated Hospital of Xuzhou Medical University (Ethics Approval: No. XYFY2024-KL017-01). The fixed samples were dehydrated, cleaned, and degreased before antigen retrieval using heat-induced epitope retrieval. Primary and secondary antibodies were incubated, followed by washing and drying. Images were acquired using a microscope.
Scratch assay
Cells were seeded into a 6-well plate and cultured for 24 h until 80% confluence was reached. A sterilized ruler was used to create a scratch with a 200 µl pipette tip through the cell monolayer. After the cells were washed with PBS and photographed under a microscope, 2 ml of fresh medium was added, and the cells were incubated for an additional 24–48 h. Changes in the scratch area were measured using ImageJ software.
Cell proliferation assay
Each well of a 96-well plate was filled with 100 µl of cell suspension. Six replicate wells were established for both the control and experimental groups. The cells (8 × 103 cells/well in 96-well plates) were cultured for 4, 8, 12, 24, or 48 h. Afterward, the culture medium was discarded, and 100 µl of fresh medium containing CCK8 reagent (10 µl of CCK8 reagent added to 100 µl of medium) was added. The plate was incubated for 1–4 h before the OD was measured at 450 nm to plot cell proliferation curves.
Statistical analysis
ImageJ, GraphPad Prism 9.4.1 and SPSS 27.0 software were used for data processing. The results are presented as the mean ± standard deviation (SD). Normality and homogeneity of variance tests were performed first. Differences between two groups were compared using two-tailed Student’s t tests, while differences among multiple groups were analyzed using one-way ANOVA. The significance level α was set at 0.05, with P < 0.05 considered to indicate statistical significance.
Results
Identification of the gastric epithelial cell intestinal metaplasia model
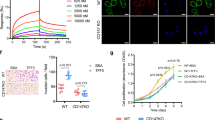
GES-1 cells were treated with varying concentrations of NaCl (20–100 mM), followed by RNA extraction and PCR to detect intestinal marker expression. The highest expression levels of CDX2, VIL1, KLF4, and MUC2 were observed after treatment with 80 mM NaCl for 24 h (Fig. 1A,B), indicating that NaCl induced the expression of CDX2 and downstream intestinal markers. This effect was comparable to that in the bile acid-induced gastric epithelial cell intestinalization model (Fig. 1C,D), with 80 mM NaCl treatment for 24 h being the optimal condition for high salt-induced intestinal marker expression.
Construction of the gastric mucosal epithelial cell intestinalization model. (A) Expression levels of CDX2, VIL1, KLF4, and MUC2 after 24 h treatment with different NaCl concentrations (20, 40, 80, and 100 mM) in GES-1 cells; (B) Expression levels after treatment with 80 mM NaCl for varying durations (4 h, 8 h, 12 h, 24 h, and 48 h); (C) Expression levels after 24 h treatment with different concentrations of CDCA (50, 62.5, 75, 87.5, and 100 µM); (D) Expression levels after treatment with 100 µM CDCA for varying durations (4 h, 8 h, 12 h, and 24 h). CDCA Chenodeoxycholic acid, NC Negative control, ns not significant; *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
TFF3 selection
Intestinal metaplasia datasets (GSE60662, GSE78523, and GSE106656) were filtered from the GEO database, and DEGs were identified. A heatmap (Fig. 2A) and volcano plot (Fig. 2B) were generated. Further selection by LASSO (Fig. 2C) and SVM-RFE (Fig. 2D) machine learning models yielded an intersection of seven DEGs: TFF3, FABP1, CCL25, MUC2, GIP, ITLN1, and MTTP. TFF3, FABP1, and CCL25 were significantly upregulated in intestinalized cells (Fig. 2E). In the established high-salt-induced intestinalization model, PCR analysis revealed significant upregulation of TFF3 (Fig. 2F) and CCL25 (Fig. 2G) compared to those in normal gastric epithelial cells, while FABP1 was not induced. TFF3, the most differentially expressed gene, was selected for subsequent experiments.
Differential gene expression in intestinalization (A) Heatmap of differential gene expression; (B) Volcano plot of upregulated and downregulated genes in intestinalization; (C) LASSO regression for differential gene selection; (D) SVM regression for differential gene selection; (E) Venn diagram of the seven differential genes common to both LASSO and SVM-RFE in intestinalization; (F,G) qRT-PCR detection of TFF3 (F) and CCL25 (G) expression levels in gastric epithelial and high-salt-induced intestinalized cells. **P < 0.01, ****P < 0.001.
TFF3 expression in various gastric specimens
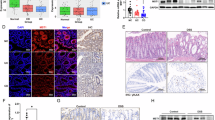
To investigate TFF3 expression patterns in clinical gastric specimens, we analyzed its distribution in normal gastric mucosa versus intestinal metaplasia tissues. Consistent with theoretical expectations, TFF3 expression was negligible in normal gastric epithelium but markedly elevated in IM lesions, particularly within goblet cells and other secretory cell populations. Immunohistochemical analysis demonstrated significant TFF3 upregulation in intestinal metaplasia tissues (Fig. 3), underscoring its clinical relevance to IM pathogenesis and highlighting its potential as a therapeutic target for precancerous intervention.
TFF3 expression in gastric tissues. (A) Representative immunohistochemical images of TFF3 in normal and intestinal metaplasia tissues. Scale bar: 50 μm; (B) Quantitative analysis of representative images; (C) Quantitative analysis of TFF3 immunohistochemistry results in 15 cases of normal and intestinal metaplasia. ***P < 0.001, ****P < 0.0001.
TFF3 promotes cell proliferation and migration
CCK8 assays showed that high-salt treatment significantly enhanced the proliferation of GES-1 gastric epithelial cells compared to the control. Elevated TFF3 expression under high-salt conditions further increased proliferation, while reduced TFF3 decreased it (Fig. 4A–C). Scratch assays also demonstrated that high-salt treatment improved cell migration, which was further enhanced by TFF3 overexpression and diminished by TFF3 knockdown (Fig. 4D–F).
Impact of NaCl and TFF3 on cell proliferation and migration. (A) Line graph of CCK8 results; (B,C) Analysis and group difference statistics of the effects of high-salt and overexpression or knockdown of TFF3 on GES-1 cell proliferation; (D) Wound healing assay for cell migration; (E,F) Ratios of migration were calculated. *P < 0.05, **P < 0.01, ***P < 0.001.
TFF3 regulates CDX2 expression via the JAK2/STAT3 pathway
Upregulation of TFF3, JAK2, STAT3, and CDX2 following high-salt treatment
To investigate the mechanistic role of TFF3 in intestinal differentiation, we hypothesized that TFF3 modulates CDX2 expression through the JAK2/STAT3 signaling pathway. Compared with control cells, GES-1 cells treated with 80 mM NaCl for 24 h exhibited increased expression of TFF3, JAK2, STAT3, and CDX2 (Fig. 5A). Knockdown of TFF3 in the high-salt-induced intestinal model decreased the expression of JAK2, STAT3, and CDX2, whereas TFF3 overexpression increased the expression of these genes (Fig. 5B–E). These results indicate that TFF3 regulates the expression of these genes.
To further elucidate the interplay between JAK2, STAT3, and CDX2, gene knockdown and overexpression studies were conducted. The silencing efficiency of siRNAs targeting JAK2, STAT3, and CDX2 was validated in the high-salt-induced GES-1 cell model, with siJAK2-1542, siSTAT3-450, and siCDX2-404 showing the highest efficiency (Fig. 6). These sequences were selected for subsequent experiments.
JAK2 modulates the expression of STAT3 and CDX2
The TFF3, JAK2, STAT3, and CDX2 genes were significantly upregulated in GES-1 cells following high-salt treatment; this effect was further amplified by TFF3 overexpression under high-salt conditions. Knockdown of JAK2 under high-salt and TFF3 overexpression conditions did not alter TFF3 expression but reduced STAT3 and CDX2 levels (Fig. 7A–D). Conversely, TFF3 knockdown reversed the downregulation of JAK2, STAT3, and CDX2 expression (Fig. S1).
STAT3 regulates CDX2 expression
In GES-1 cells, high-salt treatment alone or combined with TFF3 overexpression led to increased expression of TFF3, JAK2, STAT3, and CDX2. Moreover, STAT3 knockdown under these conditions did not affect TFF3 or JAK2 but reduced STAT3 and CDX2 expression (Fig. 8A–D). Moreover, overexpression of STAT3 significantly reversed the downregulation of STAT3 and CDX2 expression induced by TFF3 knockdown (Fig. S2).
Neither the knockdown nor the overexpression of CDX2 affects TFF3, JAK2, or STAT3 expression
TFF3, JAK2, STAT3, and CDX2 expression increased in GES-1 cells after high-salt treatment. CDX2 knockdown under high-salt and TFF3 overexpression conditions did not significantly alter the expression of TFF3, JAK2, or STAT3 (Fig. 9A–D). Moreover, overexpression of CDX2 did not significantly reverse the downregulation of JAK2 and STAT3 caused by TFF3 knockdown (Fig. S3).
TFF3 modulates CDX2 expression via JAK2/STAT3 pathway
To confirm the presence of the TFF3-JAK2/STAT3-CDX2 axis, protein samples from various cell groups were collected for Western blot analysis. The results showed a significant upregulation of these proteins in GES-1 cells treated with high salt compared to the control group. Overexpression of TFF3 in cells led to increased levels of the JAK2, STAT3, and CDX2 proteins. Inhibition of JAK2 under TFF3 overexpression conditions resulted in decreased STAT3 and CDX2 protein levels but had no effect on TFF3 expression. Simultaneous suppression of STAT3 and TFF3 overexpression reduced CDX2 expression without affecting TFF3 or JAK2 protein levels. Suppressing CDX2 expression while overexpressing TFF3 did not change the protein levels of TFF3, JAK2, or STAT3 (Fig. 10A). Further knockdown of TFF3 and overexpression of JAK2/STAT3/CDX2 supported the primary regulation of CDX2 expression by TFF3 via the JAK2/STAT3 pathway (Fig. 10B). (Protein quantification is detailed in Fig.S4 and Fig.S5)
To elucidate the mechanism of TFF3’s interaction with JAK2, IL-6 expression levels were assessed, showing significant upregulation under high salt and TFF3 conditions (Fig. 11A), suggesting TFF3’s regulatory effect on IL-6 expression. IL-6 treatment of GES-1 cells followed by protein extraction for Western blot experiments demonstrated that increasing concentrations of IL-6 significantly upregulated phosphorylation levels of JAK2 and STAT3, along with CDX2 expression (Fig. 11B,C), indicating that IL-6 may enhance JAK2/STAT3 phosphorylation and subsequently increase CDX2 expression (Fig. 11D).
TFF3 regulates the JAK2/STAT3 pathway via IL-6. (A) IL-6 expression levels in GES-1 cells treated with NaCl and TFF3, determined by ELISA; (B) Levels of JAK2, p-JAK2, STAT3, p-STAT3, and CDX2 in GES-1 cells treated with 100 ng/mL and 200 ng/mL of IL-6, analyzed by Western blot; (C) Quantification of western blot results, normalized to GAPDH; (D) Schematic of the TFF3-JAK2/STAT3-CDX2 signaling pathway.
The TFF3-JAK2/STAT3-CDX2 axis promotes cell proliferation and migration
The proliferation of GES-1 cells cultured in high-salt medium was significantly greater than that of GES-1 cells cultured in low-salt medium, and TFF3 overexpression further increased cell proliferation. However, reducing the expression of JAK2, STAT3, or CDX2 in a high-salt environment with TFF3 overexpression weakened cell proliferation (Fig. 12A–D). Conversely, overexpression of JAK2, STAT3, or CDX2 reversed the detrimental effects on cell proliferation caused by TFF3 knockdown (Fig. S6).
Scratch assays revealed that high salt treatment significantly increased GES-1 cell migration, which was further enhanced by TFF3 overexpression. Cell migration decreased when JAK2, STAT3, or CDX2 was knocked down under high salt and TFF3 overexpression conditions (Fig. 13). Conversely, overexpressing JAK2, STAT3, or CDX2 reversed the decrease in migration caused by TFF3 knockdown (Fig. S7).
The TFF3-JAK2/STAT3-CDX2 pathway characterizes IM
Under normal conditions, TFF3 is expressed in secretory granules and released onto the mucosal surface of the gastrointestinal tract when needed, while JAK2 and STAT3 are predominantly expressed in the cytoplasm, and CDX2 is expressed in the nucleus. To assess the clinical relevance of these regulatory mechanisms in gastric cells, 15 samples each of normal gastric tissue and intestinal metaplasia were analyzed via immunohistochemistry (IHC) for gene expression. Positive cells typically exhibited brown to brownish-yellow staining under a microscope. Compared to those in normal gastric tissues, TFF3, JAK2, STAT3, and CDX2 expression was significantly upregulated inintestinal metaplasia tissues (Fig. 14), indicating the activity of the TFF3-JAK2/STAT3-CDX2 pathway. Correlation analysis revealed positive gene expression relationships among TFF3, JAK2, STAT3, and CDX2 (Fig. 15).
The TFF3-JAK2/STAT3-CDX2 pathway as a feature of intestinal metaplasia. (A) Representative IHC images of TFF3, JAK2, STAT3, and CDX2 in normal gastric tissue and intestinal metaplasia tissue. Scale bar: 50 μm; (B) Quantitative analysis of IHC results for TFF3, JAK2, STAT3, and CDX2 across 15 samples of normal gastric tissue and intestinal metaplasia tissue. ****P < 0.0001.
Discussion
The diagnosis rate of gastric cancer varies significantly with geography and disease stage. Early-stage gastric cancer often has a lower diagnosis rate due to nonspecific symptoms. Due to advances in medical technology and increased awareness of screening, the diagnosis rate of early-stage gastric cancer has improved in some countries and regions. In contrast, late-stage gastric cancer has a higher diagnosis rate due to more obvious symptoms, with medical imaging and endoscopic techniques being key to diagnosis11. Thus, increasing public awareness and screening for gastric cancer is crucial for increasing early diagnosis rates and patient survival12. Intestinal metaplasia, a precancerous lesion of gastric cancer, can be triggered by factors such as a high-salt diet. In China, the average daily salt intake far exceeds the World Health Organization’s recommendation of 5 g13,14, and long-term high-salt consumption is closely associated with an increased risk of gastric cancer15.
This study pioneers a model for salt-induced intestinal metaplasia in gastric epithelial cells, laying the groundwork for investigating the role of high salt intake in gastric carcinogenesis. A bioinformatics approach identified the differentially expressed gene TFF3, which was found to be upregulated in clinical gastric samples. Functional assays confirmed the role of TFF3 in promoting cell proliferation and migration. However, the precise mechanism of TFF3 in IM remains unclear. Further investigation suggested that TFF3 primarily activates the JAK2/STAT3 pathway, regulating CDX2 expression and thereby promoting metaplasia. This finding offers new insights into the treatment and prevention of IM. And this study has addressed the lack of research regarding the connection between TFF3 and the JAK2/STAT3/CDX2 pathway in IM.
The trefoil factor family includes TFF1, TFF2, and TFF3. These members, composed of small peptides enriched with cysteines that form a trefoil structure through six conserved cysteine residues and three intramolecular disulfide bonds, maintain their biological activity in complex environments. TFF3 was initially discovered in the jejunum of rats by Suemori in 1991 and is predominantly secreted by goblet cells in the small intestine and colon16. TFF3 is also expressed in tissues that secrete mucous, such as the hypothalamus, thyroid, salivary glands, respiratory tract, mammary glands, and endometrium17,18,19. Although TFF3 expression in normal gastric tissue is minimal, its expression significantly increases during gastric cancer development20, and TFF3 has been identified as an important biomarker for precancerous mucosal changes in gastric cancer21. Previous studies have shown that the expression of TFF3 in IM patients is increased22. However, some other studies suggest that the expression of TFF3 is not related to IM23. Our research revealed that TFF3 expression increased after high-salt treatment, and we further investigated the mechanism by which TFF3 promotes intestinal epithelial metaplasia under high-salt conditions. These results suggest that TFF3 primarily regulates CDX2 and downstream intestinal markers through the JAK2/STAT3 signaling pathway, thereby promoting the occurrence of IM. This also explains why the expression of TFF3 is increased in human intestinal-type gastric tumors24, and serum TFF3 has shown good diagnostic capabilities in detecting intestinal-type gastric cancer25.
The JAK/STAT signaling pathway is an essential pathway for cytokine signal transduction and is involved in inflammatory responses, cell proliferation, differentiation, apoptosis, and immune modulation26,27. This pathway consists of cytokine receptors, the JAK family, and the STAT family. Among the STAT family members, STAT3 is closely related to carcinogenic processes28. The binding of various cytokines to their receptors activates JAK, which in turn phosphorylates STAT, allowing it to enter the nucleus and bind to specific genes, affecting cell proliferation, differentiation, and migration. The JAK/STAT3 pathway is often aberrantly activated in various malignancies; regulates the expression of proteins such as Bax, Bcl-2, and MMPs; and is involved in the abnormal apoptosis, proliferation, and migration of tumors29,30,31. Our experiments revealed that the JAK2/STAT3 pathway can upregulate CDX2 expression in gastric epithelial cells, thereby promoting the process of gastric epithelial intestinal metaplasia.
CDX2 is an intestine-specific nuclear transcription factor primarily expressed in the small intestine and colon and is involved in the differentiation and formation of intestinal epithelial cells. CDX2 is almost unexpressed in the normal gastric mucosal epithelium, but its ectopic expression is often associated with gastric intestinal metaplasia and adenocarcinoma. CDX2 is usually positive in biopsy samples of intestinal metaplasia and continues to be expressed as metaplasia transitions to dysplasia32. When gastric cancer occurs, the overexpression of CDX2 may promote the migration and invasion of cancer cells33. Therefore, studying the expression and regulation of CDX2 during intestinal metaplasia of the gastric mucosa is highly important for treating intestinal metaplasia and preventing gastric cancer.
The progression from intestinal metaplasia (IM) to gastric cancer represents a complex, multifaceted process influenced by inflammation, genetic mutations, epigenetic alterations, and interactions with the surrounding microenvironment34,35. This transition involves the dysregulation of several critical pathways, including Wnt/β-catenin36, TP53 mutations37, and the Notch pathway38. Additionally, some studies have highlighted that TFF3 is significantly upregulated in both gastric cancer and IM39,40, while the JAK2/STAT3 signaling pathway plays a pivotal role in gastric cancer development41. In this research, it was demonstrated that TFF3 activates the JAK2/STAT3 signaling pathway, thereby promoting high-salt-induced intestinal metaplasia in gastric mucosal epithelial cells. These findings suggest that TFF3 could hold potential significance in understanding the progression from IM to gastric cancer. Nevertheless, the precise mechanism by which TFF3 triggers the JAK2/STAT3/CDX2 pathway to facilitate the malignant transformation from IM to gastric cancer remains an area requiring further investigation.
The animal model used in this study has not yet been perfected, as the intestinalization effect of high salt intake on murine gastric mucosa is time consuming, and animal experiments are ongoing. Future research will aim to refine related animal studies.
Conclusion
In summary, this study is important for understanding the challenges of early gastric cancer diagnosis and advancing early detection and treatment strategies. Enhancing the early diagnosis rate of gastric cancer is critical for improving patient survival. By establishing a high-salt-induced intestinalization model of gastric epithelial cells and analyzing the TFF3-JAK2/STAT3-CDX2 signaling pathway, our understanding of the relationships among a high-salt diet, intestinalization, and gastric cancer development can be deepened. This study enhances our understanding of the biological mechanisms underlying intestinalization, potentially revealing new molecular targets for early diagnosis and treatment and providing a scientific rationale for preventative and intervention strategies against intestinalization and gastric cancer induced by high salt intake.
Data availability
The authors confirm that the data supporting the findings of this study are available within the article and its supplementary materials.
References
Sung, H. et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence andmortality worldwide for 36 cancers in 185 countries. Cancer J. Clin. 71, 209–249. https://doi.org/10.3322/caac.21660 (2021).
Du, S. et al. Gastric cancer risk of intestinal metaplasia subtypes: A systematic review and meta-analysis of cohort studies. Clin. Transl. Gastroenterol. 12, e00402. https://doi.org/10.14309/ctg.0000000000000432 (2021).
Dias-Neto, M. et al. Salt intake and risk of gastric intestinal metaplasia: systematic review and meta-analysis. Nutr. Cancer. 62, 133–147. https://doi.org/10.1080/01635580903305391 (2010).
Li, T. et al. MicroRNA-92a-1-5p increases CDX2 by targeting FOXD1 in bile acids-induced gastric intestinal metaplasia. Gut. 68, 1751–1763. https://doi.org/10.1136/gutjnl-2017-315318 (2019).
Wang, N. et al. Bile acids increase intestinal marker expression via the FXR/SNAI2/miR-1 axis in the stomach. Cell. Oncol. 44, 1119–1131. https://doi.org/10.1007/s13402-021-00622-z (2021).
Sugano, K. et al. Gastric intestinal metaplasia: real culprit or innocent bystander as a precancerous condition for gastric cancer?? Gastroenterology. 165, 1352–1366. https://doi.org/10.1053/j.gastro.2023.08.028 (2023).
Berman, A. G. & Et Quantification of TFF3 expression from a non-endoscopic device predicts clinically relevant Barrett’s oesophagus by machine learning. EBioMedicine. 82 https://doi.org/10.1016/j.ebiom.2022.104160 (2022).
Kohal, R. et al. Targeting JAK2/STAT3 for the treatment of cancer: A review on recent advancements in molecular development using structural analysis and SAR investigations. Bioorg. Chem. 143, 107095. https://doi.org/10.1016/j.bioorg( (2024).
Gu, J. et al. TFF3 and HER2 expression and their correlation with survival in gastric cancer. Tumour Biol. 36, 3001–3007. https://doi.org/10.1007/s13277-014-2933-6 (2015).
Xu, M. et al. Berberine inhibits gastric cancer development and progression by regulating the JAK2/STAT3 pathway and downregulating IL-6. Life Sci. 290, 120266. https://doi.org/10.1016/j.lfs.2021.120266 (2022).
Xia, J. Y. & Aadam, A. A. Advances in screening and detection of gastric cancer. J. Surg. Oncol. 125, 1104–1109. https://doi.org/10.1002/jso.26844 (2022).
Yoon, H. & Kim, N. Diagnosis and management of high risk group for gastric cancer. Gut Liver. 9, 5–17. https://doi.org/10.5009/gnl14118 (2015).
Guideline Sodium Intake for Adults and Children (World Health Organization, 2012).
Hipgrave, D. B. et al. Salt and sodium intake in China. JAMA. 315(7), 703–705. https://doi.org/10.1001/jama.2015.15816 (2016).
Wu, B. et al. Dietary salt intake and gastric cancer risk: A systematic review and meta-analysis. Front. Nutr. 8, 801228. https://doi.org/10.3389/fnut.2021.801228 (2021).
Suemori, S. et al. Identification and characterization of rat intestinal trefoil factor: tissue- and cell-specific member of the trefoil protein family. Proc. Natl. Acad. Sci. U. S. A.. 88 (24), 11017–11021. https://doi.org/10.1073/pnas.88.24.11017 (1991).
El-Balat, A. et al. TFF3 expression as stratification marker in borderline epithelial tumors of the ovary. Pathol. Oncol. Res. POR. 24, 277–282. https://doi.org/10.1007/s12253-017-0240-4 (2018).
Mhawech-Fauceglia, P. et al. Trefoil factor family 3 (TFF3) expression and its interaction with Estrogen receptor (ER) in endometrial adenocarcinoma. Gynecol. Oncol. 130, 174–180. https://doi.org/10.1016/j.ygyno.2013.03.030 (2013).
Lin, X. et al. TFF3 contributes to epithelial-mesenchymal transition (EMT) in papillary thyroid carcinoma cells via the MAPK/ERK signaling pathway. J. Cancer. 9, 4430–4439. https://doi.org/10.7150/jca.24361 (2018).
Leung, W. K. et al. Expression of trefoil peptides (TFF1, TFF2, and TFF3) in gastric carcinomas, intestinal metaplasia, and non-neoplastic gastric tissues. J. Pathol. 197, 582–588. https://doi.org/10.1002/path.1147 (2002).
Hadjinicolaou, A. V. et al. Cytosponge-TFF3 testing can detect precancerous mucosal changes of the stomach. Clin. Gastroenterol. Hepatol. 20, 1411–1412. https://doi.org/10.1016/j.cgh.2021.07.047 (2022).
Zan, X. et al. The association of trefoil factors with gastric Cancer and premalignant lesions: A Cross-Sectional Population-Based cohort study. Cancer Epidemiol. Biomark. Prev. 31, 625–632. https://doi.org/10.1158/1055-9965 (2022).
Latorre, G. et al. Evaluation of trefoil factor 3 as a non-invasive biomarker of gastric intestinal metaplasia and gastric cancer in a high-risk population. Gastroenterol. Hepatol. 46, 411–418. https://doi.org/10.1016/j.gastrohep.2022.04.004 (2023).
Inagaki-Ohara, K. et al. Enhancement of leptin receptor signaling by SOCS3 deficiency induces development of gastric tumors in mice. Oncogene. 33, 74–84. https://doi.org/10.1038/onc.2012.540 (2014).
Lee, J. Y. et al. Comparison of serum trefoil factor 3 with the pepsinogen test for the screening of diffuse-type gastric cancer. Clin. Exp. Med. 17, 403–410. https://doi.org/10.1007/s10238-016-0426-1 (2017).
Wang, Y. et al. SOCS1/JAK2/STAT3 axis regulates early brain injury induced by subarachnoid hemorrhage via inflammatory responses. Neural Regener. Res. 16, 2453–2464. https://doi.org/10.4103/1673-5374.313049 (2021).
Xu, Y. et al. CTR9-mediated JAK2/STAT3 pathway promotes the proliferation, migration, and invasion of human glioma cells. J. Clin. Lab. Anal. 35, e23943. https://doi.org/10.1002/jcla.23943 (2021).
Li, Y. J. et al. STAT proteins in cancer: orchestration of metabolism. Nat. Rev. Cancer. 23, 115–134. https://doi.org/10.1038/s41568-022-00537-3 (2023).
Tang, H. et al. Delayed recanalization after MCAO ameliorates ischemic stroke by inhibiting apoptosis via HGF/c-Met/STAT3/Bcl-2 pathway in rats. Exp. Neurol. 330, 113359. https://doi.org/10.1016/j.expneurol.2020.113359 (2020).
Guha, P. et al. STAT3 inhibition induces bax-dependent apoptosis in liver tumor myeloid-derived suppressor cells. Oncogene. 38, 533–548. https://doi.org/10.1038/s41388-018-0449-z (2019).
Xuan, L. et al. Ceramide induces MMP-9 expression through JAK2/STAT3 pathway in airway epithelium. Lipids Health Dis. 19 https://doi.org/10.1186/s12944-020-01373-w (2020).
Camilo, V. et al. Differentiation reprogramming in gastric intestinal metaplasia and dysplasia: role of SOX2 and CDX2. Histopathology. 66, 343–350. https://doi.org/10.1111/his.12544 (2015).
Chai, D. et al. CDX2 and reg IV expression and correlation in gastric cancer. BMC Gastroenterol. 21 https://doi.org/10.1186/s12876-021-01678-9 (2021).
Huang, K. K. & Et al. Spatiotemporal genomic profiling of intestinal metaplasia reveals clonal dynamics of gastric cancer progression. Cancer Cell. 41, 2019–2037, https://doi.org/10.1016/j.ccell.2023.10.004 (2023).
Tong, Q. Y. et al. Gastric intestinal metaplasia: progress and remaining challenges. J. Gastroenterol. 59, 285–301. https://doi.org/10.1007/s00535-023-02073-9 (2024).
Liao, W. et al. Gallic acid alleviates gastric precancerous lesions through Inhibition of epithelial mesenchymal transition via Wnt/β-catenin signaling pathway. J. Ethnopharmacol. 302 https://doi.org/10.1016/j.jep.2022.115885 (2023).
Battista, S. et al. Molecular alterations in gastric preneoplastic lesions and early gastric cancer. Int. J. Mol. Sci. 22, 6652. https://doi.org/10.3390/ijms22136652 (2021).
Loe, A. K. H. et al. YAP targetome reveals activation of SPEM in gastric pre-neoplastic progression and regeneration. Cell. Rep. 42, 113497. https://doi.org/10.1016/j.celrep.2023.113497 (2023).
Ding, A. et al. Impact of NPM, TFF3 and TACC1 on the prognosis of patients with primary gastric cancer. PLoS One. 8, e82136. https://doi.org/10.1371/journal.pone.0082136 (2013).
Huang, R. J. Et al. A spatially mapped gene expression signature for intestinal stem-like cells identifies high-risk precursors of gastric cancer. BioRxiv. https://doi.org/10.1101/2023.09.20.558462 (2023).
Yang, Y. et al. Gancao Xiexin Decoction inhibits gastric carcinoma proliferation and migration by regulating the JAK2/STAT3 signalling pathway. J. Ethnopharmacol. 319 https://doi.org/10.1016/j.jep.2023.117241 (2024).
Acknowledgements
The computations were particularly performed at the Affiliated Hospital of Xuzhou Medical University.
Funding
This work was supported by the Key Development Project of Science and Technology Bureau in Xuzhou (Social development) [Grant number KC22233]; the Key Laboratory of Jiangsu Province Open project [Grant number XZSYSKF2021029]; the Project supported by the Affiliated Hospital of Xuzhou Medical University [Grant number 2023ZL19] and Postgraduate Research & Practice Innovation Program of Jiangsu Province [Grant number KYCX25_3282].
Author information
Authors and Affiliations
Contributions
Q.F. provided the cells; W.Y. and P.X. designed experiments; Q.F., S.B., Z.L., Z.J., C.S., Y.M., L.S., C.X. performed experiments; Q.F., P.X., W.Y. and F.S. analyzed data; Q.F. and W.Y. wrote and revised the manuscript. P.X. and F.S. supervised the experiment. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Experiments were approved by the Affiliated Hospital of Xuzhou Medical University (Ethics No. XYFY2024-KL017-01). In addition, I confirm that all methods were performed in accordance with the relevant guidelines and regulations by including a statement in the methods section.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Wang, Y., Qian, F., Shan, B. et al. TFF3 initiates gastric intestinal metaplasia by activating JAK2 and STAT3 under high salt conditions. Sci Rep 15, 23576 (2025). https://doi.org/10.1038/s41598-025-09045-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-025-09045-4