Abstract
Neprilysin (NEP) cleaves active forms of A- and B-type natriuretic peptides (ANP and BNP) at multiple sites, reducing their compensatory effects in heart failure. Therapeutic inhibition of NEP by sacubitrilat may potentiate the beneficial effects of natriuretic peptides. Neprilysin 2 (NEP2), a close homolog of NEP, has been described in humans but remains poorly characterized. This study aimed to investigate NEP2 expression in the cardiovascular system, its ability to cleave natriuretic peptides, and its inhibition by sacubitrilat. NEP and NEP2 expression profiles were assessed using reverse transcription PCR. The efficiency of in vitro cleavage of ANP, BNP, and the BNP precursor proBNP by NEP and NEP2 recombinant soluble domains was evaluated using sandwich-type immunoassays using antibodies specific for novel proteolytic epitopes. Expression of both NEP2 and NEP was observed at the mRNA level in cardiomyocytes and endothelial cells. NEP2 cleaved ANP and BNP; however, it was insensitive to sacubitrilat at studied concentration. In contrast to NEP, we observed proBNP proteolysis by NEP2, which resulted in the formation of a truncated form (amino acid residues 5–32). Our results show that sacubitrilat-insensitive NEP2 mediates ANP and BNP proteolysis and therefore might reduce compensatory effects of natriuretic peptides in heart failure.
Similar content being viewed by others
Introduction
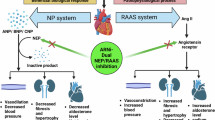
Heart failure (HF) is a highly prevalent disorder worldwide, with high morbidity and mortality rates, despite the availability of therapy1. Among the treatment options, angiotensin receptor-neprilysin inhibitors (ARNIs) have proven to be effective in patients with chronic HF with reduced ejection fraction2,3. ARNIs comprise two components: the angiotensin receptor II blocker valsartan and sacubitril, which is the precursor of the specific neprilysin (NEP) inhibitor sacubitrilat3.
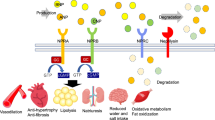
HF is accompanied by increased production of A- and B-type natriuretic peptides (NPs), ANP and BNP, by cardiomyocytes, along with the precursor forms, proANP and proBNP, and their release into the bloodstream in response to ventricular wall distension4,5,6. NPs have a similar primary amino acid structure and contain a 17-residue disulfide ring. NPs exhibit cardiovascular and renal effects, including a reduction in blood pressure, vasodilation, natriuresis, and diuresis, as well as anti-hypertrophic and anti-fibrotic effects7,8. ANP and BNP are removed from the circulation by binding to NP receptor-A (NPR-A), which mediates the physiological effects of NPs, and to NP receptor-C (NPR-C), which was initially considered to be a clearance receptor but later shown to be involved in signaling9. NPs metabolism is also affected by excretion from the kidneys and proteolytic degradation. One of the proteases responsible for NPs cleavage is NEP10. NEP cleaves NPs at multiple sites, and proteolysis occurring within the ring structure leads to loss of hormonal activity. Therefore, by cleaving the active forms of NPs, NEP reduces their compensatory effects, whereas NEP inhibition is thought to potentiate their beneficial effects11.
According to recent biomarker-based mechanistic studies, the cleavage of NPs, especially BNP, is moderately inhibited by sacubitril-based therapy, suggesting that other sacubitril-insensitive proteases may be involved in NP cleavage12,13. Among the candidate proteases, neprilysin 2 (NEP2) is of special interest for detailed analysis. This enzyme has been found in humans as a NEP homolog; the enzymes have 55% sequence identity and similarity in the active center structure, as well as substrate specificity14,15,16. NEP2 cleaves many NEP substrates, such as substrate P, bradykinin, and other vasoactive peptides in vitro17.
NEP2 was initially identified as an enzyme capable of degrading endothelin in endothelin converting enzyme-1 knockout mice. It was subsequently identified as a secreted peptide and designated a soluble-secreted endopeptidase (SEP)14. Alternative names such as neprilysin-like protein (NEPLP and Nl1) in mice and membrane-bound metalloendopeptidase-like enzyme 1 or 2 (MMEL1/2) in humans have been used15,18,19. NEP and NEP2 exist in soluble and membrane-bound forms. Similarly to NEP, NEP2 is also involved in clearance of amyloid β (Aβ), which is associated with Alzheimer’s disease pathogenesis, from the central nervous system. Aβ was shown to be elevated in NEP2-knockout mice; mRNA-based and immunochemical studies suggest that NEP2 is downregulated in Alzheimer’s disease 20. Unlike NEP, NEP2 is abundantly expressed in the testes and brain15 and NEP2 knockout mice show reduced sperm function21. However, the role and expression profile of NEP2 in other tissues and organs remain poorly understood. Furthermore, NEP2 expression in the cardiovascular system has not yet been demonstrated.
NEP and NEP2 are sensitive to phosphoramidon inhibition, whereas thiorphan, a specific NEP inhibitor, inhibits NEP2 less potently22. However, the sensitivity of NEP2 to sacubitrilat, a specific NEP inhibitor, remains unclear. Despite the high degree of similarity between NEP and NEP2, the contribution of NEP2 to regulation of the NP system remains unknown.
In this study, we investigated NEP2 expression in the cardiovascular system, its ability to cleave mature ANP and BNP, and its sensitivity to sacubitrilat. We also investigated NEP2-mediated proteolysis of the BNP precursor, proBNP, as the latter plays a crucial role as a biomarker in HF diagnostics.
Materials and methods
Materials
Expi293F (RRID: CVCL_D615) and ExpiCHO-S (RRID: CVCL_5J31) cells were obtained from Thermo Fisher Scientific, USA. Synthetic ANP (28 aa, sequence corresponding to amino acid residues (aar) 124–151 of UniProt P01160, peptide PRO_0000449729) and BNP (32 aa, sequence corresponding to aar 103–134 of UniProt P016860, peptide PRO_0000001532) were purchased from Bachem, Switzerland. Recombinant proBNP (108 aa, sequence corresponding to UniProt P16860, aar 27–134, with an additional N-terminal Met residue, Escherichia coli) was provided by HyTest, Finland. The primers were synthesized by Evrogen, Russia. A calibrator for the immunochemical detection of cleaved BNP – BNP(5–17)–(18–32) – was synthesized by Peptide 2.0 Inc, USA. Numbers in brackets correspond to aar numbering in mature 32-aar BNP.
Monoclonal antibody (mAb) 23/1 specific to the ANP ring structure (epitope aar 8–15 of ANP) was obtained from Bio-Rad, USA. mAb 50E1 (epitope aar 26–32 of BNP, corresponding to aar 102–108 of proBNP) and mAb BNP130 (epitope aar 14–21 of BNP, corresponding to aar 90–97 of proBNP) were obtained from HyTest, Finland. Here and below, aar are numbered according to the corresponding peptide: 28 aar ANP, 32 aar BNP, or 108 aar proBNP. Sacubitril (calcium salt, 98%) was purchased from MedCoo Biosciences, USA. Single Cell RNA Purification Kit was obtained from Norgen Biotek Corp., Canada, RevertAid First Strand cDNA Synthesis kit, GeneRuler 50 bp DNA ladder and DreamTaq™ Green DNA polymerase – from Thermo Scientific, USA. ZipTip C18 desalting columns were obtained from Merck Millipore, USA. Protino Ni-NTA Agarose was obtained from Macherey-Nagel, Germany.
Methods
Cell cultures
Human umbilical vein endothelial cells (HUVECs) were initially purchased from Cell Applications (#200-05n, USA) and provided by Dr. O.D. Lopina. Cardiomyocyte and neuron cultures derived from human pluripotent cells were provided by Dr. O.S. Lebedeva and Dr. M.A. Lagarkova, respectively. The induced pluripotent stem cell (IPSC) line FD5S was differentiated into cardiomyocytes using a STEMdiffTM Ventricular Cardiomyocyte Differentiation Kit (Stem Cell Technologies, Canada) according to the manufacturer’s instructions. The IPSC line RG4S23 was differentiated into GABAergic neurons according to the previously published protocol24. All cell lines used in this study were routinely tested for mycoplasma contamination. Cells found to be contaminated with mycoplasma were immediately excluded from the experiment. For more information about cell cultures maintenance please refer to Supplement 3.
Reverse transcription PCR (RT-PCR) analysis
Total RNA was isolated and purified with on-column DNase digestion using the Single Cell RNA Purification Kit according to the manufacturer’s instructions. The first strand cDNA was synthesized from 1 µg of total RNA with the RevertAid First Strand cDNA Synthesis kit using Oligo(dT)18 primers according to the kit manual. PCR was performed with 1 µl of cDNA and DreamTaq Green PCR Master Mix, using the following conditions: 94 °C for 2 min, 40 cycles of 94 °C for 10 s, 60 °C for 10 s and 72 °C for 30 s, final extension at 72 °C for 10 min, and analysed by electrophoresis in 1.5% agarose gel with GeneRuler 50 bp DNA ladder. The following primers were used for amplification: RPL13A (NM_012423.3), 185 bp, forward CTCAAGGTCGTGCGTCTGAA, reverse ACGTTCTTCTCGGCCTGTTT; NEP (NM_007289.4), 184 bp, forward GCCGAACCTACAAGGAGTCC, reverse GCAATCAAATCCTCGACCACATG; NEP2 (NM_033467.4), 165 bp, forward TGCGTTCTACTCCCCAAACC, reverse TCGAAGTTCCGGCCATTGTC.
Production of Recombinant enzymes
The soluble extracellular domains of human NEP (UniProt P08473, aar 56–750) and NEP2 (UniProt Q495T6, aar 74–779) both containing N-terminal 6xHis-tag were expressed in Expi293F and ExpiCHO-S cells, respectively. For more information about cell cultures maintenance please see Supplement 3. For recombinant protein purification, the conditioned media were dialyzed against buffer A (20 mM Tris-HCl, 500 mM NaCl, pH 8.0) and applied to an Ni-NTA agarose column pre-equilibrated with buffer A. The column was then washed with buffer A containing 10 mM imidazole. The target proteins were eluted with buffer A, containing 250 mM imidazole, and dialyzed against storage buffer (50 mM Tris-HCl, 150 mM NaCl, 1 µM Zn-acetate, pH 7.5).
The enzymatic activities of both recombinant proteins were confirmed using the fluorogenic substrate 7methoxycoumarin-4-yl)-acetyl-Arg-Pro-Pro-Gly-Phe-Ser-Ala-Phe-Lys(2,4-dinitrophenyl)-OH (R&D Systems, USA).
In vitro NP cleavage
NPs (ANP, BNP, and proBNP) at a concentration of 1.45 µM were incubated in 75 µL of reaction buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, and 0.1% Triton-X100) with 9.4 nM NEP or 113 nM NEP2. Enzyme concentrations were normalized to specific activity using the fluorogenic substrate 7-methoxycoumarin-4-yl)acetyl-Arg-Pro-Pro-Gly-Phe-Ser-Ala-Phe-Lys(2,4-dinitrophenyl)-OH. For inhibition studies, we used phosphoramidon as a non-specific inhibitor of both NEP and NEP2 and sacubitrilat (LBQ657) as a specific inhibitor of NEP activity. Phosphoramidon and sacubitrilat were used at a final concentration of 50 µM. Sacubitrilat was obtained from its inactive precursor sacubitril (LCZ696) by esterase hydrolysis as previously described25. Each sample was analyzed in triplicate. To stop the enzymatic reaction, the samples were rapidly diluted in 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 0.01% Tween-40, 0.5% BSA, and 0.05% sodium azide containing 10 mM EDTA.
Sandwich immunoassays (s-IA)
mAbs labeled with a stable Eu3+ chelate were used as detection antibodies. Two-step sandwich immunofluorescence assays were performed as previously described26,27.
To measure ANP with an intact ring structure, an intact-ring ANP s-IA was used, comprising mAb 23/1 as a capture antibody and a polyclonal antibody (pAb) specific to the C-terminal part of the ANP ring structure, aar 15–23 of ANP (see Supplement 1 for details of rabbit pAbs development), as a detection antibody. The s-IA was linear in the range of 184–5000 pg/mL (R2 = 0.9940), limit of detection (LoD) = 267 pg/mL, and intra-assay imprecision (coefficient of variation, CV) < 15% (assessed at two antigen concentrations in 11 replicates). LoD was calculated as mean(blank) + 3 x standard deviation (blank). CV was calculated as standard deviation/sample mean x 100.
To detect the uncleaved ring structures of BNP and proBNP, we used intact-ring BNP s-IAs, which comprised mAb 50E1 (capture antibody) and mAb BNP130 (detection antibody). This assay detects ring structures of BNP and proBNP, intact within aar 14–21 of BNP (90–97 of proBNP), which includes a known NEP cleavage site located between aar R17 and I18 of BNP (aar R93 and I94 of proBNP). The intact-ring BNP s-IA linear range was 11–2700 pg/mL (R2 = 0.9938), LoD = 17 pg/mL, and CV < 15% (assessed at two antigen concentrations in 19 replicates).
The neo-epitopes of BNP neo5 and neo17, formed as a result of the proteolytic cleavage of BNP at aar 4–5 (corresponding to aar 80–81 of proBNP) and aar 17–18 (corresponding to aar 93–94 of proBNP), were previously described25. To measure the BNP forms containing neo5, we performed neo5 s-IA using pAb BNPneo5 as the capture antibody (see Supplement 1) and mAb 50E1 as the detection antibody. The s-IA was linear in the range of 11–2700 pg/mL (R2 = 0.9978), LoD = 83 pg/mL, and CV < 10% (assessed at two antigen concentrations in 18 replicates). BNP forms containing the neo17-epitope were measured using neo17 s-IA, as described previously. Briefly, neo17 s-IA comprised of mAb BNP503 (for development, see Supplement 2), which was used as a capture antibody, and mAb 50E1 was used as the detection antibody. The assay linear range was 3.7–2700 pg/mL (R2 = 0.9939), LoD = 2.9 pg/mL, and CV < 15% (assessed at two antigen concentrations in 16 replicates). The designs of all the s-IAs are summarized in Table 1.
Mass-spectrometry (MS) analysis of NP fragments
For the MS analysis of the NPs after in vitro proteolysis, peptide fragments were extracted using reverse-phase ZipTip® C18 desalting columns. MS was performed on an ultraflex™ II MALDI-TOF/TOF mass spectrometer (Bruker Daltonics) with a SmartBeam™-II laser (Nd: YAG, 355 nm) in reflector and linear modes.
Statistical analysis
Data were analyzed using R statistical language and environment28. For each peptide at each experimental time point, comparisons were made between (1) enzyme-containing samples without inhibitors against those with inhibitors, and (2) enzyme-containing samples (with or without inhibitors) against no-enzyme baseline samples at time point zero. The Kruskal-Wallis test was used to determine the presence of significant variations within each comparison set. The P-values for samples with the same peptide were adjusted for multiple comparisons using Holm’s method. Where significant variations were detected, the Wilcoxon-Mann-Whitney U test was subsequently used to assess specific pairwise differences. P-values within the same comparison set were adjusted for multiple comparisons using Holm’s method29.
Ethics and consent declaration
Animal Experiments
The development of polyclonal antibodies using rabbits (see Supplement 1) was approved by Bioethics committee of Institute of Gene Biology Russian Academy of Sciences, protocol №13, December the 15th, 2020 and carried out in accordance with the European Communities Council Directive 2010/63/EU. The ARRIVE guidelines (Animal Research: Reporting of In Vivo Experiments) were followed to report animal experiments.
Results
NEP and NEP2 expression in various human tissues
We used RT-PCR to analyze NEP and NEP2 expression in HUVECs, neurons, and cardiomyocytes derived from pluripotent stem cells. Neurons were used as positive controls for NEP2 transcript detection, as its expression in these cells has been previously reported14,30,31. Specific PCR fragments indicating the presence of NEP and NEP2 transcripts, as well as the housekeeping gene RPL13A, were obtained from all tested samples (Fig. 1).
Electrophoretic analysis of RT-PCR products obtained from RNA isolated from HUVECs, neurons, and cardiomyocytes. RPL, NEP, and NEP2 RT-PCR products were obtained using primers specific to the RPL13A, NEP, and NEP2 genes, respectively. The expected lengths of the reaction products were 185 bp for RPL13A, 184 bp for NEP, and 165 bp for NEP2.
ANP and BNP proteolysis by NEP2
To test whether human NEP2 can cleave ANP and BNP, we incubated ANP and BNP with NEP2 in the presence or absence of phosphoramidon, and without the enzyme (Fig. 2). The NPs were also incubated with NEP for a comparative analysis. As here and in further experiments NEP and NEP2 were used at markedly disparate concentrations normalized by activity of enzymes towards standard fluorescent substrate, (see Methods section for details), any activities comparison should be approached with caution. ANP and BNP immunoreactivity after incubation at different time points was assessed using intact-ring ANP s-IAs (Fig. 2A) and intact-ring BNP s-IAs (Fig. 2B), respectively, as these s-IAs are specific to regions within the ring structures of ANP and BNP.
Analysis of ANP and BNP cleavage by NEP and NEP2 in vitro. The NPs were incubated with NEP, NEP2 or without enzymes in the presence or absence of inhibitors: phosphoramidon (probes NEP + Ph. and NEP2 + Ph.) and sacubitrilat (NEP + Sac. and NEP2 + Sac.). (A and C). ANP immunoreactivity was assessed using intact-ring ANP-s-IA. BNP immunoreactivity assessed with intact-ring BNP s-IA. Error bars represent SD. In all cases, concentrations are expressed relative to the initial concentration of the peptide in the absence of the enzyme at each time point. Asterisks mark points where concentration difference from no enzyme probe was statistically significant (P < 0.05).
Incubation with NEP2 decreased both ANP (Fig. 2A) and BNP (Fig. 2B) immunoreactivity, presumably by affecting their peptide ring structures. The immunoreactivity of the corresponding NP in the presence of NEP2 was significantly lower (P < 0.05) than that without the enzyme at all time points for ANP, and at 2, 6, and 16 h for BNP. Phosphoramidon inhibited ANP and BNP proteolysis by NEP or NEP2 and the NP immunoreactivity with the enzyme and inhibitor significantly differed (P < 0.005) from that with the enzyme only at all time points for ANP and at 1, 2, 6, and 16 h for BNP, supporting that NP cleavage was specific and enzymatically induced.
To determine whether NEP2 was susceptible to sacubitrilat inhibition, we tested its ability to cleave ANP, a substrate that is more prone to proteolysis in the presence and absence of sacubitrilat, using NEP-mediated cleavage as a positive control (Fig. 2C). We observed a similar degree of ANP proteolysis by NEP2, with or without sacubitrilat at 50 μM concentration, at all time points (P > 0.05), whereas sacubitrilat inhibited ANP cleavage by NEP (P < 0.005 at all time points).
ProBNP proteolysis by NEP2
The BNP precursor proBNP was previously shown to be resistant to NEP-mediated cleavage32. To test whether its ring structure was also resistant to proteolysis by NEP2, we incubated proBNP with NEP or NEP2, with or without phosphoramidon. The immunoreactivity of proBNP at different time points was assessed with the intact-ring BNP s-IA and compared with that of the enzyme-free sample (Fig. 3А). The reaction mixtures were analyzed by MS after 2 h of incubation (Fig. 3B).
Analysis of proBNP cleavage by NEP2. ProBNP was incubated with NEP, NEP2 or without enzymes in the presence or absence of phosphoramidon (probes NEP + Ph. and NEP2 + Ph.). (A) Immunoreactivity of proBNP, assessed with intact-ring BNP s-IA, shown as the percentage of peptide immunoreactivity in the enzyme-free sample. Error bars represent SD. In all cases, concentrations are expressed relative to the initial concentration of the peptide in the absence of the enzyme at each time point. Asterisks mark points where concentration difference from no enzyme probe was statistically significant (P < 0.005). (B) MS analysis of cleavage samples before (left panel) and after 2 h incubation (right panel).
The immunoreactivity of proBNP measured using intact-ring BNP s-IA decreased during incubation with NEP2 to 27% of that of the control sample (without enzymes) at 16 h. The differences were significant at 0.5, 0.75, 1, 2, and 6 h (P < 0.005). Phosphoramidon inhibited this decrease in immunoreactivity (P < 0.005 at 2 and 6 h), indicating that proBNP cleavage was enzymatically induced. No decrease in proBNP immunoreactivity was observed during NEP-mediated proteolysis. The fluctuations in proBNP immunoreactivity observed during the first hour of incubation may have been due to analytical issues.
We observed peaks with m/z ratios of 9034 and 4517, indicating a peptide with a mass of 9034 Da (its mono- and double-charged ions) generated by NEP2-mediated cleavage. This mass corresponded to a 1–80 aar proBNP fragment, which was apparently generated during NEP2-mediated proteolysis, but not during NEP-mediated proteolysis. The peaks with m/z ratios of 12,035 and 6017 corresponded to intact proBNP mono- and double-charged ions, respectively.
Site-specificity of BNP and probnp proteolysis by NEP2
Next, we analyzed the site specificity of NEP- and NEP2-mediated NP proteolysis. One of the known BNP cleavage sites for NEP is between aar 17 and 18 of BNP (corresponding to aar 93–94 of proBNP)25,32. To determine whether NEP2 cleaved BNP and proBNP ring structures at this site, we used neo17 s-IA, specific to the neo17 epitope of BNP (Table 1). Another well-described NEP proteolytic site, between aar 4 and 5 of BNP (corresponding to aar 80–81 of proBNP), lies outside the BNP ring structure (Table 1). We used neo5 s-IA to determine whether NEP2 was capable of BNP or proBNP cleavage at a site located towards the N-terminus from the ring structure to form the neo5 epitope. The results of neo5 and neo17 measurements of BNP and proBNP after incubation with NEP and NEP2, with or without phosphoramidon, are summarized in Fig. 4.
Analysis of the formation of neo-epitopes during BNP and proBNP cleavage by NEP and NEP2. The NPs were incubated with NEP, NEP2 or without enzymes in the presence or absence of inhibitors: phosphoramidon (probes NEP + Phosphoramidon and NEP2 + Phosphoramidon). (A and B) BNP proteolysis, neo5 and neo17 form generation, respectively. (C and D) proBNP proteolysis, neo5 and neo17 form generation, respectively. Error bars represent SD. Asterisks mark points where concentration difference from no enzyme probe was statistically significant (P < 0.005).
We observed neo5-epitope formation during BNP proteolysis by NEP2 and NEP (Fig. 4A), which was inhibited by phosphoramidon. The proportion of neo5-containing BNP forms significantly differed between those in the probes without enzyme in NEP samples (P < 0.005 at 0.25, 0.5, 1, 2, 6, and 16 h) and NEP2 samples (P < 0.005 at 0.25, 0.5, 1, 2, 6, and 16 h). Similarly to NEP, the neo17-epitope was generated during BNP proteolysis by NEP2 (Fig. 4B). The percentage of Neo17-containing forms differed significantly from samples without enzymes in the NEP and NEP2 samples (P < 0.005 at 0.25, 0.5, 1, 2, 6, and 16 h); however, the same was not observed in probes with phosphoramidon.
In the case of proBNP proteolysis by NEP2, our results clearly indicated that both neo5 (Fig. 4C) and neo17 (Fig. 4D) were formed (P < 0.005 when compared to no enzyme probes at 0.5, 1, 2, 6, and 16 h), although with an evidently lower efficacy than BNP. Our results show that a small amount of neo17 epitope is formed during proBNP proteolysis by NEP (P < 0.005 when compared to no enzyme probes at 0.5, 1, 2, 6, and 16 h). These data support our previous finding that NEP2 mediates proBNP ring structure proteolysis and indicate that, unlike NEP, NEP2 cleaves proBNP outside of the ring structure at position 80–81 aar (4–5 aar in BNP).
Discussion
Although NEP is involved in various physiological processes, it has received considerable attention due to its role as a therapeutic target in HF3,33. In the past decade, its cardiovascular effects have resulted in paradigm shift therapies for HF that combine NEP inhibition and angiotensin 2 type 1 receptor blockade. Potentiation of NPs, ANP, and BNP is considered to be the rationale behind pharmacological NEP inhibition33,34. NEP inhibition is thought to reduce fibrosis and hypertrophy, while increasing natriuresis and diuresis by increasing the levels of bioactive NPs. Due to this interplay, both NEP and NPs remain a scientific focus.
This hypothesis of NP potentiation by NEP inhibition assumes that NEP is a major NP-degrading enzyme and that no other enzymes with the same or similar activity towards NPs exist. However, subsequent analyses of the PARADIGM-HF trial and other trials have provided new information regarding the mechanism of NEP inhibition. The previously discovered moderate suppression of NP cleavage by sacubitrilat may indicate the involvement of other proteases capable of NP degradation. NEP belongs to the M13 family of zinc-dependent metalloproteases35. The closest homolog of NEP is NEP2, which displays > 50% overall protein sequence identity with NEP, suggesting a common function.
Compared to NEP, NEP2 has been studied much less, with data currently available only on NEP2 expression in the central nervous system and testes15,36. Apart from the role of NEP2 in Aβ clearance in the brain, little is known about its functional role in other organs and systems. Currently, specific clinical data substantiating the predictive or biomarker roles of NEP2 in cardiovascular disease is lacking. While NEP2 has been identified as one of the proteins associated with cardiovascular disease in certain proteomic studies37 comprehensive clinical data supporting its predictive or biomarker roles are currently limited to these observations. Moreover, existing immunoassays specific to the circulating form of NEP have not been validated to distinguish between NEP and NEP2 soluble forms, which additionally interferes with the establishment of the prognostic and diagnostic value of NEP2.
In this study, we identified NEP2 transcripts in cardiomyocytes and endothelial cells that were similar to those detected in neurons. This finding is in good agreement with the data reported by Bonvouloir et al.15 who were able to detect NEP2 expression in the heart on mRNA level. Although our results indicate the potential expression of NEP2 in these tissues, the presence of NEP2 at the protein level requires further confirmation. This represents a limitation of the current study, and future work should aim to validate NEP2 protein expression in cardiomyocytes and endothelial cells utilizing antibodies-based approaches or MS-based proteomics.
The substrate specificities of NEP and NEP2 have previously been shown to have a high degree of similarity, although they are not identical, suggesting that NEP and NEP2 play distinct physiological roles in humans22. Although the ability of mouse NEP2 to cleave human ANP has been demonstrated previously, whether human ANP and BNP are prone to human NEP2-mediated proteolysis remains unclear14. In ourin vitrocleavage experiments, we found that both ANP and BNP are susceptible to cleavage by NEP2. ANP is more susceptible to proteolysis by NEP than BNP; our results show that this is also true for NEP2. These data clearly demonstrate that NEP2 and NEP are capable of ANP and BNP ring structure cleavage in vitro, indicating that both peptides are possible NEP2 substrates in vivo. However, a potential limitation is the lack of possibility of using NPs and enzymes at concentrations corresponding to in vivo conditions. Both NEP and NEP2 are present in vivo as membrane-bound and soluble forms, with soluble form generation being a result of different mechanisms38,39,40. The proportions of these forms can vary between individuals, and assessing the concentration and activity of membrane-bound enzymes remains technically challenging. In addition, available data on tissue levels — particularly for NEP2 — are sparse and somewhat inconsistent41. To navigate these uncertainties, we chose to normalize enzyme input by activity rather than concentration, using a standard fluorogenic substrate (see the Methods). While extrapolating in vitro results to physiological conditions remains challenging and certainly requires further studies, this approach allowed us the meaningful comparison under controlled experimental settings.
A three-dimensional model of the active site of NEP2 revealed 97% sequence identity with NEP, with differences located within the S′2 subsite of NEP2 [16]. This results in a similar inhibitory profile for both enzymes, and both NEP and NEP2 are sensitive to the metalloendopeptidase inhibitor, phosphoramidon. However, the NEP inhibitor thiorphan has 30-times lower affinity towards NEP2 [22]. In the present study, we found that, in contrast to NEP, NEP2 was not significantly inhibited by 50 µM sacubitrilat — a specific NEP inhibitor — under the experimental conditions. This makes NEP2 unresponsive to inhibition by sacubitrilat-based therapies such as Entresto®. To the best of our knowledge, this is the first study to show that sacubitrilat does not inhibit NEP2 at a physiologically relevant concentration. Despite high similarity in the proteolytic and S1’ subsites, the differences observed in the region of the S′2 subsite where Ser133 and Leu739 in NEP2 replace two glycine residues in NEP might explain the difference in sensitivity to sacubitrilat between these two close homologs16.
Although NEP and NEP2 substrates are diverse, one might speculate that the role of NEP2 may become more important in patients undergoing sacubitrilat-based treatment. For instance, as both NEP and NEP2 play crucial roles in Aβ clearance, prolonged treatment with sacubitrilat may lead to more pronounced effect of NEP2 in Aβ catabolism.
Site-specific immunoassays and MS-based analyses revealed that NEP2 cleaved BNP at the same major cleavage sites as NEP, that is, between aar 4 and 5 and between aar 17 and 18. As the neo-epitope concentration at each time point results from two processes, neo-epitope formation during initial proteolysis and its degradation due to further cleavage of the peptide, the described difference in neo5-form levels could potentially be explained by the difference in the rates of these two reactions with NEP versus NEP2.
Although NEP2 was not significantly inhibited by sacubitrilat in given experimental conditions and may contribute to the production of BNP5-32, it does not have a pronounced effect on BNP as a biomarker. Additionally, Nougue et al. showed that BNP5-32 was below the limit of detection after the initiation of sacubitril/valsartan treatment, indicating that NEP2 plays a moderate, if any, role in BNP cleavage between amino acids 4 and 542.
In this study, we demonstrated that NEP2 cleaved the ring structure of proBNP. Apparently proBNP proteolysis by NEP2 led to the formation of nearly 5-times lower amount of neo-epitope-containing peptide forms compared with BNP cleavage. We suggest that proBNP ring structure cleavage by NEP2, as demonstrated by the intact-ring BNP s-IA, occurs after proBNP1–80 is cleaved, which is supported by MS studies.
The ability of NEP2, unlike NEP, to cleave proBNP is somewhat peculiar. Basing on the tertiary structure analysis from X-rays Pankow et al. hypothesized that proBNP, which has a large molecular weight, cannot enter the pocket of the catalytic site of NEP and thus cannot be degraded by NEP43. Despite high overall similarity between NEP and NEP2 described before, subtle structural differences exist that may impact substrate binding. In particular, the abovementioned variations in the S2’ subsite and adjacent ligand-binding regions could alter how larger substrates like proBNP interact with NEP2 compared to NEP. Furthermore, studies in related NEP2 homologs, such as in Drosophila, demonstrated distinct substrate specificity and reduced inhibition by typical NEP inhibitors, suggesting functional divergence even among structurally related enzyme44. Although crystallographic studies have characterized peptide binding to NEP, similar high-resolution data for NEP2 are lacking, making it plausible that NEP2 could accommodate substrates like proBNP through mechanisms slightly different from NEP. Detailed structural and mutagenesis studies will be necessary to determine whether NEP2 employs a distinct mechanism from NEP for proBNP processing.
Currently, commercially available BNP assays are heterogeneous and measure both proBNP and multiple breakdown products of BNP including BNP5-32. Some of these immunoassays are based on antibodies that have binding epitopes within the ring structure of proBNP and BNP. As both NEP and NEP2 are found to cleave the ring structure, one might suggest that changes in both NEP and NEP2 activity might impact the concentration of intact proBNP and BNP, available for immunochemical measurement. The potential effect of sacubitril-based therapy on BNP measurements is discussed in literature45. However, as NEP2-mediated cleavage might also contribute to BNP and proBNP ring structure degradation, one can speculate that the impact of the sacubitril therapeutic use on biomarker profile is more complex.
The ability of NEP2 to cleave proBNP may indicate the potential involvement of NEP2 in the regulation of the biological activity of BNP, not only through the degradation of the active BNP form, but also through the cleavage of proBNP, which can also affect the level of the active hormone in the bloodstream. Interestingly, the presence of alternative, longer N-terminal proBNP (NT-proBNP) forms (1–78 and 1–80), in addition to canonical NT-proBNP1–76, in plasma was previously shown by Amplatz et al.46. These forms may arise from the cleavage of NEP2 between aar 4 and 5. However, proBNP in circulation exists as a mixture of various glycosylated forms. As is well established for furin-mediated processing47 glycosylation at certain sites (Thr71 for furin) prevents proteolysis. However, how proBNP glycosylation affects its potential cleavage by NEP2 is yet to be elucidated.
HF treatment with ARNIs, including the NEP inhibitor sacubitrilat, generally results in NEP inhibition, leading to decreased NP degradation. The present findings show that sacubitrilat-insensitive NEP2 also contributes to ANP and BNP degradation. One might speculate that NEP2 might diminish their compensatory effects. However, the true effect of NEP2 on NP-dependent signaling must be investigated through its effect on cGMP levels. While data exist that prove that NEP can indirectly influence cGMP levels through its involvement in the degradation of natriuretic peptides, the specific influence of NEP2 on cGMP levels has not yet been investigated. For example, NEP inhibition with sacubitrilat increased circulating cGMP levels in a canine model of renin–angiotensin–aldosterone system48 and NEP inhibition has been associated with increased cGMP circulation in the blood49. However, the effect of NEP2 on cGMP levels remains an area that requires further investigation.
To fully understand the contribution of NEP2 to NP proteolysis, the relationship between NEP and NEP2 at the protein level in various tissues needs to be investigated, along with comprehensive research elucidating NEP2 involvement in the NP signaling cascade. We believe that these data will shed light on the exact mechanisms underlying the therapeutic benefit of NEP inhibition in HF and should be considered when interpreting the results of NP measurements in patients undergoing sacubitril-based therapy and developing novel therapeutic approaches.
Data availability
Data generated during this study will be available from the corresponding author upon reasonable request.
References
McDonagh, T. A. et al. ESC Guidelines for the diagnosis and treatment of acute and chronic heart failure. European heart journal 42, 3599–3726, (2021). https://doi.org/10.1093/eurheartj/ehab368 (2021).
McMurray, J. J. et al. Dual angiotensin receptor and Neprilysin Inhibition as an alternative to angiotensin-converting enzyme Inhibition in patients with chronic systolic heart failure: rationale for and design of the prospective comparison of ARNI with ACEI to determine impact on global mortality and morbidity in heart failure trial (PARADIGM-HF). Eur. J. Heart Fail. 15, 1062–1073. https://doi.org/10.1093/eurjhf/hft052 (2013).
McMurray, J. J. et al. Angiotensin-neprilysin Inhibition versus Enalapril in heart failure. N. Engl. J. Med. 371, 993–1004. https://doi.org/10.1056/NEJMoa1409077 (2014).
Seferian, K. R. et al. The brain natriuretic peptide (BNP) precursor is the major immunoreactive form of BNP in patients with heart failure. Clin. Chem. 53, 866–873. https://doi.org/10.1373/clinchem.2006.076141 (2007).
Mukoyama, M. et al. Increased human brain natriuretic peptide in congestive heart failure. N. Engl. J. Med. 323, 757–758. https://doi.org/10.1056/NEJM199009133231114 (1990).
Burnett, J. C. Jr. et al. Atrial natriuretic peptide elevation in congestive heart failure in the human. Science 231, 1145–1147. https://doi.org/10.1126/science.2935937 (1986).
Boerrigter, G., Costello-Boerrigter, L. C. & Burnett, J. C. Jr. Natriuretic peptides in the diagnosis and management of chronic heart failure. Heart Fail. Clin. 5, 501–514. https://doi.org/10.1016/j.hfc.2009.04.002 (2009).
Potter, L. R., Abbey-Hosch, S. & Dickey, D. M. Natriuretic peptides, their receptors, and Cyclic Guanosine monophosphate-dependent signaling functions. Endocr. Rev. 27, 47–72. https://doi.org/10.1210/er.2005-0014 (2006).
Potter, L. R., Yoder, A. R., Flora, D. R., Antos, L. K. & Dickey, D. M. Natriuretic peptides: their structures, receptors, physiologic functions and therapeutic applications. Handb. Exp. Pharmacol. 341–366. https://doi.org/10.1007/978-3-540-68964-5_15 (2009).
Potter, L. R. Natriuretic peptide metabolism, clearance and degradation. FEBS J. 278, 1808–1817. https://doi.org/10.1111/j.1742-4658.2011.08082.x (2011).
D’Elia, E. et al. Neprilysin Inhibition in heart failure: mechanisms and substrates beyond modulating natriuretic peptides. Eur. J. Heart Fail. 19, 710–717. https://doi.org/10.1002/ejhf.799 (2017).
Nougue, H. et al. Effects of sacubitril/valsartan on Neprilysin targets and the metabolism of natriuretic peptides in chronic heart failure: a mechanistic clinical study. Eur. J. Heart Fail. 21, 598–605. https://doi.org/10.1002/ejhf.1342 (2019).
Packer, M. et al. Angiotensin receptor Neprilysin Inhibition compared with Enalapril on the risk of clinical progression in surviving patients with heart failure. Circulation 131, 54–61. https://doi.org/10.1161/CIRCULATIONAHA.114.013748 (2015).
Ikeda, K. et al. Molecular identification and characterization of novel membrane-bound metalloprotease, the soluble secreted form of which hydrolyzes a variety of vasoactive peptides. J. Biol. Chem. 274, 32469–32477. https://doi.org/10.1074/jbc.274.45.32469 (1999).
Bonvouloir, N., Lemieux, N., Crine, P., Boileau, G. & DesGroseillers, L. Molecular cloning, tissue distribution, and chromosomal localization of MMEL2, a gene coding for a novel human member of the neutral endopeptidase-24.11 family. DNA Cell Biol. 20, 493–498. https://doi.org/10.1089/104454901316976127 (2001).
Voisin, S., Rognan, D., Gros, C. & Ouimet, T. A three-dimensional model of the Neprilysin 2 active site based on the X-ray structure of Neprilysin. Identification of residues involved in substrate hydrolysis and inhibitor binding of Neprilysin 2. J. Biol. Chem. 279, 46172–46181. https://doi.org/10.1074/jbc.M407333200 (2004).
Whyteside, A. R. & Turner, A. J. Human neprilysin-2 (NEP2) and NEP display distinct subcellular localisations and substrate preferences. FEBS Lett. 582, 2382–2386. https://doi.org/10.1016/j.febslet.2008.05.046 (2008).
Ghaddar, G. et al. Molecular cloning and biochemical characterization of a new mouse testis soluble-zinc-metallopeptidase of the Neprilysin family. Biochem. J. 347, 419–429. https://doi.org/10.1042/0264-6021:3470419 (2000).
Shirotani, K. et al. Neprilysin degrades both amyloid beta peptides 1–40 and 1–42 most rapidly and efficiently among thiorphan- and phosphoramidon-sensitive endopeptidases. J. Biol. Chem. 276, 21895–21901. https://doi.org/10.1074/jbc.M008511200 (2001).
Marr, R. A. & Hafez, D. M. Amyloid-beta and alzheimer’s disease: the role of neprilysin-2 in amyloid-beta clearance. Front. Aging Neurosci. 6, 187. https://doi.org/10.3389/fnagi.2014.00187 (2014).
Carpentier, M. et al. Reduced fertility in male mice deficient in the zinc metallopeptidase NL1. Mol. Cell. Biol. 24, 4428–4437. https://doi.org/10.1128/MCB.24.10.4428-4437.2004 (2004).
Rose, C., Voisin, S., Gros, C., Schwartz, J. C. & Ouimet, T. Cell-specific activity of Neprilysin 2 isoforms and enzymic specificity compared with Neprilysin. Biochem. J. 363, 697–705. https://doi.org/10.1042/0264-6021:3630697 (2002).
Holmqvist, S. et al. Creation of a library of induced pluripotent stem cells from parkinsonian patients. NPJ Parkinson’s Disease. 2, 16009. https://doi.org/10.1038/npjparkd.2016.9 (2016).
Nekrasov, E. D. et al. Manifestation of huntington’s disease pathology in human induced pluripotent stem cell-derived neurons. Mol. Neurodegeneration. 11, 27. https://doi.org/10.1186/s13024-016-0092-5 (2016).
Feygina, E. E. et al. Detection of Neprilysin-Derived BNP fragments in the circulation: possible insights for targeted Neprilysin Inhibition therapy for heart failure. Clin. Chem. 65, 1239–1247. https://doi.org/10.1373/clinchem.2019.303438 (2019).
Seferian, K. R. et al. Immunodetection of glycosylated NT-proBNP Circulating in human blood. Clin. Chem. 54, 866–873. https://doi.org/10.1373/clinchem.2007.100040 (2008).
Tamm, N. N. et al. Novel immunoassay for quantification of brain natriuretic peptide and its precursor in human blood. Clin. Chem. 54, 1511–1518. https://doi.org/10.1373/clinchem.2007.100545 (2008).
Team, R. C. (R Foundation for Statistical Computing, (2018).
Holm, S. A simple sequentially rejective multiple test procedure. Scand. J. Stat. 6, 65–70 (1979).
Facchinetti, P., Rose, C., Schwartz, J. C. & Ouimet, T. Ontogeny, regional and cellular distribution of the novel metalloprotease Neprilysin 2 in the rat: a comparison with Neprilysin and endothelin-converting enzyme-1. Neuroscience 118, 627–639. https://doi.org/10.1016/s0306-4522(02)01002-3 (2003).
Huang, J. Y., Hafez, D. M., James, B. D., Bennett, D. A. & Marr, R. A. Altered NEP2 expression and activity in mild cognitive impairment and alzheimer’s disease. J. Alzheimer’s Disease: JAD. 28, 433–441. https://doi.org/10.3233/JAD-2011-111307 (2012).
Semenov, A. G. & Katrukha, A. G. Different susceptibility of B-Type natriuretic peptide (BNP) and BNP precursor (proBNP) to cleavage by neprilysin: the N-Terminal part does matter. Clin. Chem. 62, 617–622. https://doi.org/10.1373/clinchem.2016.254524 (2016).
Feygina, E. E., Katrukha, A. G. & Semenov, A. G. Neutral endopeptidase (Neprilysin) in therapy and diagnostics: Yin and Yang. Biochem. Biokhimiia. 84, 1346–1358. https://doi.org/10.1134/S0006297919110105 (2019).
Bozkurt, B. et al. Neprilysin inhibitors in heart failure: the science, mechanism of action, clinical studies, and unanswered questions. JACC Basic. Translational Sci. 8, 88–105. https://doi.org/10.1016/j.jacbts.2022.05.010 (2023).
Turner, A. J., Isaac, R. E. & Coates, D. The Neprilysin (NEP) family of zinc metalloendopeptidases: genomics and function. BioEssays: News Reviews Mol. Cell. Dev. Biology. 23, 261–269. https://onlinelibrary.wiley.com/doi/10.1002/1521-1878(200103)23:3%3C261::AID-BIES1036%3E3.0.CO;2-K (2001).
Hafez, D. et al. Neprilysin-2 is an important beta-amyloid degrading enzyme. Am. J. Pathol. 178, 306–312. https://doi.org/10.1016/j.ajpath.2010.11.012 (2011).
Larson, A. et al. Plasma proteomic profiling in hypertrophic cardiomyopathy patients before and after surgical myectomy reveals Post-Procedural reduction in systemic inflammation. Int. J. Mol. Sci. 22 https://doi.org/10.3390/ijms22052474 (2021).
Raharjo, S. B. et al. Alternative splicing regulates the Endoplasmic reticulum localization or secretion of soluble secreted endopeptidase. J. Biol. Chem. 276, 25612–25620. https://doi.org/10.1074/jbc.M101703200 (2001).
Kuruppu, S., Rajapakse, N. W., Minond, D. & Smith, A. I. Production of soluble Neprilysin by endothelial cells. Biochem. Biophys. Res. Commun. 446, 423–427. https://doi.org/10.1016/j.bbrc.2014.01.158 (2014).
Komada, Y. et al. Shedding of leukemia-associated P24 antigen by lymphoblastoid cell lines. Jpn J. Clin. Oncol. 17, 333–342 (1987).
Rawlings, N. D. & Salvesen, G. Handbook of proteolytic enzymes. Third edition. edn.
Nougue, H. et al. Deconvolution of BNP and NT-proBNP immunoreactivities by mass spectrometry in heart failure and sacubitril/valsartan treatment. Clin. Chem. 69, 350–362. https://doi.org/10.1093/clinchem/hvac225 (2023).
Pankow, K. et al. Structural substrate conditions required for neutral endopeptidase-mediated natriuretic peptide degradation. J. Mol. Biol. 393, 496–503. https://doi.org/10.1016/j.jmb.2009.08.025 (2009).
Thomas, M. A. et al. A review of troponin assay performance in wales: can the same (method-dependent) decision limits be used in different sites? Ann. Clin. Biochem. 42, 351–356. https://doi.org/10.1258/0004563054890006 (2005).
Semenov, A. G. & Katrukha, A. G. Analytical issues with natriuretic Peptides - has this been. Overly Simplified? Ejifcc. 27, 189–207 (2016).
Amplatz, B. et al. Exposing the high heterogeneity of Circulating pro B-Type natriuretic peptide fragments in healthy individuals and heart failure patients. Clin. Chem. 66, 1200–1209. https://doi.org/10.1093/clinchem/hvaa130 (2020).
Semenov, A. G. et al. Processing of pro-B-type natriuretic peptide: Furin and Corin as candidate convertases. Clin. Chem. 56, 1166–1176. https://doi.org/10.1373/clinchem.2010.143883 (2010).
Mochel, J. P. et al. Sacubitril/valsartan (LCZ696) significantly reduces aldosterone and increases cGMP Circulating levels in a canine model of RAAS activation. Eur. J. Pharm. Sci. 128, 103–111. https://doi.org/10.1016/j.ejps.2018.11.037 (2019).
Ishii, M. et al. Cardioprotective effects of LCZ696 (Sacubitril/Valsartan) after experimental acute myocardial infarction. JACC Basic. Translational Sci. 2, 655–668. https://doi.org/10.1016/j.jacbts.2017.08.001 (2017).
Acknowledgements
We are grateful to Professor Dr. Maria A. Lagarkova and Dr. Olga S. Lebedeva for providing access to necessary cell lines. We also thank Dr. Petr N. Datskevich and Dr. Anfisa S. Popova for their constructive criticism and helpful comments during the preparation of this manuscript.
Funding
This study was supported by the HyTest Ltd., Turku, Finland.
Author information
Authors and Affiliations
Contributions
Elizaveta M. Selezneva and Evgeniya E. Feygina contributed equally to this work and share first authorship. The manuscript was written with the contributions of all authors. All authors have approved the final version of the manuscript and gave consent to publish this manuscript in the present form.
Corresponding author
Ethics declarations
Competing interests
The authors declare the following competing interests: This study was supported by the HyTest Ltd., Turku, Finland. The authors Elizaveta M. Selezneva, Evgeniya E. Feygina, Liudmila V. Ageeva, Fedor N. Rozov, Evgeny P. Altshuler and Alexander G. Semenov were employees of HyTest during the conduction of this research and preparation of the manuscript. All the remaining authors declare no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Selezneva, E.M., Feygina, E.E., Ageeva, L.V. et al. Neprilysin 2 catalyses the degradation of natriuretic peptides despite sacubitrilat Inhibition. Sci Rep 15, 27401 (2025). https://doi.org/10.1038/s41598-025-10166-z
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-10166-z