Abstract
Gestational diabetes mellitus (GDM) and preeclampsia (PE) are common pregnancy complications, with the rising incidence of GDM and the pronounced heterogeneity of PE leading to an increased prevalence of their coexistence, presenting a unique and complex clinical condition. However, the disease characteristics and maternal-neonatal outcomes of this comorbidity remain largely unknown. In this study, we collected peripheral blood samples from 42 pregnant women, including pregnant controls without complications (Control, n = 10), GDM (n = 12), PE (n = 10), and those with coexistence of GDM and PE (PG, n = 10), as well as partial neonatal cord blood samples. Through integrated lipidomic and transcriptomic analyses, we identified distinct lipid and gene expression features associated with PG state and explored their correlations with clinical maternal characteristics and neonatal outcomes. Our findings indicate that PE and PG groups exhibit more pronounced lipidomic alterations, which are associated with adverse neonatal outcomes, while GDM and PG groups show more significant transcriptomic changes related to maternal neuronal and immune dysfunction, potentially increasing the risk of postpartum depression (PD). The elevated risk of PD was further supported by independent analyses of the UK Biobank and FinnGen cohorts. Despite the limitation of a modest sample size, the study provides evidence that subclinical alterations in maternal peripheral blood lipidomic and transcriptomic profiles, along with aberrations in cord blood gas (CBG) parameters, may exert adverse effects on both maternal and neonatal health. These findings highlight the need for further mechanistic and longitudinal investigations to elucidate the underlying pathophysiological processes.
Similar content being viewed by others
Introduction
Pregnancy is a complicated physiological state that requires precise coordination of maternal and fetal systems to ensure a healthy outcome. However, complications during pregnancy such as gestational diabetes mellitus (GDM) and preeclampsia (PE), may disrupt this balance, contributing to maternal and perinatal morbidity and mortality1. GDM, characterized by impaired glucose tolerance with onset or first recognition during pregnancy2, affects approximately 9–25% of pregnancies worldwide3, with a prevalence in China as high as 16.7%4. This condition is associated with maternal metabolic dysregulation, increased long-term risks of type II diabetes, cardiovascular diseases and adverse fetal outcomes5. PE, a hypertensive disorder defined by the new-onset hypertension after 20 weeks of gestation, is often accompanied by proteinuria, endothelial dysfunction and multi-organ damage6,7, affecting 6–8% of pregnancies globally8. The pronounced heterogeneity of PE severely impacts fetal development and is accompanied by immune dysregulation and excessive maternal inflammation9,10, further increasing health risks for both mother and child11,12.
With the rising prevalence of GDM13 and previous studies identifying GDM as an independent risk factor for PE, the coexistence of these two conditions is becoming increasingly common. The comorbidity of GDM and PE (PG) further complicates clinical management, yet the risks associated with their coexistence have not received sufficient attention. While previous studies have focused on individual complications, they often fail to capture the shared pathways and distinct molecular mechanisms across GDM, PE and PG. Currently, the disease characteristics of PG and its impact on neonatal outcomes are not well understood, and the limited available literature reports inconsistent findings. Some studies suggest that GDM comorbidity may partially mitigate the progression of PE14, while others report higher rates of adverse neonatal outcomes in PG15. Placental studies indicate similarities between PG and PE16, whereas research on immune responses at the maternal–fetal interface suggests distinct mechanisms for PE compared to GDM and PG17. These incomplete similarities and differences highlight the complex mechanisms underlying PG comorbidity.
Emerging multi-omics technologies provide powerful tools to dissect the molecular underpinnings of pregnancy complications. Lipidomics, which profiles a wide range of lipid species, has revealed that abnormal lipid metabolism plays a key role in both GDM18 and PE19,20,21, contributing to adverse outcomes. Transcriptomics, meanwhile, uncovers gene expression changes, shedding light on regulatory pathways and immune responses that underpin disease pathogenesis. Integrating these approaches enables the identification of distinct molecular signatures and shared dysregulated pathways, enhancing our understanding of these pregnancy complications. In this study, we analyzed peripheral blood samples from GDM, PE, PG, and control (CTL) groups using multi-omics approaches, including lipidomics and transcriptomics, to explore both unique and shared molecular features of single and comorbid conditions, and to relate these molecular profiles to neonatal outcomes and CBG indices. This highlights the need for comprehensive analyses that integrate multiple layers of data to better understand the pathogenesis and progression of these conditions.
Notably, PE exhibited the most significant lipidomic alterations, while GDM showed the most extensive transcriptomic changes. PG displayed intermediate characteristics, with lipidomic profiles resembling PE and transcriptomic profiles more similar to GDM. Importantly, PG also demonstrated unique changes, particularly in gene expression related to neuronal deficits, which may be associated with an increased risk of postpartum depression (PD). Analyses of UK Biobank (UKB) and FinnGen data supported significant associations between GDM, PE, and PD, although the association for PG did not reach statistical significance (p-value = 0.065), possibly due to limited sample size.
PE was associated with the most pronounced lipidomic changes, including lipid metabolic disturbances linked to adverse neonatal outcomes such as low birth weight. While the PE group showed a higher incidence of low birth weight, PG did not exhibit more severe outcomes, suggesting a partial mitigation of adverse effects; however, cord blood gas (CBG) indices indicated potentially more severe disturbances in PG, reflecting complex compensatory mechanisms. From a transcriptomic perspective, GDM exhibited broader gene expression changes than PE, while PG combined features of both conditions and showed unique alterations, including genes related to neuronal deficits, potentially linking PG to more severe PD. UKB and FinnGen analyses further supported close associations between GDM, PG, and PD, although the small sample size for PG limited statistical power.
Our findings provide important insights into the distinct and overlapping mechanisms of GDM, PE, and PG, and underscore the value of lipidomic and transcriptomic analyses in understanding these conditions. Further studies integrating clinical and molecular data are warranted to clarify the impact of PG on neonatal outcomes and to validate the long-term effects of PG on maternal health.
Results
Clinical and biochemical characteristics of the participants
A total of 42 participants were categorized into four groups (CTL, GDM, PE, PG), with PB samples used to explore systemic changes in lipid and immune dysregulation. Lipidomic profiling was performed on all samples, and transcriptomic analysis was conducted on a subset of 17 samples (Fig. 1A and B).
The clinical characteristics are summarized in Table 1 (Baseline Characteristics), Table S1 (Other Clinical Characteristics), and Table S2 (Complete Blood Count [CBC] Results). While parameters like gestational age, Apgar scores, and CBC results were comparable, significant differences were found in maternal age, body mass index (BMI), blood pressure, oral glucose tolerance test (OGTT) results, parity, neonatal birth weight, and delivery mode (p-value < 0.05). Pairwise comparisons were conducted to further explore these differences (Table S3). Notably, higher maternal age in the PG group indicates an increased risk of coexisting complications. Newborns in the PE group had significantly lower birth weights and higher hospitalization and cesarean delivery rates, reflecting the severity of PE and its impact on maternal and neonatal outcomes.
Lipidomic profiling reveals distinct downregulated lipids in different pregnancy-associated complications, but shared upregulated patterns
Lipidomic profiling was conducted using mass spectrometry to understand lipid changes associated with pregnancy-related complications. A total of 763 lipid metabolites were identified using the LipidSearch library (Thermo Fisher Scientific), all of which passed quality control. The identified lipids were primarily distributed across the glycerophospholipids (GPs), followed by sphingolipids (SPs) and glycerolipids (GLs) categories (Fig. 2B). Hierarchical clustering revealed high stability and reproducibility of the lipidomic data, as evidenced by low Intra-group variation (Figure S1A and S1B). PCA demonstrated more similar lipidomic profiles for the CTL and GDM groups, while the PE and PG groups exhibited distinct profiles (Fig. 2A), suggesting that PE-related pregnancy complications may have higher association with lipid metabolism compared to GDM-related ones.
Lipidomics data analysis. (A) PCA plot of lipidomics data, colored by pregnancy complication groups: CTL (green), GDM (blue), PE (orange) and PG (red). (B) Pie chart showing the percentage distribution of major lipid categories based on the763 identified lipids. (C) Volcano plots displaying DELs between GDM, PE, and PG groups and the CTL group. Up-regulated lipids (FDR < 0.25 & log2(FC) > 0) are shown in red and down-regulated lipids (FDR < 0.25 & log2(FC) < 0) are shown in blue. (D) Bubble plots illustrating subclasses of DELs identified in (C). (E) Heatmap of scaled median lipid expression levels within each group, with an additional cutoff of |log2(FC)| > 1 to identify 43 DELs with higher confidence. Hierarchical clustering was performed using the ward.D2 method based on correlation distance.
Pairwise differential lipid expression analysis identified disease-associated lipids (Fig. 2C). Compared to the CTL group, 12 (3 upregulated and 9 downregulated), 26 (15 upregulated and 11 downregulated) and 19 (6 upregulated and 13 downregulated) lipids were differentially expressed in the GDM, PE and PG groups, respectively. Upregulated lipids were shared among the disease groups and mainly comprised sphingomyelins (SMs) within the SPs category (Fig. 2D, E). In contrast, the groups exhibited distinct patterns of downregulated lipids: the GDM group showed lower levels of phosphatidylethanolamines (PEs), the PE group was characterized by reduced phosphatidylcholines (PCs) and the PG group exhibited a significant reduction in lysophosphatidylcholines (LPCs).
Signature lipids associated with neonatal outcomes highlight PE as the most severe group
Both GDM and PE are associated with adverse birth outcomes. CBG analysis is considered a sensitive method for assessing the newborn’s acid-base status, oxygenation and overall condition during delivery. Key CBG parameters include pH, HCO3-(std) and BE(ecf) for acid-base balance; pO2, sO2(est) and tHb(est) for oxygen transport and gas exchange; mOsm for osmotic balance and RI for respiratory and circulatory function.
CBG data were conducted for 20 out of 42 infants (47.6%) shown in Table S4. All lipids were screened, from which seventeen lipids were identified as significantly correlated with CBG parameters (Fig. 3A and Figure S1D), seven of which were changed in disease conditions, predominantly in the PE group (Fig. 3B). Notably, PC(18:1/24:0) and dMePE(18:2/18:2) associated with acid-base balance and oxygenation, were reduced in PE and PG (Fig. 3D), suggesting a high likelihood of perinatal asphyxia or hypoxia, which can lead to more severe outcomes, such as neurological damage. Lipids associated with oxygenation and vascular function, including PC(32:1)(rep), PI(16:0/16:1), dMePE(16:0/16:1) and PC(16:0/16:1), were significantly increased in the PE group (Fig. 3E), indicating compromised oxygenation and hypoxemia. Hypoxemia can impair brain and organ development, potentially causing complications such as cerebral palsy or persistent pulmonary hypertension. Both PE and PG exhibited increased PC(40:4), a generalized signature metabolite indicative of compromised oxygenation, acid-base balance, osmolarity and vascular function.
Correlation of lipid profiles with CBG parameters. (A) Heatmap showing lipids significantly correlated with CBG parameters (pearson correlation, p-value < 0.05). The parameters are grouped into:1) Osmotic Balance and Circulatory (mOsm and RI); 2) Oxygen Transport and Gas Exchange (tHb(est), pO2 and sO2(est)) and 3) Acid-Base Balance (BE(ecf), HCO3-(std) and pH). mOsm: milliosmoles, tHb(est): total hemoglobin (estimated), RI: respiratory index, pO2: partial pressure of oxygen, sO2(est): estimated oxygen saturation, BE(ecf): base excess of extracellular fluid, HCO3-(std): standard bicarbonate concentration, pH: potential of hydrogen. Lipid clusters are indicated as Cluster 1 (gold) and Cluster 2 (purple), where reduced levels in Cluster 1 and elevated levels in Cluster 2 are associated with worse outcomes. (B) Volcano plots highlighting significantly correlated lipids from Cluster 1 (down-worse, gold) and Cluster 2 (up-worse, purple). (C) Violin plot showing birth weight distributions across the four groups. (D and E) Violin plots displaying the expression levels of lipids associated with adverse birth outcome: (D) upregulated and (E) downregulated. Statistical significance was determined using the Wilcoxon rank test (*: p-value < 0.05 and ns: non-significant).
Interestingly, despite the PE group showing the worst CBG results, the coexistence of GDM in the PE appeared to mitigate the severity of PE to some extent at the lipid level. This is supported by the birth weight findings, where the birth weights of PG group were comparable to the CTL group, while the PE group exhibited significantly lower birth weights (Fig. 3C).
Lipid risk score for assessment of PE risk and severity
Unlike GDM, which can be diagnosed using fasting blood glucose (FBG) test, PE is characterized by high individual heterogeneity and is often asymptomatic or presents with only mild symptoms in the early stage, making it difficult to assess its severity and to observe its progression effectively. To address this, we developed a novel lipid-based risk score for PE based on lipidomic data collected prior to delivery.
To accurately assess the risk and severity of PE not only in patients with PE alone but also in those with co-existing GDM, we extracted key features from DELs shared between the PE and PG groups, resulting in 8 candidate lipids (Fig. 4A and Figure S1E). Ridge regression was subsequently employed to compute robust weights, and a PE-related lipid risk score (PLRS) was calculated based on these key lipid features. Receiver operating characteristic (ROC) curve analysis demonstrated (Fig. 4B and C) the discriminative ability of each lipid, as well as the overall PLRS, in distinguishing PE and PG. PLRS were significantly lower in GDM, but remained higher in PG, indicating high efficiency in distinguishing PE patients, even with GDM association (Fig. 4E).
Construction of a generalized PE predictive model applicable to PG. (A) Venn diagram showing eight overlapping DELs between the PE and PG groups. (B) Table summarizing the ROC analysis for the eight DELs, including their AUC values and 95% CI. (C) ROC curves for the eight DELs, showing their predictive performance. (D) Scatter plots demonstrating the positive correlation between the PLRS and SBP/DBP, with correlation coefficients (R) and p-values indicated. (E) Box plot showing PLRS distribution across the four groups. Statistical significance between groups were assessed using the Wilcoxon test (****: p-value < 0.0001, ***: p-value < 0.001, **: p-value < 0.01 and ns: non-significant).
Correlation analysis revealed that PLRS was significantly positively correlated with SBP and DBP prior to delivery (p-value < 0.05) (Fig. 4D), as well as with OGTT-1 h glucose. No significant correlations were observed with other clinical characteristics (Table S5).
Transcriptomic profiling reveals more pronounced changes in GDM-associated pregnancy complications compared to PE
mRNA profiling offers valuable insights into early molecular changes before protein-level alterations, making it highly sensitive for identifying signature profiles and pathways associated with pregnancy-related complications, such as GDM, PE and PG.
PCA of RNA-Seq data demonstrated more distinct transcriptomic profiles between the CTL and GDM-associated pregnancy complications compared to PE, suggesting that GDM is associated with more striking mRNA-level changes (Fig. 5A and Figure S2A). Pairwise differential expression analysis between the GDM, PE, PG groups and the CTL group identified a total of 1503 DEGs in the GDM group, 69 DEGs in the PE group and 1683 DEGs in the PG group (Fig. 5B and Figure S2B). Heatmap of all DEGs revealed six distinct gene clusters based on median expression levels within each group (Fig. 5C, D).
Transcriptomic data analysis. (A) PCA plot of RNA-seq data colored by pregnancy complications groups: CTL (green), GDM (blue), PE (orange) and PG (red). (B) Volcano plots displaying DEGs between GDM, PE, and PG groups and CTL. Up-regulated genes (FDR < 0.25 and log2(FC) > 0) are shown in red and down-regulated genes (FDR < 0.25 and log2(FC) < 0) are shown in blue. (C) Heatmap showing scaled median expression levels of 776 DEGs for protein coding function across all groups, clustered into six distinct gene clusters. Hierarchical clustering was performed using ward.D2 method based on correlation distance. (D) Bar plots displaying the selected significantly enriched pathways (FDR < 0.1) for each DEG cluster. (E) Violin plots showing expression levels of previously reported genes that associated with GDM and PE. (F) Violin plots highlighting novel DEGs identified in this study as being specifically associated with GDM, PE, or PG. Statistical significance was assessed using the Wilcoxon test (**: p-value < 0.01, *: p-value < 0.5 and ns: non-significant).
Clusters C1 and C5 were characterized by genes that were generally downregulated in disease groups compared to CTL. These genes were primarily associated with inflammation-related pathways, including the TNF-alpha signaling pathway, Interferon Gamma response and the p53 pathway. Cluster C3, which showed genes downregulated specifically in GDM and PG, was enriched in pathways related to both inflammation and cardiovascular diseases, such as pulmonary hypertension, cholesteatoma, myocarditis and atherosclerosis. While fewer upregulated genes were identified, they tended to be more disease-specific. Cluster C4, specific to PG, was enriched in myeloid-related genes, such as MPO and ELANE, affecting pathways implicated in Duchenne Muscular Dystrophy. Cluster C6, upregulated in PE, included genes associated with arterial hypertension and lymphoid activities, which align with known features of PE. To be noted, genes involved in Prostaglandin Synthesis and Regulation, such as ANXA1 and S100A10, as well as genes associated with fatty acid oxidation, such as PDK422, were observed, suggesting a link between the pathogenesis of PE and lipid metabolism - consistent with our lipidomics findings. Cluster C2 contained genes upregulated in GDM. Although no significant pathways were enriched due to the limited number of protein-coding genes, this cluster contains genes associated with abnormal cholesterol homeostasis, including VSIG4 and ITM2C.
Several genes previously reported to be associated with GDM, such as CAMK1D and IL1B, as well as genes linked to PE, such as RGL3 and PREX123 (related to blood pressure regulation), and MME and GPX3 (associated with severe PE), were also differentially expressed in our PE and GDM groups (Fig. 5E). However, since other studies have typically compared each disease to healthy controls individually, these genes are not disease-specific, as they were generally downregulated across most pregnancy complications in our study. Here, by including GDM, PE and PG in our analysis, we were able to identify disease-specific marker genes, which were predominantly upregulated when compared to the CTL group (Fig. 5F). ENOSF1, KLRC3 and B3GNT7 were uniquely enriched in the GDM group, while MXRA7, CD163 and MERTK were predominately upregulated in the PE group. Most of the PE-associated genes exhibited expression patterns resembling those of either GDM or PE, reflecting the coexistence of these two conditions in PG. Nevertheless, two marker genes for PG were identified: ZNF683 and PPDPF.
Gene clusters identified linking to adverse neonatal and maternal outcomes
To investigate the potential impact of gene expression on adverse birth outcomes, we explored the expression patterns of genes correlated with CBG parameters across the different groups. Due to the limited sample size, correlation analysis was not feasible; therefore, we instead examined the trends in CBG parameters across the groups. PG was generally associated with lower oxygenation and higher acidity (Fig. 6A and B). Based on these trends, we further categorized the CBG parameters into four distinct clusters and compared them to gene clusters with similar expression patterns.
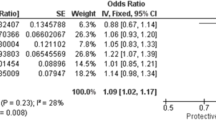
Association of transcriptomic data with adverse birth outcomes and maternal defects. Heatmap displaying scaled median values of blood gas parameters across the four groups. Hierarchical clustering was performed using the ward.D2 method based on correlation distance. (B) Trend plots showing the fitted trends of blood gas parameters across the four groups, with colors representing different function categories of the parameters. (C) Enlarged heatmap highlighting genes Cluster 4, which is specifically up-regulated in the PG group. (D) Violin plots showing the expression levels of genes associated with muscle weakness (SPSB2 and LRRC56), neuronal defects (PDZD4 and GFRA2), and birth outcomes (TTN and MPO). Statistical significance was determined using the Wilcoxon test (*: p-value < 0.5 and ns: non-significant). (E) Forest plot summarizing the association between GDM, PE, PG and PD using data from in the UKB and FinnGen databases. Results are presented as odds ratios (OR) with 95% CI (UKB) and hazard ratios (HR) with 95% CI (FinnGen), along with p-values.
CBG cluster C1, associated with impaired oxygenation, closely matched gene cluster C5, while CBG cluster C2, which reflected vascular dysfunction and osmolarity imbalances, was similar to gene cluster C4 (Fig. 6C). Cluster C4 has 22 genes and cluster C5 contains 60 genes, both of which were likely associated with adverse birth outcomes. Notably, gene cluster C5 includes TTN, a gene previously associated with peripartum cardiomyopathy24. In addition, genes in C4 - LRRC5625; SPSB226; MPO, ELANE, TNNT127 and SIK1B28 are linked to muscle weakness, such as myopathy and muscle dystrophy (Fig. 6D). These findings suggest that these genes may contribute to complications affecting fetal development and adverse birth outcomes.
Intriguingly, several genes in cluster C4, including EPHA429,30, GFRA231 and PDZD432, are associated with neuronal defects (Fig. 6D). This provides potential mechanistic insights into the link between pregnancy complications and PD at the maternal side. To validate the association, we analyzed large human cohorts from FinnGen and the UKB (Fig. 6E). Positive associations were detected between pregnancy complications and PD: GDM [OR 1.59, 95% CI 1.03–2.32; HR 2.32, 95% CI 2.02–2.67], PE [OR 1.54, 95% CI 1.26–1.85; HR 1.65, 95% CI 1.39–1.96] and PG [HR 2.61, 95% CI 0.79–6.42], While the association for PG was marginally non-significant (p-value = 0.065), likely due to limited sample size, the results indicate a compelling trend that warrants further investigation. These findings collectively highlight the interplay between pregnancy complications, adverse birth outcomes, and maternal mental health.
Discussion
Pregnancy complications pose significant challenges to maternal and neonatal health, often contributing to adverse outcomes that require better diagnostic tools and therapeutic strategies. While most previous studies focus on a single complication, such as GDM or PE, these conditions frequently share overlapping pathophysiological features, such as inflammation, endothelial dysfunction and metabolic disturbances. In this study, we incorporated multiple pregnancy complications—GDM, PE, and their coexistence (PG)—within a unified analytical framework. By directly comparing these three groups, we were able to better distinguish the molecular alterations common to all conditions from those that are disease-specific, thereby deepening our understanding of the underlying molecular mechanisms. This approach is particularly valuable for the PG group, as comorbidity is unlikely to represent a simple additive effect of individual diseases; instead, it may involve distinct pathogenic mechanisms and disease characteristics. Our comparative analysis thus provides a broader perspective on the molecular transitions associated with these complications and offers new insights into the unique features of comorbid pregnancy disorders.
A key strength of this study lies in the use of multi-omics integration, combining lipidomics and transcriptomics to provide a holistic view of the molecular alterations in pregnancy complications. Lipidomics reveals alterations in lipid species, which are critical components of cellular membranes, signaling pathways and metabolism processes. Transcriptomics, on the other hand, provides insights into gene expression changes, uncovering regulatory pathways, immune responses, and cellular functions. This dual approach allowed us to reveal distinct patterns across the two omics layers: lipidomic analyses showed more pronounced changes in PE and PG compared to GDM relative to the CTL group, while transcriptomic analyses uncovered more extensive changes in GDM and PG than in PE.
From a Lipidomics perspective, our findings highlighted significant changes in key lipid species across the three groups. Elevated levels of SMs were observed in GDM, PE and PG. SMs are known to affect plasma membrane signaling, cholesterol efflux and endothelial dysfunction33,34,35,36, which align with the observed pathophysiological features of these complications. PEs, which are crucial for maintaining cell membrane integrity and fluidity, exhibited divergent patterns. While some studies reported increased PEs in GDM37,38, we observed a dramatic reduction in specific PEs, including PE(40:5), PE(16:0/20:5), PE(36:4p), PE(40:3)(rep) and PE(42:6) in GDM and PG, but not in PE, showing consistency to39. These reductions may reflect impaired trophoblast function and placental development, underscoring their potential as specific metabolites for GDM and its associated complications.
Diacylglycerols (DGs), such as DG(34:2) and DG (32:1), were identified as potential indicators of PE, with higher intensity in PE compared to CTL40. Higher DGDG(30:3) intensity in PE was also identified in our data, consistent with previous findings. PCs, the most abundant phospholipid in mammalian cellular membranes, were notably downregulated in both PE and PG, including PC(35:2p), PC(34:3p), PC(14:0/20:4), PC(32:1p) and PC(40:7)(rep). Given the critical role of PCs in triacylglycerol transport and cardiovascular function, their reduction may be linked to impaired endothelial dysfunction, arterial stiffness and other cardiovascular abnormalities commonly observed in PE, such as atherosclerosis41,42. Additionally, LPCs and lysophosphatidylethanolamines (LPEs), known as inflammatory lipids43, who marked reductions in PG, suggesting persistence of high-grade inflammation in this group. This aligns with their roles in regulating inflammatory activity, oxidative stress and immune responses in multiple inflammatory-associated diseases, including rheumatoid arthritis44 and Alzheimer disease45.
The transcriptomic findings complemented the lipidomic results by offering insights into mRNA expression changes associated with pregnancy complications. A majority of the genes linked to these complications were downregulated across GDM, PE and PG groups compared to CTLs, suggesting overlapping pathophysiological mechanisms between these conditions. This was supported by the enrichment of inflammatory pathways, such as the Interferon Gamma response and TNF-alpha signaling via NF-kB, in gene clusters C1, C3 and C5. These findings align with the well-established understanding that both GDM and PE are inflammatory conditions. Moreover, enriched pathways, such as fatty acid metabolism, estrogen response (late), hypoxia and hypertrophic cardiomyopathy, further underscore the critical role of lipid metabolism in these conditions, emphasizing the link between disrupted lipid profiles, placental dysfunction, and higher cardiovascular risk.
Notably, GDM was characterized by the downregulation of genes associated with pulmonary hypertension. This aligns with previous reports indicating that women with GDM are at an increased risk of developing PE late in pregnancy4, hinting at a potential molecular connection between the two conditions. While several previously reported disease-specific markers, such as CAMK1D46 and IL1B47 for GDM, and GPX3, MME48, PREX1 and RGL323 for PE were differentially expressed in our data, they were not exclusive to a single disease but rather shared across all conditions.
Our findings, however, identified disease-specific genes that were predominantly upregulated. GDM specially upregulated genes were linked to abnormal cholesterol homeostasis. PE-specific genes were associated with arterial hypertension, as expected and prostaglandin synthesis, consistent with the lipidomics data, showing pronounced lipid changes in PE49. In PG, upregulated genes were linked to more severe conditions, such as muscle weakness, reflecting the compounded nature of coexisting GDM and PE, which is likely to further increase the risk of cardiovascular and metabolic diseases (CMD)14,50,51. Specifically, we identified signature genes for unique disease condition, which represent valuable candidates for disease prediction and further mechanistic exploration.
Our study also shed light on the association between lipidomic and transcriptomic changes and neonatal outcomes. Previous research suggests conflicting outcomes for women with comorbid GDM and PE. Some studies reported higher rates of adverse perinatal outcomes, such as macrosomia, respiratory distress syndrome, and jaundice52. However, other studies suggested that adverse outcomes in patients with PE and GDM comorbidity are significantly lower compared to those with PE alone53. To further investigate these associations, we integrated CBG results with lipidomic and transcriptomic data. Seventeen lipids were found to be highly correlated with critical CBG parameters linked to newborn outcomes, including acid-base balance, oxygenation and vascular function. At the gene expression level, we observed similar trends involving myeloid-related genes, such as MPO and ELANE. Additionally, we noted that the incidence of low birth weight was higher in the PE group, whereas the PG group did not exhibit more severe outcomes in this regard. However, some CBG indices were more abnormal in the PG group, suggesting that while comorbidity does not necessarily exacerbate clinical manifestations such as growth restriction, there may be complex interactions between compensatory mechanisms. These findings indicate that, beyond observable clinical outcomes, it is important to consider the potential impact of underlying pathological mechanisms on neonatal health in the PG group.
From a lipidomics perspective, PE appears to be associated with the most striking lipid changes and the worse newborn outcomes, evidenced by the lowest birth weight in this group. Low birth weight and small-for-gestational-age fetuses are often linked to restricted fetal growth54, aligning with the pathophysiology of fetal growth restriction in PE. This underscores the significant impact of dysregulated lipid metabolism on birth weight. Interestingly, PG, representing the coexistence of PE and GDM, appeared to partially mitigate the severity of adverse outcomes. This is consistent with previous findings suggesting that the presence of GDM might reduce the severity of PE-related pregnancy outcomes14, potentially through metabolic alterations.
At the transcriptomic level, direct correlation analysis with neonatal outcomes was limited by the number of samples. However, gene clusters exhibiting trends similar to the CBG results were extracted, as gene cluster C4 and C5. These clusters included immune related genes as well as genes associated with muscle atrophy or dystrophy, such as CACNB4, IGTA1, ATP2A1, TTN, TNNT127, SIK1B28, which may influence cardiac function. In addition, genes associated with neuronal degeneration, including EPHA429,30, GFRA231 and PDZD432 or higher depression risk, such as ELANE55 were differentially expressed in PG compared to the CTLs. These findings suggest not only a potential link between these genes and adverse birth outcomes but also possible associations with long-term neuronal effects, such as increased risk of PD in mothers. Previous studies have briefly explored the association between individual pregnancy complications, such as GDM56 or PE57,58, using relatively small cohorts. In this study, we leveraged two large-scale human cohorts, FinnGen and the UKB, to validate these associations. We not only confirmed positive associations between GDM and PE with PD but also extended the analysis to PG, the coexistence of GDM and PE. Notably, PG exhibited the highest OR for PD, although the statistical significance was marginal, likely due to limited sample size. These findings underscore the need for early prevention and management of pregnancy complications to mitigate adverse maternal mental health outcomes, particularly in cases of combined complications such as PG.
Our study offers several key advancements over previous research. First, integrating lipidomic and transcriptomic profiling provides a comprehensive multi-omics perspective on the pathogenesis of pregnancy complications, identifying both disease-specific and shared molecular pathways. Second, while prior studies focused on individual complications like GDM or PE, our investigation includes GDM, PE, and PG in a unified framework, revealing interactions and potential synergies between these conditions. Third, we established robust links between multi-omics findings and neonatal outcomes, using objective metrics like CBG analysis and birth weight to overcome the subjectivity of parameters like Apgar scores. Additionally, our transcriptomic analyses suggest associations between GDM, PE, and PD, with PG showing the strongest potential link. Validation using large human cohort data from the UKB and FinnGen further strengthens the results and highlights their translational potential.
However, several aspects could be improved. The small sample size for each group, particularly in lipidomic and transcriptomic analyses, required using a higher FDR (< 0.25) to identify significant findings. PB samples were collected late in pregnancy, limiting the ability to assess early gestational molecular changes that could improve predictive models for PE and other complications. As a single-center study, the findings may not fully represent the racial and ethnic diversity needed for broader generalizability. Additionally, severe PE cases leading to preterm birth were excluded, and subgroup analyses like early-versus late-onset PE were not performed due to sample size limitations. Despite these challenges, the study offers novel insights into the interplay of coexisting pregnancy complications and their downstream effects, providing a strong foundation for future research aimed at precision diagnostics and therapies.
Conclusion
This study provides a comprehensive multi-omics analysis of pregnancy complications, including GDM, PE and PG, revealing both shared and condition-specific molecular alterations that enhance our understanding of their distinct yet interconnected pathophysiology. Lipidomic profiling identified significant changes in SMs, PCs/PEs, and LPCs/LPEs, with the PE group showing most pronounced lipidomic disruptions, while transcriptomics revealed pathways linked to inflammation, cholesterol homeostasis, hypertension, and cardiovascular risks. These findings were further linked to neonatal outcomes, where altered lipid species correlated with birth weight and CBG parameters, and transcriptomic data uncovered genes associated with immune regulation, muscle atrophy, and neurodevelopmental defects. Overall, this study underscores the importance of a multi-omics approach and the inclusion of diverse pregnancy complications, providing novel insights into their molecular mechanisms, and offering translational tools to improve maternal and neonatal health outcomes.
Materials and methods
Study design and participants
This study was conducted with the approval of the Ethics Committee of Shenzhen Guangming District People’s Hospital (Approval Number: LL-KT-2023033). All research procedures were performed in accordance with relevant guidelines and regulations, and in accordance with the Declaration of Helsinki. Informed consent was obtained from all participants prior to their inclusion in the study. A final cohort of 42 pregnant women were recruited at the Shenzhen Guangming District People’s Hospital between June 2020 and January 2023. Participants were categorized to four groups based on their pregnancy conditions: the GDM group consisted of 12 individuals, while each of the other groups had 10 participants. All participants enrolled were enrolled at the time of diagnosis and monitored through delivery to assess neonatal outcomes. The inclusion and exclusion criteria were detailed in the supplementary methods.
Measurement of clinical and biochemical characteristics
Participants’ information was collected prior to labor and at delivery, encompassing a comprehensive range of maternal and neonatal parameters. PB samples were collected in late-stage pregnancy and separated serum for further testing. Neonatal umbilical artery blood was collected immediately after birth for CBG analysis. Information and blood sample collection were performed as detailed in supplementary methods.
Inclusion and exclusion criteria
The inclusion criteria for PE cases were aligned with the guidelines established by the International Society for the Study of Hypertension in Pregnancy (ISSHP)59,60. GDM was diagnosed between 24 and 28 weeks of gestation using OGTT test. Diagnosis was confirmed if any of the following plasma glucose levels met or exceeded the thresholds recommended by the International Association of Diabetes and Pregnancy Study Groups (IADPSG)61. The inclusion and exclusion criteria were detailed in the supplementary methods.
Sample preparation for lipidomics and RNA sequencing
Forty-two samples (CTL: n = 10; GDM: n = 12; PE: n = 10; PG: n = 10) were used for lipidomics profiling. RNA sequencing was performed on 17 samples (CTL: n = 5; GDM: n = 5; PE: n = 3; PG: n = 4), using 150 bp paired-end sequencing. Lipidomics and RNA-seq samples preprocessing were performed as described in the supplementary methods.
Statistical analysis
Mean differences between groups were evaluated using tableone (v0.13.2) package in R, employing either the Chi-squared test for categorial variables or the Kruskal-Wallis test for continuous variables. For post-hoc pairwise comparisons between groups, the Wilcoxon rank sum test was utilized, with p-values adjusted for multiple testing using the Benjamini-Hochberg (BH) method to control the false discovery rate (FDR).
Lipidomics analysis
Principal component analysis (PCA) was utilized to visualize sample distributions across groups using stats (v4.2.3) package. Differential expression analysis of lipids between disease groups (PE, GDM, PG) and the CTL group was performed using a linear model with empirical Bayes moderation, implemented via the limma (v3.54.2) package in R. Multiple testing correction was applied using the Benjamini-Hochberg FDR, with a significance threshold set at FDR < 0.25. To ensure high-confidence targets, differentially expressed lipids (DELs) were further filtered based on a |log2(fold change (FC))| > 2. Ridge regression analysis, implemented using the glmnet (v4.1.8) package, was employed to predict PE severity. PLRS were computed by assigning regression-derived weights to significant lipids. The relationship between neonatal blood gas parameters and lipids was evaluated using pearson correlation analysis via stats (v4.2.3) package, with significance defined as p-value < 0.05. Hierarchical clustering was subsequently performed using the ward.D2 method via pheatmap (v1.0.12) package.
RNA-Seq analysis
Differential expression analysis of RNA-Seq data was conducted using the edgeR (v3.40.2) package. Data normalization was performed using the TMM (Trimmed Mean of M-values) method. A generalized linear model (GLM) was fitted to the data using the quasi-likelihood (QL) approach via the glmQLFit function. Differentially expressed genes (DEGs) were identified using quasi-likelihood F-tests implemented in the glmQLFTest function, with adjustment for multiple testing using the FDR. Genes were considered differentially expressed if they met the criteria of FDR < 0.25 and |log2(FC)| > 1. Pathway enrichment analysis for DEGs was performed using Enrichr62,63,64 with a significance threshold of adjusted p-value < 0.1.
Selection and validation of public datasets
The validation of the associations between pregnancy complications and PD was carried out using population-wise data from the UKB and FinnGen. The UKB study was approved by the North west Multi-Centre Research Ethics Committee and all participants provided written informed consent to participate in the UKB study. This study was granted under Application Number 99,001. The correlation between the pregnancy complications and PD was assessed using logistic regression analysis using stats (v4.2.3) in R, with results presented as odds ratios (OR) and 95% confidence intervals (CI). The population in the UKB was selected according to the inclusion and exclusion criteria detailed in the supplementary methods. The FinnGen database was used with clinical endpoint via the RISTEYS website (https://r8.risteys.finngen.fi/). Prior exposure (GDM or PE) and outcome (PD) were included to calculate hazard ratios (HR) and its 95% CI using the Cox regression with default parameters without considering the lagged data.
Data availability
Lipidomic data can be accessed through the zenodo through https://zenodo.org/records/14498742. RNA-seq data have been deposited in the GEO under the accession codes GSE284329. Code is available at: https://github.com/Chenxiao-Zhang/Scripts.git.
References
McElwain, C. J., Tuboly, E., McCarthy, F. P. & McCarthy, C. M. Mechanisms of endothelial dysfunction in Pre-eclampsia and gestational diabetes mellitus: windows into future cardiometabolic health? Front. Endocrinol. (Lausanne). 11, 655 (2020).
Chorell, E. et al. Pregnancy to postpartum transition of serum metabolites in women with gestational diabetes. Metabolism 72, 27–36 (2017).
Jiang, L. et al. A global view of hypertensive disorders and diabetes mellitus during pregnancy. Nat. Rev. Endocrinol. 18 (12), 760–775 (2022).
Xie, Y., Zhou, W., Tao, X., Lv, H. & Cheng, Z. Early gestational blood markers to predict preeclampsia complicating gestational diabetes mellitus. Diabetes Metab. Syndr. Obes. 16, 1493–1503 (2023).
Diboun, I. et al. Metabolic profiling of pre-gestational and gestational diabetes mellitus identifies novel predictors of pre-term delivery. J. Transl Med. 18 (1), 366 (2020).
Cayci, T. et al. Cord blood and maternal serum neopterin concentrations in patients with pre-eclampsia. Clin. Chem. Lab. Med. 48 (8), 1127–1131 (2010).
Liu, L. et al. Association of elevated cord blood oxidative stress biomarkers with neonatal outcomes in mothers with Pre-Eclampsia: A case-control study. Gynecol. Obstet. Invest. 86 (4), 361–369 (2021).
Sun, X. et al. Screening of differentially expressed proteins from syncytiotrophoblast for severe early-onset preeclampsia in women with gestational diabetes mellitus using tandem mass Tag quantitative proteomics. BMC Pregnancy Childbirth. 18 (1), 437 (2018).
Li, J., Huang, L., Wang, S. & Zhang, Z. The prevalence of regulatory T and dendritic cells is altered in peripheral blood of women with pre-eclampsia. Pregnancy Hypertens. 17, 233–240 (2019).
Magee, L. A., Nicolaides, K. H., von Dadelszen, P. & Preeclampsia N Engl. J. Med. 386(19):1817–1832. (2022).
Campello, E. et al. Circulating microparticles in umbilical cord blood in normal pregnancy and pregnancy with preeclampsia. Thromb. Res. 136 (2), 427–431 (2015).
Deniz, R. et al. Evaluation of elabela, Apelin and nitric oxide findings in maternal blood of normal pregnant women, pregnant women with pre-eclampsia, severe pre-eclampsia and umbilical arteries and venules of newborns. J. Obstet. Gynaecol. 39 (7), 907–912 (2019).
Dugalic, S. et al. Trends of the prevalence of Pre-gestational diabetes in 2030 and 2050 in Belgrade cohort. Int. J. Environ. Res. Public. Health 19(11). (2022).
Pankiewicz, K. et al. The impact of coexisting gestational diabetes mellitus on the course of preeclampsia. J. Clin. Med. 11, 21 (2022).
Onuoha, C. et al. The simultaneous occurrence of gestational diabetes and hypertensive disorders of pregnancy affects fetal growth and neonatal morbidity. Am. J. Obstet. Gynecol. 231 (5), 548 (2024).
Mor, L. et al. Improved neonatal outcomes in pregnancies with coexisting gestational diabetes and preeclampsia in normal birthweight neonates- insights from a retrospective cohort study. Placenta 149, 1–6 (2024).
Fei, H. et al. Deciphering the preeclampsia-specific immune microenvironment and the role of pro-inflammatory macrophages at the maternal-fetal interface. Elife 13. (2025).
Aziz, F., Khan, M. F. & Moiz, A. Gestational diabetes mellitus, hypertension, and dyslipidemia as the risk factors of preeclampsia. Sci. Rep. 14 (1), 6182 (2024).
Nobakht, M. G. B. F. Application of metabolomics to preeclampsia diagnosis. Syst. Biol. Reprod. Med. 64 (5), 324–339 (2018).
Huang, Y. et al. Lipidomic signatures in patients with early-onset and late-onset preeclampsia. Metabolomics 20 (4), 65 (2024).
Alahakoon, T. I., Medbury, H. J., Williams, H. & Lee, V. W. Lipid profiling in maternal and fetal circulations in preeclampsia and fetal growth restriction-a prospective case control observational study. BMC Pregnancy Childbirth. 20 (1), 61 (2020).
Pettersen, I. K. N. et al. Upregulated PDK4 expression is a sensitive marker of increased fatty acid oxidation. Mitochondrion 49, 97–110 (2019).
Tyrmi, J. S. et al. Genetic risk factors associated with preeclampsia and hypertensive disorders of pregnancy. JAMA Cardiol. 8 (7), 674–683 (2023).
Goli, R. et al. Genetic and phenotypic landscape of peripartum cardiomyopathy. Circulation 143 (19), 1852–1862 (2021).
van der Burgt, I. et al. Myopathy caused by HRAS germline mutations: implications for disturbed myogenic differentiation in the presence of constitutive HRas activation. J. Med. Genet. 44 (7), 459–462 (2007).
Li, Y. et al. SPSB1-mediated Inhibition of TGF-β receptor-II impairs myogenesis in inflammation. J. Cachexia Sarcopenia Muscle. 14 (4), 1721–1736 (2023).
Pellerin, D. et al. Novel recessive TNNT1 congenital Core-Rod myopathy in French Canadians. Ann. Neurol. 87 (4), 568–583 (2020).
Berdeaux, R. et al. SIK1 is a class II HDAC kinase that promotes survival of skeletal myocytes. Nat. Med. 13 (5), 597–603 (2007).
Dennys, C. et al. EphA4 targeting agents protect motor neurons from cell death induced by amyotrophic lateral sclerosis -astrocytes. iScience 25 (9), 104877 (2022).
Van Hoecke, A. et al. EPHA4 is a disease modifier of amyotrophic lateral sclerosis in animal models and in humans. Nat. Med. 18 (9), 1418–1422 (2012).
Sandmark, J. et al. Structure and biophysical characterization of the human full-length neurturin-GFRa2 complex: A role for Heparan sulfate in signaling. J. Biol. Chem. 293 (15), 5492–5508 (2018).
Khan, H. et al. Biallelic variants identified in 36 Pakistani families and trios with autism spectrum disorder. Sci. Rep. 14 (1), 9230 (2024).
Fakhr, Y., Brindley, D. N. & Hemmings, D. G. Physiological and pathological functions of sphingolipids in pregnancy. Cell. Signal. 85, 110041 (2021).
Enthoven, L. F. et al. Effects of pregnancy on plasma sphingolipids using a metabolomic and quantitative analysis approach. Metabolites ;13(9). (2023).
Pyne, S., Lee, S. C., Long, J. & Pyne, N. J. Role of sphingosine kinases and lipid phosphate phosphatases in regulating Spatial sphingosine 1-phosphate signalling in health and disease. Cell. Signal. 21 (1), 14–21 (2009).
Rahman, M. L. et al. Plasma lipidomics profile in pregnancy and gestational diabetes risk: a prospective study in a multiracial/ethnic cohort. BMJ Open. Diabetes Res. Care ;9(1). (2021).
Li, L. J. et al. Exploring preconception signatures of metabolites in mothers with gestational diabetes mellitus using a non-targeted approach. BMC Med. 21 (1), 99 (2023).
Zhan, Y. et al. Plasma metabolites, especially lipid metabolites, are altered in pregnant women with gestational diabetes mellitus. Clin. Chim. Acta. 517, 139–148 (2021).
Liu, T. et al. Comprehensive analysis of serum metabolites in gestational diabetes mellitus by UPLC/Q-TOF-MS. Anal. Bioanal Chem. 408 (4), 1125–1135 (2016).
Bartho, L. A. et al. Plasma lipids are dysregulated preceding diagnosis of preeclampsia or delivery of a growth restricted infant. EBioMedicine 94, 104704 (2023).
Paapstel, K. et al. Inverse relations of serum phosphatidylcholines and lysophosphatidylcholines with vascular damage and heart rate in patients with atherosclerosis. Nutr. Metab. Cardiovasc. Dis. 28 (1), 44–52 (2018).
Rong, J., He, T., Zhang, J., Bai, Z. & Shi, B. Serum lipidomics reveals phosphatidylethanolamine and phosphatidylcholine disorders in patients with myocardial infarction and post-myocardial infarction-heart failure. Lipids Health Dis. 22 (1), 66 (2023).
Sevastou, I., Kaffe, E., Mouratis, M. A. & Aidinis, V. Lysoglycerophospholipids in chronic inflammatory disorders: the PLA(2)/LPC and ATX/LPA axes. Biochim. Biophys. Acta. 1831 (1), 42–60 (2013).
Koh, J. H. et al. Lipidome profile predictive of disease evolution and activity in rheumatoid arthritis. Exp. Mol. Med. 54 (2), 143–155 (2022).
Cui, Y. et al. Lysophosphatidylcholine and amide as metabolites for detecting alzheimer disease using ultrahigh-performance liquid chromatography-quadrupole time-of-flight mass spectrometry-based metabonomics. J. Neuropathol. Exp. Neurol. 73 (10), 954–963 (2014).
Tarnowski, M., Malinowski, D., Safranow, K., Dziedziejko, V. & Pawlik, A. CDC123/CAMK1D gene rs12779790 polymorphism and rs10811661 polymorphism upstream of the CDKN2A/2B gene in women with gestational diabetes. J. Perinatol. 37 (4), 345–348 (2017).
Liu, T., Deng, J. M., Liu, Y. L., Chang, L. & Jiang, Y. M. The relationship between gestational diabetes mellitus and Interleukin 1beta gene polymorphisms in Southwest of China. Med. (Baltim). 99 (43), e22679 (2020).
Azmi, M. B. et al. Analyzing molecular signatures in preeclampsia and fetal growth restriction: identifying key genes, pathways, and therapeutic targets for preterm birth. Front. Mol. Biosci. 11, 1384214 (2024).
He, B. et al. The maternal blood lipidome is indicative of the pathogenesis of severe preeclampsia. J. Lipid Res. 62, 100118 (2021).
Gibbone, E., Huluta, I., Wright, A., Nicolaides, K. H. & Charakida, M. Maternal cardiac function at midgestation and development of preeclampsia. J. Am. Coll. Cardiol. 79 (1), 52–62 (2022).
Malek, A. M. et al. Maternal coronary heart disease, stroke, and mortality within 1, 3, and 5 years of delivery among women with hypertensive disorders of pregnancy and Pre-Pregnancy hypertension. J. Am. Heart Assoc. 10 (5), e018155 (2021).
Yang, Y. & Wu, N. Gestational diabetes mellitus and preeclampsia: correlation and influencing factors. Front. Cardiovasc. Med. 9, 831297 (2022).
Cohen, A. et al. Circulating levels of the antiangiogenic marker soluble FMS-like tyrosine kinase 1 are elevated in women with pregestational diabetes and preeclampsia: angiogenic markers in preeclampsia and preexisting diabetes. Diabetes Care. 30 (2), 375–377 (2007).
Troisi, J. et al. Placent. Metabolomics Fetal Growth Restriction Metabolites 13(2). (2023).
Mehta, D. et al. Genome-wide gene expression changes in postpartum depression point towards an altered immune landscape. Transl Psychiatry. 11 (1), 155 (2021).
Azami, M., Badfar, G., Soleymani, A. & Rahmati, S. The association between gestational diabetes and postpartum depression: A systematic review and meta-analysis. Diabetes Res. Clin. Pract. 149, 147–155 (2019).
Wang, R., Liang, X. & Su, X. Y. Analysis of risk factors for postpartum depression after Cesarean section in women with early-onset preeclampsia. World J. Psychiatry. 14 (10), 1448–1457 (2024).
Roberts, L., Henry, A., Harvey, S. B., Homer, C. S. E. & Davis, G. K. Depression, anxiety and posttraumatic stress disorder six months following preeclampsia and normotensive pregnancy: a P4 study. BMC Pregnancy Childbirth. 22 (1), 108 (2022).
Brown, M. A. et al. Hypertensive disorders of pregnancy: ISSHP classification, diagnosis, and management recommendations for international practice. Hypertension 72 (1), 24–43 (2018).
Tanner, M. S., Davey, M. A., Mol, B. W. & Rolnik, D. L. The evolution of the diagnostic criteria of preeclampsia-eclampsia. Am. J. Obstet. Gynecol. 226 (2s), S835–s43 (2022).
Diagnostic criteria and classification. Of hyperglycaemia first detected in pregnancy: a world health organization guideline. Diabetes Res. Clin. Pract. 103 (3), 341–363 (2014).
Xie, Z. et al. Gene set knowledge discovery with enrichr. Curr. Protocols 1(3). (2021).
Kuleshov, M. V. et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 44 (W1), W90–W7 (2016).
Chen, E. Y. et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinform. 128. (2013).
Acknowledgements
We would like to thank the contribution from Ms. Wanxing Xu and Prof. Xiaoqi Zheng from the Centre for Single-Cell Omics for their HPC support. Figure 1B was created in https://BioRender.com.
Funding
This work was supported by Shenzhen Science and Technology Program [grant numbers JCY20210324141008021, JCYJ20230807141516033, JCYJ20240813152051065].
Author information
Authors and Affiliations
Contributions
Conceptualization: H.Zhang and X.Wang; Methodology: C.Zhang and X.Wang; Data collection: Y.Huang, X.Mai, L.Liu, Y.Li, F.Zheng, J.Cao, C.Zou, C.Wang, J.Ran and H.Zhang; Data analysis and curation: C.Zhang and X.Wang; Writing – original draft: C.Zhang and X.Wang; Writing – editing: C.Zhang, Y.Huang, X.Mai, H.Zhang and X.Wang; Reviewing: all authors; Visualization: C.Zhang, Y.Huang, X.Mai, H.Zhang and X.Wang; Supervision: H.Zhang and X.Wang.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zhang, C., Huang, Y., Mai, X. et al. Lipidomic and transcriptomic profiling reveal alterations in the coexistence of gestational diabetes mellitus and preeclampsia impacting maternal and neonatal outcomes. Sci Rep 15, 27102 (2025). https://doi.org/10.1038/s41598-025-12077-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-025-12077-5