Abstract
This study presents a novel deep learning (DL) framework, the Deep Neural Frailty Competing Risks (DNFCR) model, which simultaneously integrates frailty and competing risks (CR) for mortality prediction in heart failure (HF). While existing models like Neural Frailty Models (NFM) address frailty and DeepHit handles CR, DNFCR is the first DL approach to combine both components, offering improved handling of censored data in healthcare applications. We evaluated DNFCR against established models (DeepSurv, CoxPH) using real-world HF data, assessing its ability to capture unobserved heterogeneity through proportional (DNFCR_PF) and non-proportional (DNFCR_NF) frailty structures. In this retrospective cohort study, we analyzed 435 HF patients (enrolled March–September 2018; 5-year follow-up until July 2023) with 57 demographic/clinical features, categorized by cause-specific mortality. The models’ performance was evaluated using the C-index, IBS, and INBLL. Our results in mortality from heart failure demonstrated marginal but consistent improvements in predictive accuracy when incorporating frailty (C-index: ~0.66). Differences in predicting other-cause mortality were minimal across the models. DNFCR-PF also performed better regarding the IBS (0.17 ± 0.01) and INBLL (0.02 ± 0.53). However, the clinical relevance of the cause-specific mortality requires further validation in both outcome categories. Comparative analysis revealed that DNFCR and traditional models achieved comparable accuracy in survival prediction, highlighting that DL’s superiority is context-dependent and influenced by data and unmeasured confounders. Strengths include DNFCR’s potential for personalized risk stratification and adaptability to other diseases with CR. The DNFCR model showed DL potential in HF survival prediction but requires clinical validation compared to traditional approaches.
Similar content being viewed by others
Introduction
Competing risk analysis (CR) is a vital area of medical research and healthcare that aims to predict the timing of specific events1. A prime example is cardiovascular disease, a leading cause of death worldwide. Among cardiovascular diseases, heart failure (HF) is associated with high morbidity and mortality. HF is the third leading cause of cardiovascular death in developed countries2 and significantly contributes to morbidity and hospital admissions3.
Accurately predicting the survival of HF patients is essential for guiding clinical decision-making, personalizing treatment plans, and, ultimately, improving patient outcomes4. Advanced analytical techniques are needed to achieve this goal, as traditional survival models often fail to fully capture the complexity of CR and its impact on patient survival5.
In many medical studies, CR arises when multiple causes of death may influence the occurrence of a specific event, such as death from a particular cause5. Traditional survival analysis methods, such as the Kaplan-Meier method, log-rank tests, and Cox models, assume noninformative censoring, where secondary causes of death are treated as censored data6,7. However, this assumption may lead to biased estimates due to the correlation between event occurrence and censoring times8. Additionally, classical survival models assume a linear relationship between independent and dependent variables9, which may not capture the complexity of CR. This highlights the need for specialized, advanced analytical techniques that account for nonindependence and nonlinear relationships to provide more accurate and reliable inferences in such analyses7,8.
This research presents a novel approach that explicitly incorporates frailty10 to account for potential correlations in CR. Frailty refers to unobserved covariates at the individual or cluster level that impact the occurrence of an event, and it also enables the modeling of nonlinear relationships within a deep learning (DL) framework10,11.
The preliminary analysis of our data revealed complex relationships among various variables, including BMI, LVEF, SPO2, temperature, heart rate, DBP, and SBP. These variables exhibited a diverse range of relationships, such as compound, power, S-shaped, growth, exponential, logistic, cubic, and quadratic patterns, all of which explained a portion of the variance in the dependent variable. Given the limitations of classical models in handling such complexity, we opted for DL methods, which offer higher accuracy and better predictive capabilities by identifying novel patterns overlooked in previous analyses. CR and frailty challenge DL models by introducing bias when correlated event types are treated as independent CR and unmeasured features create frailty. Ignoring these factors inflates prediction variance, as Wu et al. (2023) demonstrated in neural frailty networks10. These factors can substantially affect model performance and accuracy. A systematic review by Monterrubio-Gómez et al. (2022)12 examined DL based methods13for time-to-event analysis, highlighting two key advantages: (i) DL models can automatically learn complex, nonlinear interactions and temporal patterns that traditional manual feature engineering might overlook (Feature Importance). (ii) Unique characteristics include the ability to process high-dimensional data, handle missing data through embedding layers, and model time-varying effects without relying on proportional hazards assumptions. These features make DL models valuable for survival analysis in complex clinical settings12.
Despite this progress, current DL models (e.g., DeepHit) handle CR but assume frailty is negligible, while frailty models (e.g., NFM) often ignore CR. This dual oversight limits real-world applicability, as HF patients exhibit both phenomena. Frailty increases vulnerability to both HF progression and competing infections2. Methodologically, frailty induces dependence between competing events (e.g., HF patients may die sooner from “any” cause, linking HF and non-HF mortality risks). Ignoring this inflates HF mortality estimates8. This underscores the need for broader model generalization to accommodate a wider range of survival analysis scenarios.
Our DNFCR model integrates a gamma frailty component to address unobserved heterogeneity and incorporates competing risk analysis to consider multiple potential causes of mortality. We aim to compare the performance of this model against that of established methods, such as DeepSurvCR14, to comprehensively evaluate its effectiveness in real-world scenarios and its ability to improve survival predictions.
This article is structured as follows: First, we review the literature on CR, deep learning, and frailty models. Next, we outline the methodology used in this study, detailing how frailty concepts are integrated into deep neural networks for CR analysis. Finally, we describe our experimental setup and compare the performance of our model with that of DeepSurvCR.
Literature review
Recent years have seen significant advancements in DL for survival analysis, particularly in addressing CR and unobserved heterogeneity. However, critical gaps remain in how existing models integrate these complexities, often leading to biased or oversimplified predictions.
Limitations of current DL approaches
Lee et al. (2018) introduced DeepHit, a nonparametric model for survival time distribution that outperformed traditional methods but fails to account for unobserved heterogeneity (frailty) and assumes independent CR, limiting its clinical applicability15. Rietschel et al. (2018) improved feature selection for medical data but did not address how irrelevant features might interact with latent patient-specific frailty16. While DeepCompete (Huang & Liu, 2020) enhanced CR handling, it relies on proportional hazard assumptions, often violated in real-world data17. Similarly, Nagpal et al.’s (2020) Deep Survival Machines advanced parametric estimation but ignores frailty, risking biased predictions in heterogeneous populations18.
Gaps in frailty integration
Recent frailty models, such as those by Tran et al. (2020) and Mendel et al. (2022), incorporated random effects into neural networks but require prespecified frailty distributions (e.g., Gamma or Gaussian), which may not capture true underlying heterogeneity11,19.
The DNN-FM (Lee et al., 2023) and NFM (Wu et al., 2023) made strides by integrating multiplicative frailty for censored data, yet NFM’s reliance on parametric frailty limits flexibility, and neither model fully resolves the interplay between frailty and CR10,20.
Our proposed DNFCR model advances prior work by:
-
1.
Nonparametric Frailty Learning: Unlike NFM or DNN-FM, DNFCR learns frailty distributions directly from data, eliminating the need for restrictive parametric assumptions.
-
2.
Joint Modeling of Frailty and CR: While DeepHit and DeepCompete treat CR as independent processes, DNFCR explicitly models their dependence on latent frailty, reducing bias in risk estimation.
-
3.
Dynamic Covariate-Frailty Interactions: Building on Hong et al.’s (2022) contrastive learning, DNFCR captures time-varying relationships between covariates, frailty, and CR, a feature overlooked by earlier frailty models.
By unifying these innovations, DNFCR offers a more flexible and interpretable framework for survival analysis, particularly in complex clinical scenarios like HF, where unobserved heterogeneity and CR are pervasive.
Materials and methods
Study population
We retrospectively analyzed data from 529 consecutive patients with acute heart failure with reduced ejection fraction (HFrEF; LVEF ≤ 40%) who were admitted to Rajaie Cardiovascular Medical Center (RCMCH) between March and August 2018 and received standard guideline-directed medical therapy. The study inclusion criteria required: (i) confirmed HFrEF diagnosis, (ii) index hospitalization during the study period, (iii) receipt of appropriate HF treatment, and (iv) at least one documented follow-up within six months’ post-discharge.
Exclusion criteria
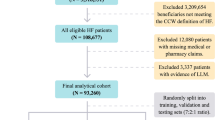
We excluded 94 patients (17.8%) for either having > 20% missing clinical data (n = 63) or not receiving HF-specific treatment (n = 31). The final analytical cohort thus comprised 435 patients with complete treatment and follow-up data.
The enrollment flowchart (Fig. 1) details 529 screened → 94 excluded (31 untreated, 63 incomplete data) → 435 analyzed. Untreated patients were excluded as their outcomes (often palliative-care-associated) would distort treated-HFrEF trajectories, consistent with ESC-HF-LT registry standards.
We followed them from 2018 to July 2023 (5 years). This dataset consists of 57 characteristics (demographic and clinical). We categorized patients by cause of death into two groups: those who died from HF and those who died from other causes.
Ethics approval
This study was approved by the ethics committee of the School of Medical Sciences, Tarbiat Modares University, under the approval ID IR.MODARES.REC.1402.012. The participants’ privacy was preserved. All participants completed an informed consent form. International agreements approved all the processes (World Medical Association, Declaration of Helsinki, Ethical Principles for Medical Research Involving Human Subjects).
Statistical analysis
Raw data was organized into a database for analysis. Continuous variables are reported as means ± standard deviations, and categorical variables are shown as frequencies and percentages. Statistical analysis was performed using Python, with the DNFCR and DeepSurvCR models implemented in PyTorch. Due to the lack of standard training/testing splits in the survival dataset, we used a 5-fold cross-validation method, allocating one fold for testing and 20% for validation8.
We employed three key metrics to evaluate model performance:
-
(i)
The concordance probability (C-index) \(\:(\mathcal{C}=\mathbb{P}\left[\widehat{S}\left({T}_{i}\mid\:{Z}_{i}\right)<\widehat{S}\left({T}_{j}\mid\:{Z}_{j}\right)\mid\:{T}_{i}<{T}_{j},{\delta\:}_{i}=1\right])\); measures how accurately the model ranks patient survival times, where 0.5 indicates random prediction and 0.7–0.8 represents clinically useful discrimination. Discrimination (c-index) indicates the model’s ability to separate patients with different outcomes.
-
(ii)
The Integrated Brier Score (IBS) \(\:\begin{array}{c}{(\mathcal{S}}_{\text{I}\text{B}\text{S}}\left(\widehat{S}\right(t\mid\:x),(z,d\left)\right)=\frac{1}{{t}_{2}-{t}_{1}}{\int\:}_{{t}_{1}}^{{t}_{2}}\:{\mathcal{S}}_{\text{Brier\:}}^{t}\left(\widehat{S}\right(t\mid\:x),(z,d\left)\right)dt)\\\:\end{array}\)quantifies prediction error through mean squared differences (0=perfect, 0.25=random), with our score of 0.17 demonstrating 32% improvement over chance. Calibration (Brier score) reflects how well predicted probabilities match observed outcomes.
-
(iii)
The Integrated Negative Binomial Log-Likelihood (INBLL) assesses prediction confidence while accounting for censored data, where values approaching zero (like our 0.02) reflect greater reliability. Together, these metrics provide complementary insights - the C-index evaluates ranking accuracy, IBS measures overall error magnitude, and INBLL gauges uncertainty, enabling a comprehensive assessment of clinical utility21
$$\begin{aligned}NBLL& =-\frac{1}{N}\sum\:_{i=1}^{N}\:\left[\frac{\text{l}\text{o}\text{g}\left(1-\widehat{S}\left(t\mid\:{x}_{i}\right)\right)I\left({T}_{i}\le\:t,{e}_{i}=1\right)}{\widehat{G}\left({T}_{i}\right)}+\frac{\text{l}\text{o}\text{g}\widehat{S}\left(t\mid\:{x}_{i}\right)I\left({T}_{i}>t\right)}{\widehat{G}\left(t\right)}\right] \\ INBLL & =\frac{1}{{l}_{2}-{l}_{1}}{\int\:}_{{l}_{1}}^{{t}_{2}}\:NBLL\left(t\right)dt.\end{aligned}$$
Survival analysis with competing risks
Survival analysis aims to estimate the time to an event (e.g., HF-related death), frequently dealing with censoring. In this study, we consider the problem of CR survival prediction when more than one event is possible and when each patient experiences only one of those events. This is a common scenario in HF prognosis, where a patient can die either of HF or other complications (e.g., cancer).
Given the training dataset \(\:D=\left\{{T}^{\left(j\right)},\:{e}^{\left(j\right)},\:{{c}^{\left(j\right)},\:x}^{\left(j\right)}\right\},\:j=1,\dots\:,N\), where \(\:{T}^{\left(j\right)}\) is the time to event for patient j,\(\:\:{e}^{\left(j\right)}\) is the event indicator, \(\:{c}^{\left(j\right)}\) is the censoring and \(\:{x}^{\left(j\right)}\) are the patient features, we want to learn to predict the probability that an event occurs before some time\(\:\:T,\:P({T}^{\left(j\right)}\le\:t,\:{e}^{\left(j\right)}=e|{x}^{\left(j\right)})\), known as the cause-specific for events. Classical analysis methods can address CR via special-cause Cox models, but implementing these methods requires an independence assumption22. When independence among CR is not confirmed, their relationship can be described via frailty as “unobserved dispersion” in the model.
This study introduces the DNFCR method, a novel DL approach for estimating time-to-event in CR in the presence of frailty.
Deep neural frailty competing risk (DNFCR) framework
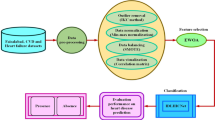
DNFCR employs two distinct deep neural network architectures to model CR in survival analysis (Fig. 2). The key idea is to utilize the censored observations appropriately in the likelihood function to obtain consistent parameter estimates despite incomplete information about event times. The incorporation of frailty into the deep neural network model allows the DNFCR structure to account for individual-specific characteristics and changes influenced by unobserved factors that may impact event risk.
Unobservable factors varying among individuals can lead to intraindividual correlation.
For the frailty variable u, we utilize the gamma density function:
We begin by integrating the conditional survival function with frailty to derive the observed likelihood function for the competing risk:
The frailty transform \(\:{G}_{\theta\:}\left(x\right)=-\text{l}\text{o}\text{g}\left({\mathbb{E}}_{{u}_{i}\sim\:{f}_{\theta\:}}\left[{e}^{-{u}_{i}\text{x}}\right]\right)\) is defined as the (\(\:-\text{l}\text{o}\text{g}\)) of the Laplace transform of the frailty distribution for each cause. Consequently, the conditional cumulative hazard function is given by \(\:{\:\text{H}\left(t\right|X)=G}_{\theta\:}\left({\int\:}_{0}^{t}\:{e}^{h\left(s,X\right)}ds\right)\).
For the PF model, we utilize two multilayer perceptrons (MLPs), denoted as \(\:{h}^{\Lambda}={h}^{\Lambda}(t;{W}^{h},{b}^{h})\) and \(\:\widehat{m}=\widehat{m}(X;{W}^{m},{b}^{m})\)), to approximate the functions h and m, parameterized by \(\:{(W}^{h},{b}^{h})\) and \(\:({W}^{m},{b}^{m})\), respectively. Here, W represents a collection of weight matrices across all layers of the MLPs, whereas b represents a set of bias vectors across all layers. Considering the standard results regarding the likelihood of censored data as presented in Eq. (2), the learning of parameters under the PF framework can be expressed as follows23:
in which\(\:{g}_{\theta\:}\left(x\right)=\frac{\partial\:}{\partial\:x}{G}_{\theta\:}\left(x\right).\).
The estimated conditional cumulative risk and survival functions are represented by Eq. (4).
For the NF model:
The estimated conditional cumulative risk and survival functions are represented by Eq. (6).
Results
This study analyzed the mortality of 435 HF patients over five years, focusing on deaths from HF versus other causes. In total, 43.96% of all patients died from HF, 26.64% died from other causes, and 29.4% survived. The median survival time was 43.40 months (Table 1).
The one-year survival rate for patients who died from HF was 80.66% (95% CI: 0.76–0.84), decreasing to 68.3% (95% CI: 0.63–0.72) at three years and 59.52% (95% CI: 0.54–0.64) at five years. For those who died from other causes, the survival rates were 91.78% (95% CI: 0.88–0.94) at one year, 79.08% (95% CI: 0.74–0.83) at three years, and 70.29% (95% CI: 0.64–0.75) at five years.
Figure 3 presents the impact of the cause of death on survival in HF patients. The analysis showed that HF itself is a major contributor to mortality in this study population, leading to a reduced probability of survival compared to deaths from other causes.
The mean age of all patients was 56.57 ± 18.11 years, ranging from 14 to 95. Among those who died from HF, the average age was 59.26 ± 1.40 years, with the highest mortality in the 56–65 age group. Of these, 63.1% were male, 89.4% were Freelancers, and 87.5% held an Undergraduate degree. Additionally, 93.1% resided in urban areas, and 89.4% were married.
In contrast, patients who died from other causes had a mean age of 62.04 ± 1.71 years, with the peak mortality rate in the 66–75 age group. Of these, 52.6% were male, 81.4% were Freelance, and 82.5% held an Undergraduate. Furthermore, 89.7% lived in Urban, and 89.7% were married (Tables 2 and 3 provide additional details on other features).
To identify the optimal learning rate for the deepsurv and DNFCR models, we conducted experiments using a range of learning rates [0.1, 0.01, 0.001, 0.0001, 0.00001, 0.003, 0.006, 0.008, 0.01, 0.02, 0.03, 0.05, 0.07], and we evaluated performance based on the IBS, INBLL, and c-index criterion. This process involved creating and deploying a new deepsurv model for each learning rate, utilizing a negative log-likelihood loss function and the Adam optimizer in 200 epochs, during which learning rate and weight decay were monitored. We systematically compared model performance across these learning rates and determined the optimal rate. Ultimately, results were categorized into two groups: mortality due to HF and mortality from other causes (Table 4).
Figure 4 depicts the structure of a 55-layer deep neural network, specifically developed for the Deepsurv and DNFCR models.
Statistical comparisons
Survival analysis revealed that the traditional CoxPH model achieved C-indices of 0.61 for HF mortality and 0.58 for other causes mortality. Then, we assessed the performance of DNFCR_PF and DNFCR_NF relative to their nonfrailty counterpart, DeepSurvCR. The results presented in Table 5 showed that the DNFCR_PF model achieved the highest c-index value of 0.66 ± 0.04 compared to other models for mortality from HF. Additionally, the DNFCR_PF model showed superior performance in both INBLL and IBS metrics. In predicting death from other causes, the DeepsurvCR model performs better regarding the C-index, while the DNFCR models exhibit similar performance in IBS.
Pairwise DeLong tests confirmed that all three advanced models (DNFCR-PF, DNFCR-NF, and DeepSurvCR) demonstrated statistically superior discriminative ability compared to the traditional CoxPH model. The DNFCR-PF model showed particular strength in predicting HF mortality, with its enhanced precision evidenced by superior IBS and INBLL relative to other models. While between-model differences for mortality from other causes predictions were non-significant (p > 0.05) (Table 6; Fig. 5).
Clinical validation through calibration analysis
The calibration plots (Fig. 6, Panel A) demonstrate that the DNFCR model achieves high accuracy in predicting mortality from HF at 1- and 3-year time horizons (showing close alignment with the ideal line), while exhibiting minor deviations in 5-year predictions, likely attributable to reduced sample size in long-term follow-ups. For non-HF mortality, while 1-year and 5-year predictions showed systematic underestimation, the mid-range (3-year) predictions maintained clinically acceptable accuracy (± 5% from ideal) (Fig. 6, Panel B).
Discussion
This study aimed to investigate the accuracy of a DL model with a frailty approach to HF data. The DNFCR method introduces a novel approach for modeling mortality in CR, utilizing two different neural structures.
Although existing models like NFM incorporate frailty and DeepHit addresses CR, the DNFCR framework represents the first DL approach to simultaneously integrate both components. Overall, the DNFCR framework provides a method to handle censored data while accounting for frailty and CR. Although its predictive accuracy is moderate (C-index ~ 0.66), it could become a valuable tool for healthcare applications where censorship frequently occurs.
Our findings showed marginal improvements when incorporating frailty. This reflects that incorporating frailty into DL models for predicting outcomes in patients with HF can improve model accuracy. Frailties are utilized in time-to-event modeling to account for unobserved heterogeneity among individuals who can impact event occurrence. In time-to-event analysis, frailty models extend traditional survival models such as the Cox model by introducing random effects, known as frailties. These frailties capture individual-specific characteristics that affect the event of interest but are not directly observed. By incorporating frailties, time event models can better address the variability in event times that the measured covariates cannot explain.
This study compared several survival models: The CoxPH, the DeepSurveCR model, the DNFCR model with proportional frailty (DNFCR_PF), and the DNFCR model with nonproportional frailty (DNFCR_NF). Despite the small sample size, the DeepSurve and DNFCR models showed comparable accuracy in predicting survival time. This preliminary comparison highlights methodological differences but does not conclusively establish superiority for clinical use.
These models capture complex, nonlinear relationships between input variables and survival outcomes14. In line with our results, in the study by Ruofan Wu et al., the authors examined the results of their real-world survival data. They used five survival datasets plus one nonsurvival dataset to evaluate NFM models. These include four small datasets, including METABRIC, RotGBSG, FLCHAIN, and SUPPORT, and a large dataset called KKBOX. The results show that the NFM model outperforms 12 other reference models and provides significant improvements, especially on the METABRIC, SUPPORT, and MIMIC-III datasets. Thus, NFM, as a robust and practical framework for survival prediction, has demonstrated better results in evaluation criteria such as IBS and INBLL than other models10.
Another study proposed combining DL techniques with feature enhancement methods to assess cardiovascular disease risk in patients, achieving a prediction accuracy of 90%24.
While the DNFCR model demonstrates innovation in integrating frailty with CR, its discriminative power (C-index: 0.66) remains moderate compared to the ideal threshold of 0.75 for clinical decision-making. This performance is consistent with existing DL models in similar clinical contexts, such as DeepSurv (C-index: 0.65) and NFM (C-index: 0.62–0.69)10,25. The modest discrimination may reflect both the inherent noise in real-world clinical data (e.g., unmeasured confounders) and the complexity of modeling correlated CR. Although larger datasets could improve precision, particularly for rare events.
Overall, these studies indicate that DL models, with their ability to model intricate relationships and employ advanced structures such as neural networks, can achieve superior performance in survival data analysis compared with traditional models.
On the other hand, some studies indicate that no definitive and consistent results demonstrate the absolute superiority of DL models over classical models26. While DL has garnered attention for its ability to model complex and nonlinear relationships, particularly in survival time prediction, its performance is not always significantly better than that of traditional models. Certain comparisons indicated that classical models can perform satisfactorily and, in some cases, even surpass DL models27.
Some studies suggest that DL models may need more data for training and fine-tuning and may not consistently outperform traditional models when data are scarce28. DNFCR and the CoxPH model are essential for analyzing CR, yet they differ fundamentally in structure and application. Additionally, a model’s suitability depends on the data’s specific characteristics and research requirements, so one cannot universally assert that DL models are superior to traditional models26,27.
While the DNFCR-PF model’s C-index of 0.66 represents moderate discriminative ability, its clinical value emerges from three key innovations: (1) simultaneous handling of CR, avoiding overestimation of HF-specific mortality prevalent in traditional models; (2) incorporation of frailty as a proxy for unmeasured patient heterogeneity, mirroring clinical assessment practices; and (3) superior calibration (IBS 0.17 ± 0.01) enabling reliable absolute risk estimation. In practice, this model could stratify patients into actionable risk tiers, identifying those with > 50% 1-year mortality risk who may benefit from advanced therapies while flagging low-risk patients for de-escalation. The modest C-index reflects the inherent complexity of HF prognostication, where competing comorbidities and treatment responses create irreducible uncertainty. Future integration of dynamic biomarkers and larger datasets may enhance performance, but the current framework already provides clinically meaningful distinctions that traditional Cox models cannot achieve.
Strengths
The DNFCR model, while initially developed for HF patients, has the potential to be applied to other patient groups, such as those with different types of heart failure or even other complex diseases, provided that the relevant data from these populations are appropriately adjusted. However, its performance may vary depending on the specific characteristics of the new dataset, such as sample size, data quality, and the presence of CR. The model’s ability to account for frailty and unobserved heterogeneity enhances survival predictions, offering more personalized and accurate estimates. While the model’s current performance is limited, future iterations with improved accuracy could potentially help identify high-risk patients, facilitating timely interventions that may reduce mortality. By incorporating frailty, the model better reflects individual patient characteristics, allowing for more targeted treatment strategies and improved resource allocation. Additionally, the model could be adapted for use in other clinical settings, improving outcomes in diseases where CR is a factor. Overall, the DNFCR model represents a theoretical approach that may, after substantial refinement and validation, contribute to clinical decision-making and improve the quality of care across diverse patient populations.
This study highlights the potential of DL models, particularly those incorporating frailty, for improving the prediction of patient outcomes in complex diseases such as HF. While these models demonstrate superior performance in capturing complex relationships between variables and survival outcomes, their practical application requires careful consideration. While DL models can significantly enhance predictive accuracy, they may necessitate large datasets and computational resources. Additionally, traditional models such as the Cox model may still be suitable for specific scenarios, especially when data are limited. Therefore, the optimal choice of model should be based on a careful assessment of the particular clinical context and the trade-off between model complexity and predictive accuracy.
Limitations
Given the stringent inclusion and exclusion criteria, our study cohort represents a specific subset of HF patients, which may limit the generalizability of our findings to broader HF populations typically examined in traditional studies.
On the other hand, analyzing CR with frailty via a DL approach in real-world settings involves several challenges that necessitate further research and advanced methods. The DNFCR model represents a preliminary step toward addressing unobserved heterogeneity in survival analysis, but its clinical applicability requires further validation. The DNFCR framework often requires extensive, accurate datasets that are frequently hard to obtain in practice. Additionally, the black box nature of neural networks complicates result interpretation and understanding of prediction mechanisms, posing significant limitations in applications where interpretability is crucial. Small sample size, internal validation, and lack of external benchmarks preclude definitive claims about clinical impact.
Understanding the strengths and limitations of each model is crucial for informed decision-making regarding adoption and implementation in practice.
Future Directions: Larger datasets, external validation in multi-center, and integration with established clinical risk scores are needed to justify real-world use.
Conclusion
This study introduces a novel approach to survival prediction in HF patients by developing a DNFCR model. The DNFCR model demonstrates a technical innovation and a proof-of-concept for integrating frailty into competing risk analysis, because it can account for unobserved heterogeneity and analyze CR, but requires further validation before clinical application. Given the similarity in performance, choosing the best model may depend on different factors, such as model complexity, training time, and computational cost. Ultimately, this research emphasizes the importance of integrating DL with frailty models and competing risk analysis to improve predictions and facilitate more precise therapeutic decisions, highlighting methodological challenges that warrant further research.
Data availability
The datasets used during the current study are available from the corresponding author on reasonable request.
Abbreviations
- DNFCR:
-
Deep neural frailty competing risk
- HF:
-
Heart failure
- Frailty:
-
Unobserved heterogeneity
- CR:
-
Competing risk analysis
- CoxPH:
-
Cox proportional hazards
- DL:
-
Deep learning
- HfrEF:
-
Heart failure with a reduced ejection fraction
- DNFCR_PF:
-
DNFCR model with proportional frailty
- DNFCR_NF:
-
DNFCR model with nonproportional frailty
- DeepSurv:
-
Deep neural survival
- DeepSurvCR:
-
Deep neural survival competing risk
- NFM:
-
Neural frailty machine
- C-index:
-
Concordance probability
- IBS:
-
Integrated Brier Score
- INBLL:
-
Integrated negative binomial log-likelihood
References
Kleinbaum, D. G. & Klein, M. Survival Analysis a Self-Learning Text (Springer, 1996).
Martin, J. F., Perin, E. C. & Willerson, J. T. Direct stimulation of cardiogenesis: a new paradigm for treating heart disease. Circul. Res. 121(1), 13–15 (2017).
Dutra, G. P. et al. Mortality from heart failure with mid-range ejection fraction. Arq. Bras. Cardiol. 118(4), 694–700 (2022).
Li, J. et al. Two machine learning-based nomogram to predict risk and prognostic factors for liver metastasis from pancreatic neuroendocrine tumors: a multicenter study. BMC Cancer. 23(1), 529 (2023).
Putter, H., Fiocco, M. & Geskus, R. B. Tutorial in biostatistics: competing risks and multi-state models. Stat. Med. 26(11), 2389–2430 (2007).
Kaplan, E. L. & Meier, P. Nonparametric estimation from incomplete observations. J. Am. Stat. Assoc. 53(282), 457–481 (1958).
Austin, P. C., Lee, D. S. & Fine, J. P. Introduction to the analysis of survival data in the presence of competing risks. Circulation 133(6), 601–609 (2016).
Austin, P. C. & Fine, J. P. Accounting for competing risks in randomized controlled trials: a review and recommendations for improvement. Stat. Med. 36(8), 1203–1209 (2017).
Zhong, Q., Mueller, J. & Wang, J-L. Deep learning for the partially linear Cox model. Annals Stat. 50(3), 1348–1375 (2022).
Wu, R. et al. Neural frailty machine: beyond proportional hazard assumption in neural survival regressions. Adv. Neural. Inf. Process. Syst. 36, 5569–5597 (2023).
Mandel, F., Ghosh, R. P. & Barnett, I. Neural networks for clustered and longitudinal data using mixed effects models. Biometrics 79(2), 711–721 (2023).
Monterrubio-Gómez, K., Constantine-Cooke, N. & Vallejos, C. A. A review on competing risks methods for survival analysis. ArXiv Preprint arXiv arXiv:221205157:221205157. (2022).
Wiegrebe, S., Kopper, P., Sonabend, R., Bischl, B. & Bender, A. Deep learning for survival analysis: a review. Artif. Intell. Rev. 57(3), 65 (2024).
Katzman, J. L. et al. DeepSurv: personalized treatment recommender system using a Cox proportional hazards deep neural network. BMC Med. Res. Methodol. 18, 1–12 (2018).
Lee, C., Zame, W., Yoon, J. & Van Der Schaar, M. (eds) Deephit: A deep learning approach to survival analysis with competing risks. In Proceedings of the AAAI Conference on Artificial Intelligence (2018).
Rietschel, C., Yoon, J. & van der Schaar, M. Feature selection for survival analysis with competing risks using deep learning. arXiv preprint arXiv:181109317 (2018).
Huang, P. & Liu, Y. (eds) Deepcompete: A deep learning approach to competing risks in continuous time domain. In AMIA Annual Symposium Proceedings (American Medical Informatics Association, 2020).
Nagpal, C., Yadlowsky, S., Rostamzadeh, N. & Heller, K. (eds) Deep Cox mixtures for survival regression. In Machine Learning for Healthcare Conference (PMLR, 2021).
Tran, M-N., Nguyen, N., Nott, D. & Kohn, R. Bayesian deep net GLM and GLMM. J. Comput. Graphical Stat. 29(1), 97–113 (2020).
Lee, H., HA, I. & Lee, Y. Deep Neural Networks for Semiparametric Frailty Models via H-likelihood. arXiv preprint arXiv:230706581 (2023).
Rindt, D., Hu, R., Steinsaltz, D. & Sejdinovic, D. (eds) Survival regression with proper scoring rules and monotonic neural networks. In International Conference on Artificial Intelligence and Statistics (PMLR, 2022).
Klein, J. P., Van Houwelingen, H. C., Ibrahim, J. G. & Scheike, T. H. Handbook of Survival Analysis (CRC, 2014).
Kalbfleisch, J. D. & Prentice, R. L. The Statistical Analysis of Failure time Data (Wiley, 2011).
García-Ordás, M. T., Bayón-Gutiérrez, M., Benavides, C., Aveleira-Mata, J. & Benítez-Andrades, J. A. Heart disease risk prediction using deep learning techniques with feature augmentation. Multimedia Tools Appl. 1–15 (2023).
Katzman, J. L. et al. DeepSurv: personalized treatment recommender system using a Cox proportional hazards deep neural network. BMC Med. Res. Methodol. 18(1), 1–12 (2018).
Wang, M. et al. Dementia risk prediction in individuals with mild cognitive impairment: a comparison of Cox regression and machine learning models. BMC Med. Res. Methodol. 22(1), 284 (2022).
Kantidakis, G., Biganzoli, E., Putter, H. & Fiocco, M. A simulation study to compare the predictive performance of survival neural networks with Cox models for clinical trial data. Comput. Math. Methods Med. 2021(1), 2160322 (2021).
Jung, W. et al. Deep learning model for individualized trajectory prediction of clinical outcomes in mild cognitive impairment. Front. Aging Neurosci. 16, 1356745 (2024).
Author information
Authors and Affiliations
Contributions
Data curation was performed by N.N. and S.N. The investigation was conducted by S.N. and M.A.J. Methodology was developed by S.N., M.A.J., and E.H. Project administration was handled by S.N., M.A.J., and E.H. Software development was done by S.N. and H.KH. S.N. and M.A.J. were responsible for writing, reviewing, and editing the manuscript. All authors reviewed the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Norouzi, S., Khormaei, H., Jafarabadi, M.A. et al. Development of a deep learning model for survival prediction in heart failure: competing risk and frailty model. Sci Rep 15, 34088 (2025). https://doi.org/10.1038/s41598-025-14715-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-025-14715-4