Abstract
The phosphatidylethanolamine binding protein (PEBP) family regulates key plant processes including growth, development, flowering, seed germination, formation, and dormancy. Despite their importance for these processes that determine agronomical important characters, PEBP genes in faba bean (Vicia faba L.) and closely related legumes remain underexplored. This study identified 11 VfPEBP genes for the first time in Vicia faba, classifying them into MFT-like, TFL-like, and FT-like subfamilies, and examining their relationships to PEBP-genes of the legumes Pisum sativum and Vicia sativa. TFL1 homologs, crucial for growth determinacy, are an important focus for breeding to achieve a more confined flowering and maturation time. Cis-element analysis suggested VfTFL1-genes are regulated by light, hormones, and abiotic stress. Amplicon sequencing of VfTFL1a, a gene linked to shoot apical meristem fate, identified novel allelic variation but none of these could discriminate determinate from indeterminate varieties, invalidating a previously reported marker. Field trials revealed that determinate varieties of faba bean flowered later but had more uniform agronomic traits compared to indeterminate ones. These findings provide new insights into the PEBP gene family, highlighting its critical role in regulating flowering time and plant architecture in Vicia faba and underscoring its potential as a target for crop improvement.
Similar content being viewed by others
Introduction
Faba bean (Vicia faba L.), a grain legume of significant agricultural importance, is renowned for its high seed protein content and adaptability to various environmental conditions, making it a valuable crop for sustainable agriculture and a promising food source for a healthy diet1. The plant architecture of faba bean, affecting traits such as determinacy, branching, lodging, flowering time, pod setting, and maturation time, plays a crucial role in its overall productivity. Understanding the genetic and molecular basis of plant architecture in faba bean can therefore lead to significant advancements in breeding and agricultural practices. Central to plant development and architecture are the phosphatidylethanolamine-binding protein (PEBP) genes. Particularly the balance between FT, which promotes flowering, and TFL1, which represses it, modulates plant architecture by determining whether the meristem adopts indeterminate or determinate growth2. Although most faba bean varieties are indeterminate, some varieties exhibit determinate growth, characterized by the formation of a terminal inflorescence at the shoot apical meristem (SAM) that halts further plant growth, leading to a limited flowering time and more condensed seed maturation.
In plants, the PEBP gene family stands out due to its highly conserved nature and multifaceted functions; the genes in this family are involved in flower induction, determination of plant architecture, embryogenesis, stress tolerance, hormone signaling, and tuberization34. The PEBP family can be divided into the three subfamilies MOTHER OF FT AND TFL1 (MFT)-like proteins, TERMINAL FLOWER1 (TFL1)-like proteins, and FLOWERING LOCUS T (FT)-like proteins5. Many plant species, especially crop plants, have experienced an expansion of the PEBP gene family through whole-genome and segmental duplications, leading to functional diversification. Extensive research on individual PEBP genes across various plant species has revealed that dicotyledons typically possess six to nine PEBP genes, while monocotyledons exhibit approximately three times this number6.
In legumes, the FT and the TFL1-like genes are conserved but exhibit a substantial expansion in number compared to Arabidopsis, with some members displaying notably divergent sequences78. For example, there are five orthologs of TFL1 identified in chickpea (Cicer arietinum)9 and four in soybean (Glycine max)10. Three orthologs of TFL1 have been described in pea (Pisum sativum), one of which was found identical to the already known gene DETERMINATE (TFL1a/DET) and another one corresponding to the late flowering gene (TFL1c/LF). While the function of PsTFL1b remains uncharacterized11,12, the corresponding GmTFL1b has been identified as the key gene responsible for determinate plant architecture in soybean13. Three distinct subclades of FT (FTa, FTb, and FTc) genes are found in legumes8. In Arabidopsis, FT, and TFL1 are relatively short proteins (175 and 177 amino acids long, respectively) showing a high degree of sequence similarity (only 39 non-conserved amino acids) despite their opposite functions in flowering. In fact, a single amino acid substitution can reverse their respective functions14. Loss-of-function mutants of TFL1 in Arabidopsis exhibit early flowering, with a terminal inflorescence15. Conversely, over-expression of TFL1 significantly prolongs all developmental phases, showing that TFL1 functions to maintain the SAM in an indeterminate state by delaying the activation of genes responsible for floral meristem identity16. Just as for Arabidopsis, a mutation in the orthologous TFL1 gene of several legumes has been shown to increase the expression of LEAFY (LFY) and APETALA (AP1), resulting in a determinate growth of the inflorescence and the development of a terminal floral meristem preventing further SAM maintenance7,12,17.
In this study, members of the PEBP gene family in Vicia faba were identified and systematically analyzed at the genome-wide level for the first time, including their phylogenetic relationships, gene structure, and comparison to other Fabaceae species. Genetic variations in VfTFL1a were examined in a panel of eight Vicia faba varieties, encompassing determinate types, characterized by terminal inflorescences, and indeterminate types, which exhibit continuous leaf development at the shoot apical meristem. Additionally, field data on phenotypic variations in determinate and indeterminate varieties were assessed. PEBP genes were also identified in Vicia sativa, the closest relative of Vicia faba with a sequenced genome18. This study aimed to provide deeper insights into the evolutionary dynamics of the PEBP family within leguminous species.
Material & Methods
Plant material and DNA extraction
For genotypic comparisons, four indeterminate varieties Aurora, Mikko, Fanfare, and Taifun, and four determinate varieties Ticol, Tinova, Tina, and Bruno were used. All seeds were kindly provided from the John Innes Centre (Norwich, England), NordGen (Alnarp, Sweden), and Lantmännen (Svalöv, Sweden).
Seeds were sown in plastic pots with nutrient soil (50% peat, pH 5.5–6.5, added per m3 soil: 5.5 kg lime, NPK 11-5-18 kg, 200 g micronutrients and 100 g iron) under greenhouse conditions: 18–21 °C light 6–22 h < 200 W/m2. Young leaves were cut from plants and stored at −80 °C, until ground to a fine powder in liquid nitrogen in steel containers with 2 mm Ø steel beads, using a mixer mill (MM 400, Retsch GmbH, Haan, Germany) at 30 Hz. Genomic DNA from frozen leaf tissue was extracted using the Plant Mini Kit (Qiagen, Valencia, CA, USA) according to the manufacturer’s protocol. Complementing plants were germinated on media, and their DNA was extracted using the Nucleospin® Plant II kit (MACHERY-NAGEL), following previously established protocols19.
Sequence isolation and characterization
Vicia faba DNA sequences corresponding to AtTFL1 were amplified by PCR (Unocycler, VWR) using specific primers designed based on the published reference genome Vf var. Hedin/2 and var. Tiffany or primers derived from Avila et al. 2006 and 2007. For primer sequence and PCR settings, see Supplementary Table S1. The amplified products were cloned into pJET1.2-vector (Thermo Scientific, Massachusetts, USA) and sequenced using Sanger sequencing (Eurofins, Ebersberg, Germany). Sequence characterization, multiple sequence alignments, and visualizations were made using the software CLC Main Workbench 21.0.3 (Qiagen, Valencia, CA, USA).
Identification of PEBP gene family members in Vicia faba
This study used genetic data from four plant species—three legumes (Vicia faba, Vicia sativa, and Pisum sativum) and the model species Arabidopsis thaliana—to perform a comparative evolutionary analysis of the PEBP gene family. Protein and coding sequences were retrieved from publicly available reference genomes of Arabidopsis thaliana (TAIR11), Pisum sativum (GCA_024323335.2), Vicia faba var. Hedin/2 (GCA_948472305.1), and Vicia sativa (GCA_021764765.1). The A. thaliana genome was obtained from The Arabidopsis Information Resource (TAIR, http://www.arabidopsis.org), P. sativum and V. faba from NCBI, and V. sativa from the GigaDB database (http://gigadb.org/dataset)18,20,21.
PEBP family proteins containing the conserved PEBP motif (Pfam ID: PF01161.24) and the Hidden Markov Model (HMM) profiles were acquired from the HMMER web server (https://www.ebi.ac.uk/Tools/hmmer/). All PEBP genes were mined using two steps. First, the HMM profile was used as a query to search the protein databases of all species using HMMER V3.0 to identify candidate genes with the default “inclusion threshold”22. The Basic Local Alignment Search Tool Protein (BLASTP) program was used to search against the Arabidopsis thaliana proteome database using retained candidate genes to confirm their identity as PEBP genes, with an E-value of 1e−5 and a minimum alignment coverage of 50%23. The nomenclature for PEBP genes in other species was based on the corresponding AtPEBP orthologues.
Phylogenetic analysis
To explore the phylogenetic relationships of PEBP family members, a maximum likelihood (ML) tree was generated using IQ-TREE v2.3.5 based on the deduced protein sequences of these genes in examined species, with the Q.plant + G4 model24. To validate the ML tree, a neighbor-joining (NJ) tree was constructed using MEGA 7.0 (Molecular Evolutionary Genetics Analysis, Tokyo Metropolitan University, Japan) based on PEBP proteins from the plant species examined. Bootstrap analysis with p-distance and pairwise deletion was conducted with 1,000 replicates25. Multiple sequence alignments of PEBP proteins were performed using MUSCLE with default parameters26. The resulting phylogenetic tree was visualized using FigTree v1.4.4 (http://tree.bio.ed.ac.uk/software/figtree/).
Protein conservative domain and gene structure analysis
The Multiple Expectation Maximization for Motif Elicitation program (MEME v5.5.7, http://alternate.meme-suite.org) suite was used to analyze the conserved motifs of PEBPs with the following settings: minimum and maximum motif width 6 and 100, respectively, and 10 number of motifs. The motifs in the protein database were applied using MAST 4.12.027. Exon-intron organization of PEBP was analyzed using Gene Display Server 2 (http://gsds.cbi.pku.edu.cn/)28.
Analysis of TFL1
A 1500 bp genomic sequence located both upstream and downstream of the transcribed region of the TFL1a and TFL1c genes was analyzed to identify potential cis-elements. Cis-acting elements were obtained using the PlantCARE online software (https://bioinformatics.psb.ugent.be/webtools/plantcare/html/) and supplemented with manually added elements known to be important in the PEBP family based on literature29. The data was visualized with TBtools v2.119. The alignment of the non-coding sequences from the different TFL1 orthologues was carried out using mVISTA (http://genome.lbl.gov/vista/index.shtml)30.
Chromosomal location analysis
The chromosomal distribution map was generated and visualized using MapChart v.2.3231, with sequence position data obtained from the Ensembl Plants database (release 45)32 and chromosome information sourced from the European Nucleotide Archive (ENA) (https://www.ebi.ac.uk/ena/browser/home).
Field trial and phenotyping data
Field experiments were conducted at the SITES Research Station Lönnstorp, Swedish University of Agricultural Sciences (SLU), in Alnarp (55.65° N, 13.06° E) over two consecutive growing seasons, 2021 and 2022. The field trials were used for a characterization of a diversity panel of faba bean with 220 different varieties33, among which the varieties used in this study were included and phenotyped in detail with regard to plant architecture traits. Each plot, measuring 1 m x 0.75 m, was hand-sown with fifty seeds of a single variety, with each variety represented in duplicate plots. For details on field experimental design, management, and weather conditions see Ohm et al. (2024). Growth behavior was assessed through visual inspection of plants towards the end of the flowering period, categorizing them as either determinate (with a terminal inflorescence on the main stem) or indeterminate (with continuous leaf development at the top of the main stem). Plant height was measured as the average height of five randomly selected plants per plot ~ 75 days after sowing. The number of days from sowing to flowering was recorded when 50% or more of the established plants in a plot had at least one open flower. Similarly, the number of days from sowing to maturity was noted when 50% or more of the established plants in a plot had filled, dry, and brown/black pods.
Seed size and thousand-grain weight (TGW) were determined using 100–200 seeds with the seed analyzer MARViN ProLine I (Marvitech, Germany). The yield parameter, expressed as grams of seeds per plant (YIELD), was calculated by dividing the total weight of harvested seeds from a plot by the number of established plants in that plot. The number of seeds per plant (SEEDS) was determined by dividing the total weight of harvested seeds from a plot by its TGW, multiplying by 1000, and then dividing by the number of established plants in that plot.
Statistical analysis of field data
Descriptive statistics were used to compare the phenotypic traits between the indeterminate and determinate varieties from the field study. Statistical analysis was conducted on the average values for each variety, calculated across two replicates and two seasons. Student’s t-test determined the statistical significance of differences; the alpha level was set to p < 0.05. Data visualization was conducted using the ggplot2 package in R34.
Results
In silico identification and characterization of PEBP family genes in Vicia faba
Through an HMMER search targeting the conserved PEBP domain and subsequent BLASTP alignment, we initially identified 47 gene members across four plant species. However, three sequences were excluded from further analysis due to having over 50% gaps or ambiguous regions in the evolutionary analysis. The remaining sequences comprised six members in Arabidopsis thaliana, 15 in Pisum sativum, and 11 members each in Vicia sativa and Vicia faba (Supplementary Table S2. The protein lengths of PEBP family members ranged from 173 to 177 amino acids in A. thaliana, 162–188 in Pisum sativum, 134–180 in Vicia faba, and 130–224 in Vicia sativa. The 11 VfPEBP genes were named according to their similarity with the orthologous Arabidopsis thaliana or Pisum sativum genes35, defining six VfFT, three VfTFL1, one, and one VfMFT. The expansion and structure of PEBP family members in several legume species as compared to the model plant Arabidopsis is highlighted in Table 1. The PEBP genes identified in this study are marked in bold; all other genes were obtained from existing literature35,36,37,38,39,40.
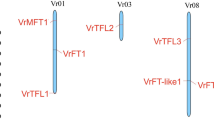
To understand the distribution patterns of the PEBP gene family across the genome, genomic annotation data from faba bean was utilized to determine the chromosomal positioning of the VfPEBP genes. As shown in Fig. 1, the 11 genes are spread across the four chromosomes, 1 S, 5, 6 (and possibly 1 L). Chromosome 1 S holds the largest number, with six VfPEBP genes, while chromosome 5 contains three. In contrast, only one VfPEBP gene is located on chromosome 6, and possibly another on chromosome 1 L.
Phylogenetic and structural analysis of PEBP family genes
To further investigate the relationship among PEBP family members in Vicia faba and other species, a phylogenetic tree was constructed using the full-length PEBP protein sequences from Vicia faba, Pisum sativum, Arabidopsis thaliana, and Vicia sativa (Fig. 2a). The analysis identified the three distinct subgroups of genes: FT-like, TFL1-like, and MFT-like. In Vicia faba, six genes were classified as FT-like, which is a higher number compared to Arabidopsis thaliana (three) and equal to Vicia sativa (six), but fewer than Pisum sativum (ten). Three genes in Vicia faba were categorized as TFL1-like, a consistent number among legumes investigated, with the addition of TFL1b compared to Arabidopsis. One gene in Vicia faba was categorized for each BFT and MFT, consistent with the numbers found in most other species, except for Pisum sativum, which possesses two BFT genes.
Phylogenetic and structural analysis of PEBP family genes. (a) Phylogenetic tree of PEBP family proteins from Vicia faba (Vf), Arabidopsis thaliana (At), Vicia sativa (Vs), and Pisum sativum (Ps), illustrating their evolutionary relationships. (b) The corresponding intron-exon structures of each DNA sequence are shown alongside the tree. (c) Conserved amino acid motifs among the PEBP sequences are depicted and color-coded (Motif 1–10), highlighting shared functional regions across species. Motif sequences are found in Supplementary Table S3.
To explore the structural diversity of VfPEBP genes, the number of exons and introns, and the distribution of conserved domains were investigated (Fig. 2b and c). The gene lengths, including introns, generally ranged from 1 kb to 4 kb, with the notable exception of the three PsFT2 genes, which extend beyond 9 kb. Most PEBP genes are composed of four exons and three introns, with substantial variation observed in the lengths of introns 2 and 3. Eight conserved amino acid motifs could be found in all PEBPs and occurring in the same order across the proteins, with a single exception (VfFTb1-2). Additionally, ten motifs were identified in the majority of the proteins (Supplementary Table S3). All Vicia faba PEBP proteins contain one or both of the highly conserved motifs, DPDxP and GxHR (part of motif 4 and 2, respectively, in Fig. 2c), which likely contribute to the ligand binding pocket conformation41 (Supplementary Figure S4). The conserved motif DPDxP is present in almost all sequences, except for the FTb-proteins in each of Pisum sativum, Vicia faba, and Vicia sativa that instead possess an alternative domain, NPDxP. Additionally, the FTc sequence exhibited a variation in the motif, changing to DADxP. The conserved motif GxHR is present across all sequences except for VsFTa2. An amino acid residue at alignment position 96, which alternates between tyrosine (Y) in FTs and histidine (H) in TFL1 proteins, are observed in our data in agreement with previous reports in other plants. This single amino acid serves as a distinguishing feature separating FT from TFL1, converting the flowering activator to a repressor14 (Supplementary Fig. S4). Interestingly, this distinction also applies to BFT, except for in Arabidopsis thaliana.
Analysis of putative cis-acting regulatory elements
The TFL1 genes TFL1a and TFL1c, also known as DET and LF in Pisum sativum, which play a critical role in inhibiting meristem inflorescence development and thus regulating plant architecture, were investigated in more detail based on reference genomes. Genomic regions of 1500 bp flanking the coding sequences, both upstream and downstream of their homologs in Vicia faba, Pisum sativum, and Vicia sativa were mapped for predicted cis-regulatory elements (Fig. 3a). These elements were further categorized by function (Fig.3b). Our analysis focused on predicted DNA transcription factor binding sites derived from conserved short motifs relevant to TFL1’s biological functions, including responses to light, hormones, stresses, and other signaling pathways. Out of 54 motifs searched, 30 were predicted to be present in the up-and downstream regions of the TFL1 genes (detailed in Supplementary Table S5). Among these, Motif Box 4, part of a conserved module involved in light responsiveness, and the CAAT box, a common cis-acting element in promoter and enhancer regions, were the most prevalent. In Vicia faba, approximately 40% of the motifs in both the upstream and downstream regions are categorized as light-responsive, highlighting the potential significance of photo response regulation in TFL1 gene expression across species. Several cis-acting elements were found to be of particular interest as they coincided between species and regional location (highlighted in Supplementary Figure S6), which suggests that these could be of relevance for the control of TFL1.
Predicted regulatory elements in TFL1 genes. (a) Distribution of predicted regulatory elements in the 1500 bp upstream and downstream genomic regions of the TFL1a and TFL1c genes in Vicia faba (two reference varieties; Hedin and Tiffany), Pisum sativum, and Vicia sativa species. Color-coded captions show element names. Sequences were aligned to the sequence of Vf Hedin and the graphical output in the background shows base pair identity to Vf Hedin (0-100% range) in a sliding window of 1500 bp, > 75% similarity in red. (b) Bar graphs showing the count of elements associated with light response (yellow), hormone response (green), abiotic stress response (orange), and others (grey) for each TFL1 gene. All cis-acting elements are further detailed in Table S5.
Determinacy by DET mutations in faba bean
To investigate mutations that potentially affect the function of TFL1a and c genes that could explain a determinate phenotype (i.e. showing terminal inflorescence) in faba bean, we isolated, cloned, and sequenced the coding region, including 500 bp up-and downstream of the gene. We analyzed eight varieties, comprising four individuals from each phenotypic group (indeterminate and determinate). The isolation and sequencing of the VfTFL1a coding sequence, including upstream and downstream regions, revealed several sequence variations among the investigated varieties (Supplementary Figure S7). We observed 1–37 single nucleotide polymorphisms (SNP) and insertion/deletions throughout the sequence, with five variations in the transcribed region of the gene confined to the first exon and intron. Notably, none of these variations could be linked to distinguishing determinate varieties from indeterminate varieties. Further, we identified 35 nucleotide variations between the two reference genomes, Tiffany and Hedin (which both are indeterminate varieties), none of which were located within the transcribed region.
All primers utilized in this study are detailed in Supplementary Table 1. Due to the significant challenges encountered in isolating the target sequences from different Vicia faba varieties, we have also included the primers that were unsuccessful, to contribute to potential future refinements in methodology.
Allelic variation and phylogenetic relationships of the VfTFL1a gene in Vicia faba varieties with different phenotypes. (a) Multiple sequence alignment of the coding sequence for VfTFL1a from 10 Vicia faba varieties, including four determinate phenotype varieties (in light green) and six indeterminate phenotype varieties, including the reference varieties Hedin and Tiffany (in red). The SNP (G/T) at the 26th bp, previously identified as a diagnostic marker distinguishing the two phenotypes is highlighted. In this dataset, the SNP differentiates only half of the determinate varieties from the indeterminate varieties. For alignment of the entire sequence (transcribed region of the gene as well as 500 bp up-and downstream of it) see Supplementary Figure S7. (b) Phylogenetic relationships among the 10 varieties based on the transcribed region of the gene and 500 bp upstream and downstream of the VfTFL1a gene. The tree shows mixed branching patterns and does not group varieties strictly by phenotype, with determinate (light green) and indeterminate phenotypes (white, including reference varieties in red). The tree was constructed using the Maximum Likelihood method with Neighbor-Joining and the Kimura 80 substitution model in CLC42,43.
Previously, a SNP at bp position 26 in the coding sequence of the VfTFL1 gene was reported by Avila et al. (2006 and 2007) to effectively distinguish determinate faba bean varieties from indeterminate ones42,43. However, our study, found that the previously reported SNP marker did not consistently correlate with the growth phenotype (Fig. 4a). Phylogenetic analysis of VfTFL1a sequences from the 10 varieties showed diverse allelic patterns with no clear clustering based on determinate versus indeterminate growth habit (Fig. 4b).Our observed genetic variations in VfTFL1a among varieties coincided with only one predicted regulatory motif, CCCAATTT (bp 149–156, upstream), which contains an A/C SNP at its first position. This variation occurs in both determinate and indeterminate varieties, suggesting possible regulatory relevance.
Field phenotypic observations
To explore phenotypic differences between indeterminate and determinate faba bean varieties, we selected a group of indeterminate and determinate varieties, which were also used in the above-mentioned DNA sequence comparisons. The varieties were characterized for several agronomic traits in two years of field trials and their distinguishing plant architecture is shown in Fig. 5a and Supplementary Figure S9. Significant differences were observed in the number of days from sowing to flowering, with determinate varieties requiring a longer time to reach flowering. Even though not statistically significant, determinate varieties also showed a trend towards a longer time from sowing to maturity compared to indeterminate varieties. Determinate varieties exhibited a more condensed interval to maturity with less variability, in contrast to the greater variation observed for this trait in indeterminate varieties (Fig. 5b and Supplementary Table S8). While other traits did not show significant differences between determinate and indeterminate varieties, trends indicated that determinate varieties tended to have shorter plant height, lower seed yield, and fewer seeds per plant. Seed size, however, showed relatively similar characteristics between the two types. Overall, determinate varieties demonstrated reduced variability across all traits observed, compared to indeterminate varieties.
Growth habits and traits of indeterminate and determinate faba bean varieties. (a) Comparison of indeterminate (left) and determinate (right) faba bean varieties, photo and illustration. Images of all determinate varieties at the flowering stage are available in Supplementary Figure S9. (b) Boxplots illustrating the distribution of plant height, days to flowering, maturity, yield, seeds per plant, and seed size for both growth habits, based on data from a two-year field trial (Supplementary Table S8). Eight faba bean varieties were evaluated, with four representing each growth type. For each variety, 10–50 plants were assessed depending on the specific trait being measured. The values are the averages of each variety’s performance across the two years and both replicates, shown as mean (horizontal line), upper and lower quartiles (box), range (whiskers) and outliers (dots). Asterisk indicates significant differences according to Students t-test (p < 0.05).
Discussion
Expansion and conservation of PEBP family genes in Vicia faba
The identification of 11 PEBP gene family members in Vicia faba—comprising six VfFT, three VfTFL1, one VfBFT, and one VfMFT gene—points to a significant expansion of this gene family in faba bean compared to Arabidopsis, particularly with respect to the FT-like genes. This expansion may underpin the more complex growth and flowering architecture of faba bean, allowing for functional specialization across gene copies. Unlike the simpler growth and flowering structure of Arabidopsis with its primary inflorescence, Vicia faba develops early lateral shoots, each with its inflorescences. The bushier growth form and more complex flowering pattern of faba bean suggest an evolutionary adaptation to optimize resource allocation, extending the flowering period and enhancing adaptability to diverse environmental conditions. The structural analysis of VfPEBP genes in this study revealed a high degree of conservation in terms of exon-intron organization, with most genes consisting of four exons and three introns. This structural consistency across species underscores the evolutionary importance of the PEBP family. However, the substantial variation observed in intron lengths, particularly in introns 2 and 3, may indicate potential regulatory differences among family members. The abundance of FT-like genes relative to MFT-like genes appears conserved across legumes, which aligns with findings by Pan et al., (2023) which suggests that species possessing numerous FT-like genes tend to exhibit fewer MFT-like genes6. This pattern may indicate an evolutionary relationship in which MFT-like genes could be ancestral to FT-like and TFL-like genes, with their divergence potentially coinciding with the origin of flowering plants. Interestingly, unlike in Arabidopsis, some of the numerous FT genes in soybean, for example, are expressed in the leaves while others are expressed in the shoot apical meristem (SAM) to regulate flowering. Su et al., (2024) suggest that genome duplication in soybean has altered the tissue-specific localization of the FT genes and their functional expression37. Future studies in faba bean could reveal a similar diversification within this and other subcategories of the PEBP gene family, with members functioning in various tissues of the plant. Alongside the ten conserved motifs identified in the PEBP individuals of Vicia faba, Vicia sativa, and Pisum sativum, the presence of specific motifs like DPDxP and GxHR in most VfPEBPs suggests functional conservation, as they are critical for ligand-binding pocket conformation. Variations in FT-like members both within different legume species and among the FT proteins themselves, such as the alternative NPDAP domain, may indicate functional specialization within this subfamily.
The uneven distribution of VfPEBP genes across four chromosomes in Vicia faba, with a notable concentration on chromosome 1 S, offers insights into the evolutionary dynamics of these genes. This pattern likely results from segmental duplication events, which are frequent in plant genomes and contribute to gene family expansion and functional diversification. The proximity of VfFTb genes mirrors the arrangement seen in Medicago truncatula and Glycine max, where FTc genes are located adjacent to FTa genes, suggesting a common origin from the same duplication event35.
In summary, the expansion and diversification of the PEBP gene family in Vicia faba, particularly the FT-like and TFL1-like genes, offer promising opportunities for targeted breeding strategies concerning flowering and plant architectural traits, which can be of high importance for agronomic performance. These findings also highlight the need for further research focused on the functional characterization of individual VfPEBP genes to elucidate their specific roles in controlling flowering time and plant architecture. Such studies will be essential for enhancing our understanding of the molecular mechanisms underlying these traits and for guiding future crop improvement efforts in Vicia faba. Additionally, it is important to address the inconsistent nomenclature of these genes, which varies not only among species, but even among studies on the same species, creating challenges in maintaining clarity and consistency. Future studies can help establish a standardized naming convention, facilitating clearer communication and more precise genetic information in this field.
Phenotypic implications of determinate growth architecture in field
Plant architecture including the timing and distribution of reproductive structures are fundamental agronomic characteristics shaped by patterns of determinate and indeterminate growth. Mutations in TFL1 could potentially lead to earliness, which is of great importance when cultivating the long-season crop faba bean in colder climate countries such as in the Nordic region. In our study, we selected eight varieties based on their growth habits: four determinate and four indeterminate. The varieties Mikko and Aurora were selected for their early maturity traits, essential for successful cultivation in the Nordic climate with its short growing season. Additionally, all varieties were chosen to ensure genetic diversity, as they are not closely related to each other. Field trials comparing determinate and indeterminate varieties revealed significant differences in earliness traits, with determinate varieties requiring a longer time from sowing until flowering. This aligns with the findings of Zhu et al. (2020), which show that TFL1 competes with FT to interact with transcription factor FD, thereby inhibiting the expression of meristem fate genes and delaying flowering44. While other traits such as plant height, maturity time, and seed yield only showed trends rather than significant differences between determinate and indeterminate varieties, the lower variability observed in determinate varieties across multiple traits suggests a more uniform growth pattern. This uniformity is particularly pronounced in the condensed maturation period (103–108 days for determinate varieties versus 96–108 days for indeterminate ones). Further, our observations aligned with previous studies indicating that determinacy often comes at the cost of yield reduction44,45.
Molecular characterization of TFL1 in Vicia faba
To elucidate the genetic basis of growth architecture in faba bean, we specifically examined the TFL1-like clade and identified three TFL1 subclade members, designated as VfTFL1a, VfTFL1b, and VfTFL1c. The analysis of predicted cis-acting regulatory elements in the TFL1 genes revealed a predominance of light-responsive elements, accounting for approximately 40% of the identified motifs. This finding highlights the probable importance of photoperiod in regulating TFL1 expression and, consequently, plant architecture and flowering time in Vicia faba. The conservation of certain regulatory elements across species suggests their functional significance and potential targets for future breeding efforts, warranting further experimental validation.
Our sequence analysis of VfTFL1a, a homolog to the determinacy gene in other plant species, did not reveal distinguishing variations that could confirm the previously reported diagnostic SNP marker at position 2642,43. This discrepancy contrasts with the marker’s original validation across 36 European inbred lines, suggesting that its applicability may be limited to specific germplasm collections. The diverse allelic inheritance patterns observed among our varieties and the lack of clear phylogenetic clustering between determinate and indeterminate phenotypes indicate that the determinate trait likely arose independently multiple times through different genetic mechanisms. This suggests that the genetic control of determinacy in Vicia faba is more complex than previously thought, possibly involving multiple genetic factors, regulatory elements beyond the VfTFL1a coding sequence, or alternative molecular pathways across different breeding lineages.
Evolutionary implications and future directions
The phylogenetic analysis of VfTFL1a sequences from different varieties did not show a clear separation between determinate and indeterminate phenotypes. This lack of clear clustering indicates that the determinate phenotype likely arose independently multiple times, possibly through different molecular mechanisms or regulatory changes. Genetic relatedness among the varieties could possibly have influenced the results for Avila et al. (2006, 2007), by limiting the genetic variability observed. Our varieties instead show a divergent background and a variation of the allelic heritage for TFL142,43.
Given the distinct roles of PsTFL1a/DET on meristem fate and PsTFL1c/LF on flowering time in pea, it raises the question of whether TFL1b in pea as well as in faba bean might play a unique or complementary role in determination cues. Investigating VfTFL1b could provide valuable insights into the regulatory mechanisms governing SAM determinacy in legumes and shed light on evolutionary adaptations within the TFL1 gene family. Previous studies in Arabidopsis have demonstrated that elements in the downstream sequence of AtTFL1 influence meristem determinacy up to 3 kb46 from the coding region. Consequently, expanding the investigation to include further up- and downstream genomic regions of Vf genes may be relevant for identifying diagnostic markers that differentiate between determinate and indeterminate faba bean varieties. Warranting further investigation, this suggests that the key gene determining determinacy in faba bean may be one of the orthologs, TFL1b or TFL1c, or that the regulatory element responsible for the trait appears further up or downstream than 1 kb. Further, since it is known that the balance between the two homologous proteins, FT and TFL1, controls the SAM fate it is also important to look into FT and its potential functional mutations or affecting expression levels14.
Challenges in gene isolation
The successful retrieval of specific gene sequences from the Vicia faba genome through PCR amplification was found challenging in this study, probably due to the high genomic repetitiveness in the species causing primer mismatches. While these repeats (categorized as transposable elements, tandem repeats, or segmental duplications) serve various evolutionary and regulatory roles, they could explain the off-target retrieval of sequences from different genomic regions, which was a frequent problem encountered in this study. In fact, the particularly large and complex genome of faba bean was estimated to have over 85% of its genome as repetitive elements20.
Conclusion and future prospects
This analysis of the PEBP gene family in Vicia faba provides valuable insights into the molecular basis of regulation of flowering time and plant architecture in this legume crop which has great potential for increased cultivation globally. The findings lay a strong foundation for future functional studies and targeted breeding efforts aimed at optimizing faba bean varieties for diverse agricultural needs and environmental conditions. Future research should investigate the regulatory networks controlling VfTFL1 expression, including the role of identified cis-acting elements. It should also explore interactions among PEBP family members and their combined effects on plant phenotypes. Additionally, developing more robust molecular markers for determinate growth habit will be valuable, especially given the complex genetic control suggested by our results.
Data availability
All data relevant to the reproduction of this study are included in the supplementary material provided or can be requested from authors. The datasets generated and analysed during the current study are available in the BankIt repository under the following accession numbers: PQ878658, PQ878659, PQ878660, PQ878661, PQ878662, PQ878663, PQ878664, PQ878665.
References
Costa, M. P. et al. Legume-modified rotations deliver nutrition with lower environmental impact. Front Sustain. Food Syst 5, 656005 (2021).
Moraes, T. S., Dornelas, M. C. & Martinelli A. P. FT/TFL1: calibrating plant architecture. Front. Plant. Sci. 10, 97 (2019).
Danilevskaya, O. N., Meng, X., Hou, Z., Ananiev, E. V. & Simmons, C. R. A genomic and expression compendium of the expanded PEBP gene family from maize. Plant. Physiol. 146, 250–264 (2008).
Zhu, Y., Klasfeld, S. & Wagner, D. Molecular regulation of plant developmental transitions and plant architecture via PEPB family proteins: an update on mechanism of action. J. Exp. Bot. 72, 2301–2311 (2021).
Jin, S., Nasim, Z., Susila, H. & Ahn, J. H. Evolution and functional diversification of flowering Locus T/Terminal flower 1 family genes in plants. Semin Cell. Dev. Biol. 109, 20–30 (2021).
Pan, C. et al. Identification and analysis of phosphatidylethanolamine-binding protein family genes in the hangzhou white chrysanthemum (chrysanthemum morifolium ramat). Agric 13, 374 (2023).
Benlloch, R. et al. Genetic control of inflorescence architecture in legumes. Front. Plant. Sci. 6, 1–14 (2015).
Weller, J. L. & Ortega, R. Genetic control of flowering time in legumes. Front. Plant. Sci. 6, 207 (2015).
Gretsova, M. et al. Transcriptomic analysis of flowering time genes in cultivated chickpea and wild cicer. Int J. Mol. Sci 24, 2692 (2023).
Wang, Z. et al. Functional evolution of phosphatidylethanolamine binding proteins in soybean and arabidopsis. Plant. Cell. 27, 323–336 (2015).
Foucher, F. et al. Determinate and late flowering are two terminal flower1/centroradialis homologs that control two distinct phases of flowering initiation and development in pea. Plant. Cell. 15, 2742–2754 (2003).
Sinjushin, A. A. Mutations of determinate growth and their application in legume breeding. Legum Perspect 14–15 (2015).
Liu, B. et al. The soybean stem growth habit gene Dt1 is an ortholog of arabidopsis terminal flower1. Plant. Physiol. 153, 198–210 (2010).
Hanzawa, Y., Money, T. & Bradley, D. A single amino acid converts a repressor to an activator of flowering. Proc. Natl. Acad. Sci. U S A. 102, 7748–7753 (2005).
Shannon, S. & Meeks-Wagner, D. R. A mutation in the arabidopsis TFL1 gene affects inflorescence meristem development. Plant. Cell. 3, 877–892 (1991).
Ratcliffe, O. J., Bradley, D. J. & Coen, E. S. Separation of shoot and floral identity in Arabidopsis. Development 126, 1109–1120 (1999).
Ávila, C. M. et al. Identification of plant architecture and yield-related QTL in Vicia faba L. Mol Breed 37, 88 (2017).
Xi, H. et al. Chromosome-level assembly of the common vetch (Vicia sativa) reference genome. Gigabyte 2022, 1–2 (2022).
Östberg, J. Swedish University of Agricultural Sciences, SLU,. Vicia faba determinate and indeterminate inflorescence genotypes-comparison of genetic variation at TFL1 locus Vicia faba determinanta och icke-determinanta genotyper-jämförelse av genetisk variation vid TFL1 locus. (2021).
Jayakodi, M. et al. The giant diploid faba genome unlocks variation in a global protein crop. Nature 615, 652–659 (2023).
Yang, T. et al. Improved pea reference genome and pan-genome highlight genomic features and evolutionary characteristics. Nat. Genet. 54, 1553–1563 (2022).
Finn, R. D., Clements, J. & Eddy, S. R. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 39, W29–W37 (2011).
Mahmood, U. et al. Comprehensive analysis of polygalacturonase genes offers new insights into their origin and functional evolution in land plants. Genomics 113, 1096–1108 (2021).
Nguyen, L. T., Schmidt, H. A., Von Haeseler, A. & Minh, B. Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274 (2015).
Kumar, S., Stecher, G. & Tamura, K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874 (2016).
Edgar, R. C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 (2004).
Bailey, T. L. et al. MEME suite: tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208 (2009).
Hu, B. et al. GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31, 1296–1297 (2015).
Zhang, M. M. et al. Genome-wide identification of Pebp gene family in two dendrobium species and expression patterns in dendrobium chrysotoxum. Int. J. Mol. Sci. 24, 17463 (2023).
Frazer, K. A., Pachter, L., Poliakov, A., Rubin, E. M. & Dubchak, I. I. VISTA: Computational tools for comparative genomics. Nucleic Acids Res. 32, W273–W279 (2004).
Voorrips, R. E. & Mapchart Software for the graphical presentation of linkage maps and QTLs. J. Hered. 93, 77–78 (2002).
Yates, A. D. et al. Ensembl genomes 2022: an expanding genome resource for non-vertebrates. Nucleic Acids Res. 50, D996–D1003 (2022).
Ohm, H. et al. Novel SNP markers for flowering and seed quality traits in faba bean (Vicia faba L.): Characterization and GWAS of a diversity panel. Front. Plant. Sci. 15, 1348014 (2024).
Wickham, H. ggplot2 elegant graphics for data analysis. (Springer Cham, (2016). https://doi.org/10.1007/978-3-319-24277-4
Hecht, V. et al. The pea GIGAS gene is a flowering locus t homolog necessary for graft-transmissible specification of flowering but not for responsiveness to photoperiod. Plant. Cell. 23, 147–161 (2011).
Ortega, R. et al. Altered expression of an FT cluster underlies a major locus controlling domestication-related changes to Chickpea phenology and growth habit. Front. Plant. Sci. 10, 824 (2019).
Su, T. et al. The critical roles of phosphatidylethanolamine-binding proteins in legumes. Plant. Cell. Environ. https://doi.org/10.1111/pce.15255 (2024).
Tribhuvan, K. U. et al. Identification and characterization of PEBP family genes reveal CcFT8 a probable candidate for photoperiod insensitivity in C. cajan. 3 Biotech10, 194 (2020).
Williams, O. et al. The genetic architecture of flowering time changes in pea from wild to crop. J. Exp. Bot. 73, 3978–3990 (2022).
Xue, R. et al. Genome-wide characterization of PEBP genes in mung bean (Vigna radiata L.) with functional analysis of VrFT1 in relation to photoperiod. Sci. Rep. 14, 26413 (2024).
Yang, Z. et al. Identification and characterization of the PEBP family genes in Moso bamboo (Phyllostachys heterocycla). Sci. Rep. 2019. 9, 1–12 (2019).
Avila, C. M., Atienza, S. G., Moreno, M. T. & Torres, A. M. Development of a new diagnostic marker for growth habit selection in faba bean (Vicia faba L.) breeding. Theor. Appl. Genet. 115, 1075–1082 (2007).
Avila, C. M., Nadal, S., Moreno, M. T. & Torres, A. M. Development of a simple PCR-based marker for the determination of growth habit in Vicia faba L. using a candidate gene approach. Mol. Breed. 17, 185–190 (2006).
Zhu, Y. et al. TERMINAL FLOWER 1-FD complex target genes and competition with FLOWERING LOCUS T. Nat Commun 11, 5118 (2020).
Stoddard, F. & Hämäläinen, K. Towards the world’s earliest maturing faba beans. Grain Legum 56, 9–10 (2011).
Serrano-Mislata, A. et al. Separate elements of the TERMINAL FLOWER 1 cis-regulatory region integrate pathways to control flowering time and shoot meristem identity. Development https://doi.org/10.1242/dev.135269 (2016).
Funding
Open access funding provided by Swedish University of Agricultural Sciences. Open access funding provided by Swedish University of Agricultural Sciences. This study was made possible by funding from SLU Grogrund ‘Future faba beans for food and feed’ (#ltv.2018.1.1.1.1067), SLU LTV faculty for supporting growth facilities, the Swedish Infrastructure for Ecosystem Science (SITES) Lönnstorp Research Station at SLU (Swedish Research Council 2017 − 00635), and The Royal Physiographic Society in Lund, Sweden, for analytical equipment.
Author information
Authors and Affiliations
Contributions
HO: Performed laboratory and field work, developed methodology, conducted formal analysis and investigation, validated results, created visualizations, and wrote the original draft of the manuscript. UM: Retrieved data, conducted formal analysis, developed software tools, created visualizations, and contributed to manuscript review and editing. JÖ: Performed laboratory work, conducted formal analysis and investigation, validated results JA: Performed laboratory work. ÅG: Secured funding, contributed to methodological development, provided resources, supervised research, and contributed to manuscript review and editing. PH: Conceived and designed the study, acquired funding, developed methodology, provided resources, supervised research, validated results, and contributed to manuscript review and editing.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
41598_2025_12864_MOESM1_ESM.xlsx
Supplementary Material 1
41598_2025_12864_MOESM2_ESM.pdf
Supplementary Material 2
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ohm, H., Mahmood, U., Östberg, J. et al. Comprehensive identification of the PEBP gene family in Vicia faba highlighting VfTFL1a variation and phenotypic insights into determinate growth. Sci Rep 15, 28687 (2025). https://doi.org/10.1038/s41598-025-12864-0
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-12864-0