Abstract
The sulfur-containing amino acids (SAAs) play a key role in the occurrence and development of tumors. However, the clinical prognostic value of SAAs metabolism (SAAM) regulators in stomach adenocarcinoma (STAD) remains unclear. We systematically evaluated the clinical and immune characteristics of SAAM-related genes in STAD. Furthermore, a SAAM score model was constructed, and patients in the low-SAAM score group had a better prognosis. As the core gene in the model, the low expression of cystathionine beta-synthase (CBS) indicated a better prognosis for patients. Interfering with CBS expression in MKN-45 cells inhibited cell proliferation, reduced the production of glutathione (GSH), and promoted cellular oxidative stress. Importantly, the downregulation of CBS heightened sensitivity to ferroptosis inducers in STAD cells, highlighting the involvement of CBS in ferroptosis. In conclusion, the utilization of SAAM for the identification and personalized scoring of patients might potentially play a significant role in evaluating prognosis, immune infiltrates, and guiding treatment for STAD.
Similar content being viewed by others
Introduction
Gastric cancer (GC) ranks among the top five leading causes of cancer-related deaths worldwide1. Patients with advanced GC, particularly stage IV stomach adenocarcinoma (STAD), face a poor prognosis, with median survival often less than 1 year despite chemotherapy2,3. The notable heterogeneity observed among STAD tumors, encompassing genetic disparities and distinct microenvironmental characteristics, creates a conducive environment for both tumor progression and the emergence of drug resistance4,5. In personalized therapy-focused clinical trials, the adoption of a scoring system based on tumor mutational burden (TMB) emerged as a potential biomarker, correlating with improved overall survival (OS) in cases of toripalimab-treated chemorefractory GC6. This underscores the potential of personalized stratification for prognostic assessment and treatment guidance, pointing towards a promising avenue of development7,8. Nonetheless, the current landscape still lacks dependable categorization and prognostic evaluation methodologies for STAD. As such, the quest for more reliable and precise prognostic biomarkers or evaluation approaches remains a frontier worth exploring.
Sulfur-containing amino acids (SAAs), primarily methionine and cysteine, are vital in human metabolism9. Methionine undergoes transmethylation to synthesize S-adenosylmethionine and homocysteine. Homocysteine is then either remethylated back to methionine or catabolized via the transsulfuration pathway by cystathionine beta-synthase (CBS) and cystathionine gamma-lyase (CTH) to synthesize cysteine and glutathione (GSH)10,11,12. CBS is a crucial enzyme in sulfur metabolism, catalyzing the essential condensation of homocysteine and serine to form cystathionine in the transsulfuration pathway13,14. This process involves the transfer of sulfur atoms between these amino acids. Cystathionine is subsequently converted into cysteine through the action of other enzymes in the pathway15. Given its central role in sulfur metabolism, any dysfunction or deficiency in CBS activity has implications for the cellular levels of GSH, potentially affecting the ability of cells to manage oxidative stress and maintain proper cellular function16. Previous studies have shown that sulfur-containing amino acids metabolism (SAAM) and related genes are associated with many diseases. For instance, high concentrations of methionine, S-adenosylmethionine and homocysteine favor the transsulfuration over the remethylation in cells, thereby reducing the excessive accumulation of homocysteine and reducing the occurrence of severe hyperhomocysteinemia17. Silencing of GSH synthesis enzymes selectively induced the cytotoxicity of cell renal cell carcinoma cells while sparing normal cells18. Similarly, cysteine (a key GSH precursor) drives hypoxia adaptation and carboplatin resistance in ovarian cancer19. However, there are few studies on SAAM and related genes in STAD. This study aims to analyze SAAM in STAD using bioinformatics, construct a prognostic and therapeutic scoring model, and identify a central SAAM gene to provide a potential therapeutic target for STAD.
In the present investigation, a significant correlation has been discerned between SAAM and STAD. Furthermore, the metabolic processes involving SAAs confer unique action in organisms. Our findings suggested that SAAM is associated with OS and clinical outcomes in STAD patients. We used the databases to explore the effect of CBS, a key gene in SAAM, on STAD prognosis and subsequently validated our findings in vitro. The results demonstrated that CBS plays a protective role against oxidative stress and ferroptosis in STAD cells. To summarise, our investigation underscored the prospective utility of the SAAM scoring model and CBS as a biomarker, with the potential to significantly contribute to the assessment of therapeutic efficacy and prognosis in the context of STAD.
Materials and methods
Data sources and pre-processing
From The Cancer Genome Altas (TCGA, https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga) database, the RNA sequencing (FPKM value) and corresponding clinical data of STAD were downloaded, including 343 STAD and 30 normal control samples. The data set GSE84437 (433 samples) was obtained from Gene Expression Omnibus (GEO, https://www.ncbi.nlm.nih.gov/geo/). We used the FPKM function of the “limma” R package to transform the RNA-seq transcriptome data into transcripts per kilobase million (TPM). The CNV (copy number variation) analysis of the SAAM regulators was performed on the eligible data sets. Following the CNV analysis, a depiction of copy number alterations across the 23 chromosomes was rendered using the R programming language and its associated packages.
Consensus clustering analysis of SAAM regulators
Patients were divided into three different subtypes based on the expression of SAAM regulators by performing consistent unsupervised cluster analysis of the Consensus Cluster Plus R package20. A stable clustering met the following conditions. Firstly, a more step-like curve and a greater area under the CDF (cumulative distribution function) were considered more stable clustering. Afterward, each group has a larger sample size. Lastly, correlations between groups decreased and within groups increased.
Assessment of the relationship between various molecular subtypes with the clinical characteristics and prognosis of STAD
The progression of STAD is closely related to the clinical characteristics of patients. To further identify the clinical value of different subtypes, we analyzed the relationship among molecular subtypes, prognosis, and clinicopathological characteristics after downloading clinical data. The corresponding clinical data covered sex, age, TNM stage, and survival status. The difference between the OS of patients identified by consensus clustering was examined through Kaplan–Meier curves of R packages. The “GSVA” R package was employed for GSVA enrichment analysis to understand the differences of SAAM regulators in each group during the biological process. We received the data for GSVA analysis from the MSigDB database.
Identification of the immune cell infiltrating feature in the disparate molecular subtypes
The infiltrating immune cell gene set covers activated CD8 T cells, activated CD4 T cells, activated dendritic cells, activated B cells, macrophages, mast cells, monocytes, natural killer T cells, regulatory T cells, and neutrophils. We performed the single-sample gene set enrichment analysis (ssGSEA) algorithm for quantitative analysis of the difference in each immune cell infiltration in STAD. The R package “ggplot2” was used to map the enrichment results. The discrepancy of disparate SAAM clusters was manifested by the immune cell infiltration analysis.
Screening of overlap genes and functional annotation
In this study, the R package “limma” was used to calculate the fold change value of gene expression level to uncover the overlapping genes of various SAAM groups. Criteria of statistical significance were set at P < 0.001 for screening the overlap genes in the SAAM pattern in STAD. Subsequently, the underlying functional attributes of overlap genes were explored by applying the Gene Ontology (GO) and the Kyoto Encyclopedia of Genes and Genomes (KEGG).
Establishment of the SAAM-related gene signature
To establish a prognostic score model, we conducted an unsupervised clustering analysis to investigate overlapping gene features in various SAAM groups, which divided the STAD patients into different groups for further study. We studied the prognosis of each crossover gene utilizing univariate Cox regression analysis. Next, the principal component analysis (PCA) was conducted to further analyze genes associated with prognosis. Finally, the result of PCA analysis in the “ggplot2” R package was used to structure the SAAM-related gene signature as well as analyze the prognosis of STAD patients21. Patients were divided into two groups by the maximally selected rank statistics as the threshold, which covered high- and low-score groups. We continuously performed Kaplan–Meier analysis of high- and low-score groups in STAD.
Correlation between the SAAM score and immune-related functions
The correlation analysis between SAAM score groups and immune cells was carried out by the Wilcoxon test. To identify the somatic mutations of patients in various groups with STAD, the TMB score was investigated by stratified analysis. We then conducted survival analysis for samples with high- or low-TMB scores. Microsatellite instability (MSI) refers to the phenomenon of MS sequence length changes caused by insertion or deletion mutations during DNA replication. Hence, we assessed the relationship between the SAAM-related gene signature and MSI to evaluate the prognostic value of immunotherapy. PD-1 is a significant immune checkpoint, therefore, we evaluated its differential expression densities employing the Wilcoxon test.
Cell lines and cell culture conditions
GES-1 cells (#SNL-304) and MKN-45 cells (#SNL-173) were purchased from Wuhan Sunncell Biotechnology and authenticated by STR. MKN-1 cells (#CBP60486) were purchased from Nanjing Cobioer Biosciences and authenticated by STR. Briefly, cells were suspended and cultured in RPMI 1640 (Gibco, #8118179) containing 10% serum (Corille, #C1015-05) and 1% penicillin-streptomycin (Beyotime, #C0222) in an incubator with 10% CO2 at 37 °C. Mycoplasma scavengers (Yeasen Biotechnology, #40607ES08) were added to the complete medium once a month for one week to exclude mycoplasma contamination. All experiments were performed using cells in the logarithmic growth phase.
Western blot
Cells were lysed with RIPA (Beyotime, #P0013B) supplemented with protease and phosphatase inhibitors (Beyotime, #P1051) to obtain total protein. The proteins were adjusted to a consistent concentration using the Pierce™ BCA Protein Assay Kits (Thermo Scientific™, #23227) and were analyzed by SDS-PAGE using a 12% polyacrylamide gel. Subsequently, proteins were transferred to PVDF membranes (Bio-Rad, #1620177) for evaluation. PVDF membranes were blocked with 5% skim milk for one hour prior to incubation with primary antibodies for 12 h. After another hour of incubation with the corresponding secondary antibody, the antibody bound to the FDbio-Pico ECL luminescent solution (FDbio science, #FD8000) was visualized the protein in Image Lab software. The antibodies contain CBS (ABclonal, #A11612), GAPDH (ABclonal, #AC033), nuclear factor erythroid 2-related factor 2 (NRF2, Proteintech, #80593-1-RR), manganese superoxide dismutase (MnSOD, Abcam, #ab68155), solute carrier family 7 (xCT, Cell signaling, #12691S), γ-H2AX (Abcam, #ab81299), thioredoxin-2 (TRX-2, Abcam, #ab133524), ferritin heavy chain (FTH, Abcam, #ab176558), glutathione peroxidase 4 (GPX4, Proteintech, #30388-1-AP), Catalase (Proteintech, #21260-1-AP), glutathione reductase (GR, Abcam, #ab124995), HRP-labeled Goat Anti-Rabbit IgG (H + L) (Beyotime, A0208) and HRP-labeled Goat Anti-Mouse IgG (H + L) (Beyotime, A0216). Unprocessed images are presented in the Supplementary Materials 2.
Clinical tissue samples
Six pairs of STAD clinical samples and adjacent tissues were obtained from STAD patients who underwent surgery at Zhejiang Provincial People’s Hospital. The postoperative tumor specimens were diagnosed as STAD by the pathologists. The STAD patients from whom the specimens were selected for this study were all voluntary participants and had signed the relevant informed consent forms. The collection process of tissue specimens has been reviewed by the ethics committee of Zhejiang Provincial People’s Hospital (approval number: ZJPPHEC2024O(087)). Clinical samples were analyzed for CBS protein levels using paired T-tests. All methods in our study were carried out in accordance with relevant guidelines and regulations.
Quantitative real-time PCR
Total RNA of gastric normal cells and cancer cells was extracted utilizing SteadyPure RNA Extraction Kit (Accurate Biology, #AG21024) and reverse transcribed to cDNA using the Evo M-MLV RT Mix Kit with gDNA Clean for qPCR Ver.2 (Accurate Biology, #AG11728) according to the instructions of the manufacturer. Quantitative real-time PCR (qRT-PCR) was performed with SYBR Green Premix Pro Taq HS qPCR Kit (Accurate Biology, #AG11701). The primer sequences were listed as below: human CBS, forward: 5′ - GCAAAGTCATCTACAAGCAG-3′, reverse: 5′ - CGAAGTTCAGCAAGTCAATG − 3′. human β-actin, forward: 5′ -CACCATTGGCAATGAGCGGTTC-3′, reverse: 5′ - AGGTCTTTGCGGATGTCCACGT − 3′.
Small interfering RNA transfection
Down-regulation of CBS in STAD cells was achieved by transfection of small interfering RNA (siRNA). Cells were subjected to transfection experiments when 50% density was reached in six-well plates. 400 µL of MEM medium (CIRENRY, #CR22600) supplemented with 5 µL Lipofectamine 3000 (GLPBIO, #GK20006) and 5 µL siRNA were added to the complete medium without antibiotics. The effect of interference with CBS was verified by Western blot after 72 h.
The growth curve and edu staining experiments
5000 cells were seeded per well in a 96-well plate (NEST Biotechnology), and 200 µL of 10% complete medium was added, which was cultured in a cell incubator. At 0, 24, 48, and 72 h, 10 µL of CCK-8 (Meilunbio, #MA0218) reagent was added to each well and incubated for 2 h. The absorbance at 450 nm was measured using a microplate reader to assess cell viability. 200,000 cells were cultured in 24-well plates (NEST Biotechnology) for 12 h with the complete medium. Prior to fixing the cells with formaldehyde, the STAD cells were incubated with 10 µM EdU (Beyotime, #C0071S) for 4 h. Subsequently, the cells were treated with 0.3% Triton for 20 min and blocked with 10% BSA for 1 h. Finally, the reaction solution was prepared according to the instructions. Representative images were captured under an M7000 microscope.
Determination of reactive oxygen species (ROS), H2S, and lipid peroxide content
ROS, H2S, and lipid peroxides in STAD cells were labeled using DCFH-DA (Sigma, #35845), BODIPY (Invitrogen, #D3861), and WSP-1 (MCE, #HY-124409) dyes, respectively. Cells were treated with RSL-3 (5 µM) for 4 h and then stained with DCFH-DA (10 µM) or BODIPY (5 µM) for half an hour, then examined by flow cytometry or typically photographed by an M7000 microscope. Before staining WSP-1 (15 µM), cells were treated with serum-free medium for 6 h and subsequently detected for changes in fluorescence by flow cytometry.
Measurement of total GSH levels
500,000 cells were collected and resuspended in protein removal reagent S. Subject the suspension to three cycles of rapid freezing and thawing, alternating between liquid nitrogen and 37 °C. GSH samples were obtained by centrifugation at 10,000 g for 15 min after 5 min of ice bath. Prepare the necessary working solutions as the instructions in the Total Glutathione Assay Kit (Beyotime, #S0052). Employ the microplate reader to continuously monitor the maximum absorbance at 412 nm of the samples for one hour. Simultaneously, prepare equivalent samples for protein concentration determination to facilitate the calculation of GSH content.
Statistical analysis
The correlation coefficient between tumor microenvironment (TME) and SAAM-related gene expression was determined by applying Spearman and differential expression analyses. We compared the differences between different groups by conducting the One-way ANOVA and the Kruskal–Wallis test. The R package “survminer” was used to determine the cutoff point for each dataset subgroup. Survival analysis of different clusters was conducted via the Kaplan–Meier method and log-rank test. The hazard ratios (HR) for SAAM regulators were analyzed by using univariate regression analyses to identify the prognostic value. The mutation landscape in patients with high and low SAAM scores was presented by the waterfall function of the maftools package. In this study, all statistical analysis was performed using R software version 4.1.0 (http://www.R-project.org) and GraphPad Prism (version 9.0). Statistical significance was defined at *P < 0.05, **P < 0.01, and ***P < 0.001 compared to the corresponding control.
Results
Landscape of SAAM-related genes in STAD
In the present study, genes for SAAM were analyzed and the summary of these genes is presented in Supplementary Table S1. We first obtained the incidence of somatic mutations in 23 SAAM-related genes. As shown in Figure S1A, the frequency of SAAM-related gene mutation in STAD was 14.09% and MTR exhibited the highest mutation rate. We further analyzed the correlation between the MTR mutation and the expression of SAAM-related genes. Surprisingly, the expression of GOT1, ENOPH1, and CTH was significantly upregulated in the MTR mutation group compared to the wild group (Figure S2). Among those SAAM-related genes, GCLC and SUOX exhibited higher gain in CNVs, while MTAP and CDO1 showed CNV loss (Figure S1B). Notably, the locations of CNV alterations in SAAM-related genes on chromosomes were presented in Figure S1C. We then compared the mRNA expression levels of SAAM-related genes between normal and STAD samples to explore whether SAAM-related genes are affected by gene variations. Our results indicated that there was no significant correlation between the CNV alterations and expression levels of the SAAM-related gene. Even some SAAM-related genes with CNV loss, most of them were evidently elevated in STAD tissues or showed no differences between tumor and normal tissues (Figure S1D). Expression of mRNA is regulated by multiple factors, not only CNV, which may explain the result.
Identification of SAAM subtypes in STAD
To investigate the relationship between SAAM-related gene expression and clinicopathological features, the k-means consensus clustering analysis was conducted in STAD samples. After adjusting the clustering variables (k) from 2 to 9, we found that patients showed the appropriate grouping status when k = 3 (Fig. 1A, Figure S3). Hence, these three subgroups (cluster A, cluster B, and cluster C) defined by k = 3 were used for further analysis. Kaplan-Meier survival analyses among the three subtypes showed that patients in cluster B had the best prognosis (Fig. 1B). Next, we explored the relationships between three subgroups and clinicopathologic characteristics including survival status (alive or dead), age ( < = 65 or > 65 years), node (0 or 1–3), gender (female or male), clinical stage (1–2 or 3–4) in GSE84437 and TCGA data sets. Patients with alive status or age < = 65 were mainly concentrated in the SAAM cluster C (Fig. 1C). In short, these analysis results indicated that the SAAM cluster is associated with the clinicopathologic characteristics and survival of patients with STAD.
TME and biological characteristics analysis of different subtypes
The TME plays a crucial role in tumor growth, invasion, metastasis, and response to therapy22. Understanding the TME is important for developing effective cancer treatments and improving patient outcomes. To estimate the TME heterogeneity of SAAM clusters, we compared the levels of infiltrating immune cells among the three groups. The results indicated that subtype A was enriched in CD56dim natural killer cells, monocytes, and neutrophils. Cluster B revealed an increased abundance of activated CD4 T cells, CD56bright natural killer cells, type 17 T helper cells, and type 2 T helper cells, while cluster C exhibited the highest level of activated B cells, activated CD8 T cells, eosinophil, immature B cells, MDSC, macrophage, mast cells, natural killer T cells, natural killer cells, plasmacytoid dendritic cells, regulatory T cells, T follicular helper cells, type 1 T helper cells than A and B (Fig. 1D).
Ulteriorly, GSVA enrichment analysis was performed to estimate the biological characteristics of SAAM clusters. As shown in Figure S4A and B, cluster A was highly enriched in the cellular processes, including the metabolism of GSH, drugs, and pyrimidines, as well as the biosynthesis of steroids, terpenoids, and N-glycans. The pathways associated with cluster B contained dilated cardiomyopathy, calcium signaling pathway, and vascular smooth muscle contraction. Evidently, cluster C was enriched in vascular smooth muscle contraction, dilated cardiomyopathy, and calcium signaling pathways. Collectively, SAAM clusters showed different biological characteristics and were closely related to the prognosis of patients.
Identification of the SAAM regulators’ molecular subtypes and clinicopathological characteristics analysis in STAD by consensus clustering. (A) Heatmap showing consensus clustering matrix for k = 3 as well as their area. (B) Kaplan–Meier curves presenting the survival rate of STAD patients based on three SAAM clusters utilizing univariate analysis. (C) Heatmap revealing relationships among clinicopathologic features, gene expression levels, and three SAAM clusters in STAD. (D) Associations between immune infiltration level of 23 immune cell types and three SAAM subtypes. Blue, green, or pink represent three various SAAM subtypes.
Differentially expressed genes (DEGs) among SAAM clusters in STAD and functional enrichment analysis
To further understand the biological features of different SAAM patterns, we tend to identify the DEGs among the three SAAM subtypes. PCA analysis exhibited the gene profile of different clusters (Fig. 2A). Next, 968 SAAM subtype-related DEGs were identified by the R package “limma” for further study (Fig. 2B). GO enrichment analysis indicated that these DEGs were evidently enriched in various cellular and biological processes, including organelle fission, nuclear division, mitochondrial matrix, mitochondrial inner membrane, and so on (Fig. 2C–D). In addition, the DEGs were closely involved in amyotrophic lateral sclerosis, biosynthesis of cofactors, and carbon metabolism signaling pathways (Fig. 2E–F). Our results suggested the momentous role of SAAM in tumorigenesis and tumor development.
DEGs among SAAM clusters in STAD and functional enrichment analysis. (A) PCA analysis based on three distinct SAAM subtypes in TCGA. STAD in the cluster A, B, or C subgroup is marked with blue, green, and pink. (B) Venn plots presenting intersection analysis of genes between SAAM subtypes in STAD. (C, D) GO enrichment analysis of intersectant genes among three SAAM clusters. (E, F) KEGG enrichment analysis of intersectant genes among three SAAM clusters.
Identification of gene cluster based on prognostic DEGs and construction of SAAM score
To comprehensively understand the molecular characteristics of various SAAM patterns, the consensus clustering analysis was executed, and the STAD patients were identified into three gene clusters (1, 2, and 3) based on 284 prognostic DEGs screened out from the DEGs of three SAAM clusters (Figure S5). Among the analyses exploring the correlation between gene clusters and clinical characteristics, there were significant differences in features such as prognosis and clinical T stages in the three clusters (Fig. 3A). However, those patients with various gene cluster profiles exhibited prognosis discrepancies. Kaplan-Meier plots showed that gene cluster 2 had the longest survival time (Fig. 3B). Whereafter, the expression level of SAAM genes was evaluated in multiple gene clusters. SAAM genes were distinct among the three gene clusters, and most of the genes were upregulated in gene cluster 2, as shown in Fig. 3C. Considering the individual heterogeneity, an optimum SAAM score based on DEGs was constructed using PCA analysis. STAD patients were divided into two groups according to the appropriate cut-off of scores. The distribution of patients in multiple groups, including SAAM subtypes, gene clusters, SAAM scores, and survival status was exhibited by Sankey plots (Fig. 3D). Meanwhile, the difference in SAAM score among SAAM clusters and gene clusters was evaluated. Gene cluster 3 presented the highest score while gene cluster 2 showed the lowest score (Fig. 3E). Similarly, the SAAM score in SAAM cluster C was the highest, and the score in SAAM cluster B was the lowest (Fig. 3F).
Identification of gene cluster based on prognostic DEGs and construction of SAAM score. (A) Heat map clarifying differences in clinicopathological parameters in various genetic patterns. (B) Kaplan–Meier curves illustrating the OS of patients in different gene clusters (log-rank tests, p-value = 0.002). (C) Boxplots of the distribution of gene expression based on three genetic patterns. The horizon axis represents distinct genes, while the vertical axis indicates gene expression levels. (D) Sankey diagram suggesting the relationship among SAAM clusters, gene clusters, SAAM score, and survival outcomes. (E) Box plots of the distribution of the SAAM score in three gene clusters. (F) Box plots displaying the distribution of the SAAM score in three SAAM clusters.
Prognostic significance and immune correlation of SAAM score in STAD
We further investigated the prognostic value of the SAAM score. The Kaplan–Meier curves showed that patients with low-SAAM score had longer survival times than those in the high-SAAM score group (Fig. 4A). This result of Kaplan–Meier analysis was consistent with the previous results that SAAM subtype C (Figs. 1B and 3F) and gene cluster 3 (Fig. 3B and E) with the highest SAAM score had the poorest prognosis. Moreover, those patients with low-SAAM scores exhibited higher live rates (Figs. 4B and 64% vs. 46%), which can explain why the SAAM score of survivors was significantly decreased (Fig. 4C). In addition, no matter whether in stage T1-2 or T3-4, the high SAAM score of patients suggested a poor prognosis (Fig. 4D–E). As shown in Fig. 4F, the SAAM score was significantly positively related to activated B cells, mast cells, and type 17 T helper cells, while negatively related to activated CD4 T cells, CD56dim NK cells, neutrophil, and type 2 T helper cells.
Prognostic significance and immune correlation of the SAAM score in STAD. (A) Kaplan–Meier analysis of the OS of patients based on SAAM score in STAD (log-rank tests, p < 0.001). The high and low SAAM scores of STAD samples are listed under the Kaplan–Meier plot. (B) Correlations of the SAAM score and the survival status of patients. (C) Box plot of the allocation between the SAAM score and the survival status of patients. (D) Survival analysis between patients in T1-2 cohorts stratified by high-SAAM score (pink) and low-SAAM score (blue). (E) Kaplan–Meier curve analysis between patients in T3-4 cohorts stratified by SAAM scores high (pink) and low (blue). (F) Heatmap revealing the correlation between the SAAM score and immune infiltrating cells. Red and blue manifest a positive correlation and a negative correlation, respectively.
Correlation between TMB, MSI, and SAAM scores in STAD
TMB has gained significant attention in cancer research and clinical practice as tumors with high TMB may be more susceptible to immune checkpoint inhibitor therapy23. Next, we intended to clarify the correlativity among TMB and SAAM scores in STAD. Spearman correlation analysis indicated a negative correlation between TMB and SAAM score (Figure S6A). As expected, patients with high SAAM scores exhibited low TMB, while those with low SAAM scores showed high TMB (Figure S6B). After proving the prognostic value of TMB, we combined TMB with the SAAM risk score in STAD to evaluate the outcomes of four novel subgroups. The subsequent stratified survival analysis showed that the SAAM score could distinguish the survival of STAD patients in both high- and low-TMB subgroups, and the trend of survival advantage in the high- and low-TMB groups was improved by the low-SAAM score (Figure S6C). Ultimately, the top 20 mutation genes in the high- and low-SAAM score groups based on the TCGA-STAD dataset were presented on the waterfall diagram. The top mutated genes in the two groups were the same, but the mutation frequencies were different. The mutation frequencies of the top three genes, including TTN, TP53, and MUC16, in the high-SAAM score group were significantly lower than those in the low SAAM score group (Figure S6D).
Accumulated evidence indicated that patients with high microsatellite instability (MSI-H) are more likely to respond to immune checkpoint inhibition and benefit from immunotherapy24,25. In the further study, we visualized the relationship between the SAAM score with MSI status. Whether in the high SAAM score group (52%) or low score group (84%), patients with microsatellite stable (MSS) account for the majority. Furthermore, patients with MSI-H in the low SAAM score group were more than those in the high SAAM score group (Figure S6E, 31% vs. 4%). Consistently, the MSI-H group had the lowest SAAM score (Figure S6F). In addition, the mRNA level of PD-L1 was downregulated in the high-score group, which may indicate that a high SAAM score shows a worse response to ICIs (Figure S6G).
CBS as a key gene in the SAAM model. (A) Interaction map of the SAAM-associated proteins. The color from light green to dark blue represents the degree value of the node from small to large. (B) Survival analysis of patients with high and low CBS expression. (C) CBS expression in STAD at different stages. (D) Differences in CBS protein between normal and STAD tissues. N and T represent normal and tumor samples, respectively. (E) CBS protein expression in normal and STAD cells. (F) Quantification of protein levels of CBS in normal and STAD cells. (G) CBS mRNA expression in normal and STAD cells.
CBS as a key gene in the SAAM model
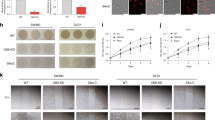
Utilizing the STRING database, an analysis of the interaction network among 23 SAAM-related proteins was conducted. CBS was confirmed to have the strongest association with other SAAM genes (Fig. 5A). Survival curve analysis revealed that low expression of CBS was associated with a favorable prognosis among STAD patients (Fig. 5B). Subsequent analysis of CBS expression in various stages of STAD demonstrated lower expression in early-stage samples and higher expression in advanced-stage samples (P = 0.0042) (Fig. 5C). We detected the expression of CBS in 15 pairs of STAD tissues and adjacent non-cancerous tissues. In the examined samples, 33.3% exhibited relatively low expression of CBS in STAD tissues, while 53.3% demonstrated elevated expression of CBS in STAD tissues (Figure S7). In addition, we collected normal gastric tissue and STAD tissue samples to analyze the expression of CBS protein, attesting that the CBS protein level was up-regulated in STAD tissues (Fig. 5D). Furthermore, we performed additional investigations into the expression of CBS in both normal gastric cells and STAD cells, which unveiled a noteworthy elevation in both CBS mRNA and protein levels in the STAD cell line MKN-45 (Fig. 5E–G).
CBS deletion induced Inhibition of STAD cell proliferation and reduced GSH production
To elucidate the role of CBS in STAD cells, siRNA was employed to specifically silence the expression of CBS protein in MKN-45 cells (Fig. 6A-B). The comparison of growth curves between the negative control (NC) and siCBS groups revealed a significant inhibition of MKN-45 proliferation following CBS silencing (Fig. 6C). Homoplastically, a comparison of EdU-positive cells between the NC and siCBS groups in MKN-45 demonstrated a marked impact of CBS silencing on STAD cell proliferation (Fig. 6D–E).
CBS undertakes the conversion of homocysteine to cystathionine and finally to cysteine in the transsulfuration pathway. Their levels were evaluated to determine the effect of CBS silence on STAD cells. The results showed that the level of homocysteine increased, while the levels of cysteine, cystathionine, and GSH decreased after interfering with CBS (Fig. 6F–I). Although H2S exhibited a decreasing trend, no significant difference was observed, indicating that the 50% efficiency of CBS silencing was inadequate to impact H2S production (Fig. 6J). The decrease in GSH content usually disrupts the balance between cellular oxidative and antioxidative systems. By examining pertinent proteins, alterations in this equilibrium can be elucidated. The downregulation of CBS expression resulted in the decrease of antioxidant proteins NRF2, MnSOD, TRX-2, and GPX4, and the increase of GR and Catalase, signifying a disturbance in the equilibrium between cellular oxidative and antioxidative systems, thereby inducing oxidative stress (Fig. 6K and Figure S8). Oxidative stress can result in various forms of DNA damage, with γ-H2AX serving as a marker for it26. The silencing of CBS induced an upregulation of γ-H2AX protein levels, indicating that CBS deletion could trigger DNA damage (Fig. 6K). In summary, targeted silencing of CBS substantially suppressed the proliferation of STAD cells, diminished GSH production, and provoked oxidative stress within the STAD cells.
CBS deletion inhibited STAD cell proliferation and induced oxidative cell damage. (A) The changes in CBS protein were detected by western blot. (B) The silencing efficiency of CBS was quantified. (C) The cell viability of MKN-45 was measured at 0, 24, 48, and 72 h to reflect its growth rate. (D) Proliferating cells from both the NC and siCBS groups were photographed by an M7000 microscope. (E) EdU-positive cells from both the NC and siCBS groups were quantified. (F–H) Relative levels of homocysteine (F), cysteine (G), and cystathionine (H) in STAD cells after interfering with CBS. (I) Changes in total intracellular GSH. (J) Effect of CBS deletion on intracellular H2S production. (K) Antioxidant proteins and markers of DNA damage were tested by western blot.
CBS silencing promotes ferroptosis in STAD cells
Besides its role in ROS scavenging, GSH also eliminates lipid peroxides, which are an important defense to ferroptosis27. Thus, it is hypothesized that CBS can heighten the susceptibility of STAD cells to ferroptosis through GSH depletion. CBS deficiency resulted in reduced xCT and FTH levels, enhancing the propensity of cells to undergo ferroptosis (Fig. 7A). The xCT protein plays a critical role in transporting cysteine for GSH synthesis, while FTH is indispensable for iron ion storage28,29. Therefore, we administered the ferroptosis inducers RSL-3 and DHA, both targeting the GPX4 protein, which uses GSH as a reducing agent to catalyze the reduction of lipid peroxides into the corresponding alcohols (Fig. 7B). Cytotoxicity assays demonstrated the enhanced sensitivity of the siCBS group to the ferroptosis inducers compared to the NC group (Fig. 7C). DCFH-DA fluorescence probe was employed to label intracellular ROS, demonstrating a notable elevation in ROS levels following RSL-3 treatment for 4 h in the siCBS group (Fig. 7D–E). Flow cytometry analysis utilizing the BODIPY fluorescence probe, a marker of lipid peroxides, revealed a substantial rise in the siCBS group treated with RSL-3 (Fig. 7F–G). These results underscored the significant role of CBS in the pathway of ferroptosis, which should not be overlooked.
Identification of the correlation between CBS and the TME
We utilized two single-cell RNA-seq datasets (STAD_GSE134520 and STAD_GSE167297) sourced from the TISCH database to investigate the expression of CBS in cells related to the TME. As depicted in Figure S9A, CBS exhibited predominant expression in immune cells. Further analysis revealed that fibroblasts and plasma cells displayed the highest levels of CBS expression (Figure S9B). These results indicated that the expression pattern of the CBS gene is different in STAD at the single-cell level, suggesting that the CBS gene may play an important role in specific cell types.
CBS silencing promotes ferroptosis in STAD cells. (A) Ferroptosis-related proteins changed with the decrease of CBS. (B) Schematic representation of GSH involvement in the ferroptosis pathway. (C) The cytotoxicity of RSL-3 (2.5 µM) and DHA (100 µM) was tested after the treatment of 20 h treatment in both NC and siCBS cells. (D) Typical pictures were captured to measure intracellular ROS levels using an M7000 microscope after the application of RSL-3 (5 µM) in MKN-45 cells. (E) DCFH-DA-labeled ROS levels were quantified. (F) After 4 h of treatment with RSL-3 (5 µM), the cells were stained with BODIPY for 30 min and the fluorescence intensity was measured by flow cytometry. (G) BODIPY-labeled lipid peroxide levels were quantified.
Discussion
STAD is a highly heterogeneous disease, even if similar pathological characteristics can lead to different outcomes. This suggests that previous staging systems may have reached the limits of predicting patient outcomes and treatment benefits30. Therefore, there is a need for a new classification of STAD patients based on genomic analysis and clinical evidence to provide preventive and therapeutic approaches. Precision oncology approaches based on molecular subtypes represent a promising frontier in cancer therapy31. For instance, Zhang et al. comprehensively evaluated m6A modification patterns based on 21 m6A regulators in GC and linked them to TME cell infiltration characteristics, further to determine the immune phenotypes of tumors and guide the more effective clinical practice32. Similarly, Chong et al. established a cuproptosis-related gene signature for prognostic risk assessment in GC33, while Lin et al. constructed an histone deacetylases score based on the expression level of histone deacetylases in GC to characterize the TME and predict immunotherapy efficacy34. This study integrated data from multiple levels including gene mutations, CNVs, and transcriptome expression from a novel perspective to comprehensively analyze the role of SAAM-related genes in STAD. We systematically evaluated their clinical and immunological relevance and constructed a SAAM scoring model as a novel prognostic and therapeutic tool. Furthermore, CBS was identified as the core gene in the SAAM model, and its mechanism of action in STAD cell proliferation, oxidative stress, and ferroptosis was explored, offering a new potential target for the treatment of STAD.
SAAM dysregulation contributes to tumorigenesis and malignant progression35,36. As important members of the antioxidant family, the increased level of SAAs was observed in the tissue metabolomic analysis of colorectal cancer37. Cysteine was upregulated in ovarian cancer patients, which helps ovarian cancer cells adapt to hypoxia stress and escape from carboplatin cytotoxicity38. Conversely, cysteine deprivation may lead to cell death. Inhibition of cysteine in retinal pigment epithelial cells induced GSH depletion, ultimately resulting in ferroptosis, autophagy, and premature senescence39. Moreover, studies have reported that methionine deficiency promoted mitophagy via lncRNA PVT1-mediated promoter demethylation of the BNIP3 in GC40. This evidence suggests that SAAM has the ability to influence tumor malignant progression. Our SAAM scoring system characterizes the integrated SAAM-mediated TME landscape in STAD, elucidating its impact on tumor development and guiding precision therapy.
We initially analyzed the mutation and expression patterns of SAAM genes in normal and STAD samples, revealing that CNVs may be the primary cause of gene alterations in tumor samples. The STAD samples were divided into three clusters, and clustering analysis was performed based on SAAM genes to investigate their clinical characteristics and TME. In comparison, SAAM cluster B showed better OS, while SAAM cluster C had the poorest prognosis. SAAM cluster B enriched activated CD4 T cells, CD56bright natural killer cells, type 17 T helper cells, and type 2 T helper cells, which play crucial roles in recognizing and attacking cancer cells41,42. Subsequently, we identified DEGs from these SAAM clusters and performed PCA to classify STAD into three refined gene clusters. These gene clusters delineated molecular characteristics of SAAM clusters more precisely, revealing key prognostic pathways and verifying SAAM cluster stability while uncovering intrinsic molecular differences between subtypes. The gene clusters exhibited distinct prognostic outcomes and TME features.
By further performing PCA analysis from prognostic DEGs, we successfully developed an SAAM scoring algorithm and discovered that STAD patients with lower scores exhibited better prognosis and improved survival status. The measurement of TMB is of significant importance in cancer research and treatment as it helps identify specific gene alterations that drive tumor growth and influence prognosis and treatment response43. Therefore, the association of SAAM scores with TMB was evaluated, which uncovered that high TMB and low SAAM scores were associated with better prognostic outcomes. Furthermore, there was a substantial increase in MSI-H in samples with low SAAM scores, hinting that immune therapy could be an effective treatment strategy for patients with low SAAM scores44. Besides, some tumor cells can evade the immune surveillance by expressing PD-L1, which interacts with PD-1 on T cells and suppresses their anti-tumor activity. This interaction between PD-L1 and PD-1 is known as an immune checkpoint pathway and serves as the target for immune checkpoint inhibitors45. Hence, we further examined the expression of PD-L1 in two different groups and found a significant increase in the low SAAM score group. In conclusion, these results indicated that the constructed SAAM scoring model can predict the prognosis of STAD patients as well as their sensitivity to immunotherapy.
Despite the extensive research on molecular biomarkers, many of the identified markers have not been validated successfully. Targeted therapy remains unavailable for the majority of patients with advanced STAD, and there are currently no diagnostic markers for secondary prevention. Interestingly, low expression of CBS was associated with a favorable prognosis in STAD patients. Single-cell analysis has identified CBS enrichment in fibroblast cells in STAD. Fibroblasts modulate subsequent immune responses when challenged with various tissue injuries, which include interactions with granulocytes and myeloid cells, as well as regulating the recruitment and retention of lymphocytes, which are crucial in connective tissue, contributing to wound healing, tissue repair, and immune regulation46,47.
CBS is an enzyme that plays a crucial role in SAAM and has been implicated in promoting cancer growth and progression through autocrine and paracrine mechanisms48,49. In 2012, CBS was first identified as an epigenetic biomarker for the molecular diagnosis of gastrointestinal cancers, sparking interest in its role in GC. In colorectal and GC patients, CBS was prone to methylation-mediated silencing, leading to the loss of its mRNA expression50. However, Zhang et al. discovered that the CBS protein is expressed and upregulated in human gastric carcinoma mucosa51. The inconsistent expression of CBS in different STAD patients may be due to inter-individual variability and tumor heterogeneity. Nevertheless, CBS was upregulated in STAD tissues compared to normal and STAD tissues in our analysis. Notably, CBS exhibited varying levels of expression across different stages of STAD, with higher expression of the CBS gene observed in more advanced stages of the tumor. Ulteriorly, we assessed the expression of CBS mRNA and protein in normal gastric cells and STAD cells through qRT-PCR and western blot experiments, respectively, revealing upregulation of CBS mRNA and protein expression in the STAD cell line MKN-45. This evidence suggested that CBS can be used as a molecular biomarker to provide new ideas for prognosis and tumor-targeted immunotherapy in STAD.
To test whether CBS affects the growth of STAD cells in vitro, we utilized siRNA to knock down CBS expression in MKN-45 cells. The growth curve and EdU staining assay results showed that CBS deletion had a significant impact on the proliferation of MKN-45 cells. CBS is responsible for the production of cystathionine, cysteine and H2S in the transsulfuration pathway, all of which are key antioxidant molecules36. CBS silencing causes the accumulation of homocysteine, decreased levels of cysteine and GSH. The lack of CBS catalysis prevents the smooth conversion of homocysteine to cystathionine. Although homocysteine forms mixed disulfides with cysteine and is excreted from the body, high homocysteine concentrations have little effect on the degree of dehydrosulfation of cysteine catalyzed by CBS or CTH. In general, the reaction of CTH is not saturated in cystathionine, and its reaction of catalyzing homocysteine to H2S, 2-ketobutyrate and NH4+ is highly sensitive to high concentrations of homocysteine10,52,53,54. Unfortunately, no significant change in H2S concentration was observed in STAD cells with CBS interference, which may be due to the low expression of CTH in gastric tissues55. Cystathionine has been considered an important intermediate in the transsulfuration pathway, and recent studies have shown that it resists endoplasmic reticulum stress-induced injury in a mouse model56. Cystathionine accumulation may occur in gastric tissues with low CTH expression, protecting gastric tissues from stress57. However, interfering with CBS reduced the production of cystathionine, and further studies are needed to investigate the changes in cystathionine and its role in STAD cells.
Previous studies have shown that cells with low expression of CBS and CTH only activate the transsulfuration pathway to maintain cysteine and REDOX homeostasis when exogenous cystine is depleted36,58,59. Most of the intracellular cysteines are derived from cystines transported from the extracellular by the xCT system60. Interestingly, xCT protein levels tended to decrease with CBS silencing. This indicated that CBS silencing exerted an inhibitory effect on the xCT system, which significantly reduced intracellular cysteine and GSH levels. However, the mechanism of the inhibition of the xCT system after CBS silencing needs to be further studied.
GSH is the most prevalent nonprotein thiol in cells. It is catalyzed by glutamate-cysteine ligase (GCL) to link cysteine to glutamate, producing γ-glutamylcysteine. Subsequently, glycine and γ-glutamylcysteine are catalyzed by GSH synthase (GSS) to produce GSH. Among them, GCL expression and cysteine concentration are the key factors for GSH synthesis61. The decreased concentration of cysteine in cells with CBS silence resulted in the lack of reaction substrates for GCL and GSS to synthesize GSH. Whether the deletion of CBS affects the expression of GCL and GSS has not been reported. Cancer cells, adapting to high levels of oxidative stress, become more sensitive to GSH deficiency62,63. After CBS deletion, resulting in decreased expression of antioxidant enzymes such as NRF2, MnSOD, and GPX4, and increased expression of GR and Catalase. These changes indicated that the antioxidant defense system of the cells is inhibited, and the activation of compensatory mechanisms is not able to fully counteract the increased oxidative stress. In addition, reduced TRX-2 expression further impaired the antioxidant capacity of the cells. These changes led to a decrease in GSH levels. The decrease in GSH disrupted the REDOX balance, preventing the consumption of intracellular ROS and lipid peroxides, and contributing to oxidative stress in cells, manifesting as a decrease in antioxidant proteins and the appearance of DNA damage.
In 2012, Dixon proposed ferroptosis as an iron-dependent programmed cell death process, which also heavily relies on GSH metabolism64. Hence, we hypothesized that CBS deficiency may render STAD cells more susceptible to ferroptosis. FTH serves as a key component in iron metabolism and storage, modulating the availability of intracellular iron65,66,67. By sequestering excess free iron, FTH helps maintain iron homeostasis and prevents the accumulation of labile iron, which is critical for initiating the lipid peroxidation process in ferroptosis68. FTH protein showed a tendency to decrease after CBS silencing. The loss of FTH will increase the intracellular free iron ion, induce more lipid peroxide production, and promote the occurrence of ferroptosis.
GPX4 catalyzes the reaction of GSH with ROS and lipid peroxides to form GSSG, which protects cells from ferroptosis11. RSL-3 and DHA were selected to confirm our hypothesis. They are both ferroptosis inducers that inhibit GPX4 protein. Consequences exhibited that ferroptosis inducers have greater toxicity against siCBS-treated STAD cells. In addition, siCBS-treated STAD cells treated with RSL-3 also revealed an increase in ROS and lipid peroxides. These results all provided evidence that CBS deletion induces cell susceptibility to ferroptosis. Based on this, studying the impact of CBS on ferroptosis will help deepen our understanding of the role of CBS-mediated resistance to oxidative stress and free radicals in the occurrence and development of STAD.
Several limitations need to be acknowledged in this study. First, although we constructed a SAAM scoring model from the first 23 genes, a number of emerging SAAM-related genes need to be enrolled in the model to optimize the accuracy of the patterns. Second, this SAAM scoring model was derived from retrospective datasets; therefore, a prospective cohort of STAD patients is required to validate our findings. Third, due to the variability and limitations of data from public databases, extensive in vitro and in vivo experiments are still required to support our findings.
Conclusions
In summary, we constructed a fascinating risk score model based on the expression patterns of SAAM in STAD. Our results revealed that the SAAM risk score can serve as a dependable indicator of clinical characterization, immune cell infiltration, and survival outcome. The downregulation of CBS, serving as the key gene in the model, led to reduced intracellular levels of cysteine and GSH, inducing oxidative stress, enhancing ferroptosis sensitivity, and inhibiting the growth of STAD cells. These findings provided evidence supporting the notion that patients with low CBS expression have a better prognosis. Our study significantly promoted the understanding of the potential mechanism of SAAM regulators in STAD, which may help to predict prognosis and even forecast the effects of immunotherapy.
Data availability
Sequence data that support the findings of this study have been deposited in the Gene-Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) and The Cancer Genome Atlas (TCGA; https://cancergenome.nih.gov/abouttcga/) with the primary accession code GSE84437.
Abbreviations
- CBS:
-
Cystathionine beta-synthase
- CDF:
-
Cumulative distribution function
- CTH:
-
Cystathionine gamma-lyase
- CNV:
-
Copy number variation
- DEG:
-
Differentially expressed genes
- FTH:
-
Ferritin heavy chain
- GC:
-
Gastric cancer
- GEO:
-
Gene expression omnibus
- GO:
-
Gene ontology
- GPX4:
-
Glutathione peroxidase 4
- GR:
-
Glutathione reductase
- GCL:
-
Glutamate-cysteine ligase
- GSH:
-
Glutathione
- GSS:
-
GSH synthase
- HR:
-
Hazard ratios
- KEGG:
-
Kyoto encyclopedia of genes and genomes
- MnSOD:
-
Manganese superoxide dismutase
- MSI:
-
Microsatellite instability
- MSI-H:
-
High microsatellite instability
- MSS:
-
Microsatellite stable
- NC:
-
Negative control
- NRF2:
-
Nuclear factor erythroid 2-related factor 2
- OS:
-
Overall survival
- PCA:
-
Principal component analysis
- qRT-PCR:
-
Quantitative real-time PCR
- ROS:
-
Reactive oxygen species
- SAAM:
-
Sulfur-containing amino acids metabolism
- SAAs:
-
Sulfur-containing amino acids
- siRNA:
-
Small interfering RNA
- ssGSEA:
-
Single sample gene set enrichment analysis
- STAD:
-
Stomach adenocarcinoma
- TCGA:
-
The cancer genome altas
- TMB:
-
Tumor mutational burden
- TME:
-
Tumor microenvironment
- TPM:
-
Transcripts per kilobase million
- TRX-2:
-
Thioredoxin-2
- xCT:
-
Solute carrier family 7
References
Bray, F. et al. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Cancer J. Clin. 74(3), 229–263 (2024).
Smyth, E. et al. Gastric cancer. The Lancet 396(10251), 635–648 (2020).
Ajani, J. et al. Gastric adenocarcinoma. Nat. Rev. Dis. Primers 3, 17036 (2017).
Cristescu, R. et al., Molecular analysis of gastric cancer identifies subtypes associated with distinct clinical outcomes. 21(5), 449–456 (2015).
Yang, J. et al. Identification of mitochondrial respiratory chain signature for predicting prognosis and immunotherapy response in stomach adenocarcinoma. Cancer Cell Int. 23(1), 69 (2023).
Wang, F. et al. Safety, efficacy and tumor mutational burden as a biomarker of overall survival benefit in chemo-refractory gastric cancer treated with toripalimab, a PD-1 antibody in phase ib/ii clinical trial NCT02915432. Ann. Oncol.: Official J. Eur. Soc. Med. Oncol. 30(9), 1479–1486 (2019).
Xu, H. et al. Development of a novel autophagy-related gene model for gastric cancer prognostic prediction. Front. Oncol. 12, 1006278 (2022).
Wang, X. et al. Characterization of pyruvate metabolism and citric acid cycle patterns predicts response to immunotherapeutic and ferroptosis in gastric cancer. Cancer Cell Int. 22(1), 317 (2022).
Bin, P., Huang, R. & Zhou, X. Oxidation resistance of the sulfur amino acids: Methionine and Cysteine. Biomed. Res. Int. 2017, 9584932 (2017).
Stipanuk, M. H. Metabolism of sulfur-containing amino acids: How the body copes with excess methionine, cysteine, and sulfide. J. Nutr. 150 (Suppl 1), 2494S–2505S (2020).
Bansal, A. & Simon, M. C. Glutathione metabolism in cancer progression and treatment resistance. J. Cell Biol. 217(7), 2291–2298 (2018).
Asantewaa, G. & Harris, I. Glutathione and its precursors in cancer. Curr. Opin. Biotechnol. 68, 292–299 (2021).
Zhu, H. et al., Cystathionine β-synthase in physiology and cancer. BioMed. Res. Int. 2018, 3205125 (2018).
Anturaniemi, J. et al. The effect of atopic dermatitis and diet on the skin transcriptome in Staffordshire bull terriers. Front. Vet. Sci. 7, 552251 (2020).
Zuhra, K. et al. Cystathionine-β-synthase: Molecular regulation and pharmacological inhibition. Biomolecules 10(5) (2020).
Li, H. et al. GSH exhaustion via Inhibition of xCT-GSH-GPX4 pathway synergistically enhanced DSF/Cu-induced cuproptosis in myelodysplastic syndromes. Free Radic. Biol. Med. 222, 130–148 (2024).
Tamanna, N. et al. Methionine restriction leads to hyperhomocysteinemia and alters hepatic H2S production capacity in Fischer-344 rats. Mech. Ageing Dev. 176 (2018).
Miess, H. et al. The glutathione redox system is essential to prevent ferroptosis caused by impaired lipid metabolism in clear cell renal cell carcinoma. Oncogene 37 (40), 5435–5450 (2018).
Nunes, S. et al. Cysteine boosters the evolutionary adaptation to CoCl mimicked hypoxia conditions, favouring carboplatin resistance in ovarian cancer. BMC Evol. Biol. 18(1), 97 (2018).
Jing, Q. et al. A novel RNA modification prognostic signature for predicting the characteristics of the tumor microenvironment in gastric cancer. Front. Oncol. 13, 905139 (2023).
Li, H. et al. Autophagy-related prognostic signature characterizes tumor microenvironment and predicts response to ferroptosis in gastric cancer. Front. Oncol. 12, 959337 (2022).
Sha, D. et al. Tumor mutational burden as a predictive biomarker in solid tumors. Cancer Discov. 10(12), 1808–1825 (2020).
Jardim, D. L. et al. The challenges of tumor mutational burden as an immunotherapy biomarker. Cancer Cell. 39(2), 154–173 (2021).
Picard, E. et al. Relationships between immune landscapes, genetic subtypes and responses to immunotherapy in colorectal cancer. Front. Immunol. 11, 369 (2020).
van Velzen, M. et al. MSI as a predictive factor for treatment outcome of gastroesophageal adenocarcinoma. Cancer Treat. Rev. 86, 102024 (2020).
Yu, X. et al. TEOA inhibits proliferation and induces DNA damage of diffuse large B-Cell lymphoma cells through activation of the ROS-dependent p38 MAPK signaling pathway. Front. Pharmacol. 11, 554736 (2020).
Li, Y. et al. Sorafenib induces mitochondrial dysfunction and exhibits synergistic effect with cysteine depletion by promoting HCC cells ferroptosis. Biochem. Biophys. Res. Commun. 534, 877–884 (2021).
Hu, W. et al. FTH promotes the proliferation and renders the HCC cells specifically resist to ferroptosis by maintaining iron homeostasis. Cancer Cell Int. 21(1), 709 (2021).
Koppula, P., Zhuang, L. & Gan, B. Cystine transporter SLC7A11/xCT in cancer: ferroptosis, nutrient dependency, and cancer therapy. Protein Cell. 12(8), 599–620 (2021).
Puliga, E. et al. Microsatellite instability in gastric cancer: Between lights and shadows. Cancer Treat. Rev. 95, 102175 (2021).
Matsuoka, T. & Yashiro, M. Biomarkers of gastric cancer: Current topics and future perspective. World J. Gastroenterol. 24(26), 2818–2832 (2018).
Zhang, B. et al. m6A regulator-mediated methylation modification patterns and tumor microenvironment infiltration characterization in gastric cancer. Mol. Cancer 19(1), 53 (2020).
Chong, W. et al. Clinical features and molecular landscape of cuproptosis signature-related molecular subtype in gastric cancer. IMeta 3 (3), e190 (2024).
Lin, Y. et al. Histone deacetylase-mediated tumor microenvironment characteristics and synergistic immunotherapy in gastric cancer. Theranostics 13(13), 4574–4600 (2023).
Ward, N. & DeNicola, G. Sulfur metabolism and its contribution to malignancy. Int. Rev. Cell. Mol. Biol. 347, 39–103 (2019).
Zhang, H-F. et al. Transsulfuration, minor player or crucial for cysteine homeostasis in cancer. Trends Cell Biol. 32(9), 800–814 (2022).
Kibi, M. et al. GC/MS and LC/MS-based tissue metabolomic analysis detected increased levels of antioxidant metabolites in colorectal cancer. Kobe J. Med. Sci. 65(1), E19–E27 (2019).
Nunes, S. et al. Cysteine allows ovarian cancer cells to adapt to hypoxia and to escape from carboplatin cytotoxicity. Sci. Rep. 8(1), 9513 (2018).
Sun, Y. et al. Glutathione depletion induces ferroptosis, autophagy, and premature cell senescence in retinal pigment epithelial cells. Cell Death Dis. 9(7), 753 (2018).
Xin, L. et al. Methionine deficiency promoted mitophagy via LncRNA PVT1-mediated promoter demethylation of BNIP3 in gastric cancer. Int. J. Biochem. Cell Biol. 141, 106100 (2021).
Sakaguchi, S. et al. Regulatory T cells and human disease. Annu. Rev. Immunol. 38, 541–566 (2020).
Freud, A. G. et al. The broad spectrum of human natural killer cell Diversity. Immunity 47(5), 820–833 (2017).
Addeo, A. et al. TMB or not TMB as a biomarker: That is the question. Crit. Rev. Oncol. Hematol. 163, 103374 (2021).
Baretti, M. & Le, D. T. DNA mismatch repair in cancer. Pharmacol. Ther. 189, 45–62 (2018).
Keir, M. E. et al. PD-1 and its ligands in tolerance and immunity. Annu. Rev. Immunol. 26, 677–704 (2008).
Buechler, M. B. et al. Cross-tissue organization of the fibroblast lineage. Nature 593(7860), 575–579 (2021).
Davidson, S. et al. Fibroblasts as immune regulators in infection, inflammation and cancer. Nat. Rev. Immunol. 21(11), 704–717 (2021).
Prudova, A. et al. S-adenosylmethionine stabilizes cystathionine beta-synthase and modulates redox capacity. Proc. Natl. Acad. Sci. U.S.A. 103(17), 6489–6494 (2006).
Szabo, C. et al. Tumor-derived hydrogen sulfide, produced by cystathionine-β-synthase, stimulates bioenergetics, cell proliferation, and angiogenesis in colon cancer. Proc. Natl. Acad. Sci. U.S.A. 110(30), 12474–12479 (2013).
Zhao, H. et al. Frequent epigenetic silencing of the folate-metabolising gene cystathionine-beta-synthase in gastrointestinal cancer. PloS One. 7 (11), e49683 (2012).
Zhang, L. et al., An anticancer role of hydrogen sulfide in human gastric cancer cells. Oxid. Med. Cell. Longev. 2015, 636410 (2015).
Chiku, T. et al. H2S biogenesis by human cystathionine gamma-lyase leads to the novel sulfur metabolites lanthionine and homolanthionine and is responsive to the grade of hyperhomocysteinemia. J. Biol. Chem. 284(17), 11601–11612 (2009).
Kabil, O. et al. Mechanism-based Inhibition of human persulfide dioxygenase by γ-glutamyl-homocysteinyl-glycine. J. Biol. Chem. 293(32), 12429–12439 (2018).
Majtan, T. et al. Biogenesis of hydrogen sulfide and thioethers by cystathionine beta-synthase. Antioxid. Redox. Signal. 28(4), 311–323 (2018).
Qu, H. et al. Identifying CTH and MAP1LC3B as ferroptosis biomarkers for prognostic indication in gastric cancer decoding. Sci. Rep. 14(1), 4352 (2024).
Maclean, K. N. et al. Cystathionine protects against Endoplasmic reticulum stress-induced lipid accumulation, tissue injury, and apoptotic cell death. J. Biol. Chem. 287 (38), 31994–32005 (2012).
Kraus, J. P. et al. Cystathionine gamma-lyase: clinical, metabolic, genetic, and structural studies. Mol. Genet. Metab. 97(4), 250–259 (2009).
Zhang, H-F. et al. Proteomic screens for suppressors of anoikis identify IL1RAP as a promising surface target in ewing sarcoma. Cancer Discov. 11(11), 2884–2903 (2021).
Sbodio, J. I., Snyder, S. H. & Paul, B. D. Regulators of the transsulfuration pathway. Br. J. Pharmacol. 176(4), 583–593 (2019).
Koppula, P. et al. Amino acid transporter SLC7A11/xCT at the crossroads of regulating redox homeostasis and nutrient dependency of cancer. Cancer Commun. (London England) 38(1), 12 (2018).
Chen, Y. et al. Glutathione defense mechanism in liver injury: Insights from animal models. Food Chem. Toxicol.: Int. J. Publ. Br. Ind. Biol. Res. Assoc. 60, 38–44 (2013).
Ren, X. et al. Overcoming the compensatory elevation of NRF2 renders hepatocellular carcinoma cells more vulnerable to disulfiram/copper-induced ferroptosis. Redox Biol. 46, 102122 (2021).
Li, Y. et al. Inhibition of CISD2 promotes ferroptosis through ferritinophagy-mediated ferritin turnover and regulation of p62-Keap1-NRF2 pathway. Cell. Mol. Biol. Lett. 27 (1), 81 (2022).
Dixon, S. J. et al. Ferroptosis: An iron-dependent form of nonapoptotic cell death. Cell 149(5), 1060–1072 (2012).
Du, J. et al. Identification of frataxin as a regulator of ferroptosis. Redox Biol. 32, 101483 (2020).
Du, J. et al. DHA exhibits synergistic therapeutic efficacy with cisplatin to induce ferroptosis in pancreatic ductal adenocarcinoma via modulation of iron metabolism. Cell Death Dis. 12 (7), 705 (2021).
Du, J. et al. DHA inhibits proliferation and induces ferroptosis of leukemia cells through autophagy dependent degradation of ferritin. Free Radical Biol. Med. 131, 356-369 (2019).
Li, Y. et al. CISD3 Inhibition drives cystine-deprivation induced ferroptosis. Cell Death Dis. 12(9), 839 (2021).
Funding
This study was supported by the National Natural Science Foundation of China (No. 82202429), the Medical and Health Science and Technology Project of Zhejiang Province (No. 2023RC228), Basic Scientific Research Funds of Department of Education of Zhejiang Province (No. KYYB2023012), Zhejiang Province Traditional Chinese Medicine Science and Technology Project (No. 2022ZB018).
Author information
Authors and Affiliations
Contributions
Z.P. and L.Y. designed the article form and wrote the paper. Z.P. and L.Y. finished the experiments. D.A., C.J., and H.H. investigated the information. W.C, D.J., L.Y and X.J. revised and finalized the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zhang, P., Liu, Y., Lu, Y. et al. Explore the role of CBS in stomach adenocarcinoma based on the sulfur-containing amino acid metabolism network. Sci Rep 15, 28829 (2025). https://doi.org/10.1038/s41598-025-12460-2
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-12460-2