Abstract
Horses and humans are among the few mammals susceptible to spontaneous atrial fibrillation (AF), both suffering from high recurrence rates after treatment. Treatment resistance is often attributed to progressive atrial remodeling, but current treatment options fail to effectively address this aspect. Here, we introduce a novel horse model of spontaneous AF to investigate the biological pathway changes in early stages of the disease. Through data-independent acquisition mass spectrometry on biopsies from the right and left atrium and left ventricular chamber of horses with early-stage persistent AF (n = 8) and controls (n = 8), we identify several differentially regulated proteins across all three chambers. Pathway enrichment analyses and histological stainings highlight a significant role of atrial extracellular matrix (ECM) remodeling in early AF. Other key proteomic changes relate to metabolism, contractility, and protein-folding, and overlap with findings from publicly available human datasets. Our results demonstrate that horses and humans share several AF-related proteomic changes, providing translational insights into the early atrial remodeling processes that are likely to contribute to treatment resistance. These protein-level changes could serve as biomarkers or pharmacological targets for preventing AF-associated atrial remodeling and improve treatment outcomes across species.
Similar content being viewed by others
Introduction
Atrial fibrillation (AF) is a highly prevalent cardiac arrhythmia characterized by rapid, irregular electrical activity in the atria, leading to inefficient atrial contraction and subsequent blood stasis. The condition can have serious clinical implications, including an increased risk of stroke, heart failure, and a reduced quality of life1. While AF is well-documented in humans2, horses are among the few other species also known to suffer from AF, with a reported prevalence of up to 4.9% in Thoroughbred racehorses3 and 6.2% in a mixed breed population4. Several studies have explored the epidemiology and pathogenesis of the disease in horses3,4,5,6,7,8,9,10,11,12, and although stroke and heart failure are rare, AF remains the most common cardiac arrhythmia affecting horse performance, often leading to early retirement or euthanasia5. Importantly, AF pathogenesis in horses shares many similarities with humans, including a similar progressive course, pulmonary vein ectopy and fibrotic changes8,10,13. Despite increasing knowledge of the mechanisms driving AF, clinical management remains challenging in both species. The human AF incidence is rising, new treatment modalities targeting the electrical disturbance have shown limited efficacy, and recurrence rates are up to 50% in both humans and horses with AF14,15,16,17. To improve treatment success, it is essential to understand the molecular pathways that drive the progressive arrhythmogenic remodeling. Targeting atrial remodeling, in addition to addressing the electrical disturbance, could constitute an “upstream” approach for more effective AF management18.
Mass spectrometry-based proteomics has been used previously to study atrial changes in samples from humans suffering from AF19,20,21. These studies have provided valuable insights into the potential atrial consequences of AF, particularly emphasizing local metabolic changes. However, these studies have been limited by small sample sizes (n = 3 to 10) and restricted to samples from either the left atrium (LA) or the left atrial appendages from patients undergoing cardiac surgery for ablation therapy or for reasons unrelated to AF. Furthermore, most of the included patients also had several other comorbidities (mitral valve stenosis, heart failure, coronary disease etc.) and had often suffered from AF for extended periods (three months to more than a year)19,20,21. This creates a highly heterogeneous study group, making it difficult to distinguish between AF-induced changes and changes caused by other conditions.
Studying AF at earlier stages in an animal model free from potential confounding variables allows us to investigate the early changes that occur in AF. We hypothesize that these early changes are associated with the transition of AF into a more treatment-resistant phenotype, and therefore could be key in AF drug discovery. Horses often develop AF at a relatively young age without underlying health issues, thus overcoming the limitations posed by confounding health factors in human studies. This study maps the protein changes associated with natural AF in horses and compares these with publicly available human datasets20,22. By studying atrial arrhythmogenic remodeling in horses at the early stages of the disease and comparing our findings with human data, we identify shared metabolic, contractile, and structural changes that could serve as potential targets for AF prevention or treatment.
Results
Horse characteristics
Eight horses with AF and eight healthy horses in sinus rhythm confirmed by surface ECG recordings were included in this study (ECG example in Supplementary Fig. 1). The two groups of horses did not differ in terms of age and weight, however, a significantly lower left atrial (LA) reservoir index was observed in the horses with AF (Table 1).
Large overlap between cardiac proteomes of healthy horses and humans
To map the protein landscape of healthy horses, we applied label-free data-independent acquisition (DIA)-MS quantification of three regions in the heart: the right atrium (RA), the LA, and left ventricle (LV). Our analysis identified a total of 3,400 proteins (Fig. 1A). To elucidate the similarity to humans, we compared the data against a human heart proteome database23, identifying an 88.8% overlap in cardiac proteins, indicating a baseline resemblance between healthy horse and human heart proteomes (Fig. 1B). When employing principal component analysis (PCA), the LV samples clustered separately from the LA and RA samples along component 1, accounting for 29.8% of the observed variance (Fig. 1C). Although no clear clustering was observed between atrial samples along component 2, we identified 134 differentially abundant proteins (DAPs; Student’s t-test; non-adj. p-value < 0.05) when comparing RA to LA (Fig. 1D). These DAPs included TRMO, CKB, THEM4, BBOX1, PKIA and DTNA. Pathway enrichment analysis of the DAPs revealed an enrichment of pathways associated with “ribosomes” and “oxidoreductase activity” in LA, while RA exhibited higher association with metabolic and mitochondrial proteins (Fig. 1E and F).
Characterization of healthy horse proteome. (A) Stacked bar representation of unique proteins identified by DIA-MS at 1% false-discovery rate (FDR). (B) Bar chart showing the number of proteins shared with a previously published study of the human heart23. (C) Principal component analysis (PCA) plot of log2-transformed intensities associated with the healthy control horse samples. Components 1 and 2 are presented. Left atrium (LA) = red, right atrium (RA) = blue, left ventricle (LV) = green. (D) Volcano plot comparing protein abundance in LA and RA regions from healthy control horses. Log2 fold change (logFC) difference and non-adjusted p-value are presented in volcano plot. Red = upregulated and blue = downregulated in LA compared to RA. (E and F) Significance score identified in enrichment analysis of upregulated or downregulated proteins in LA compared to RA. Following databases were used; GO MF (GO Molecular Function, depicted in blue) and GO CC (GO Cellular Component, depicted in grey).
Proteomic profiling of LA in horses with AF revealed changes in metabolism and contractility
To investigate the protein signatures of AF in different regions of the heart, we initially compared the LA region from control subjects to those with AF. Using PCA, we observed two distinct clusters corresponding to the groups along component 2, accounting for 18.5% variance (Fig. 2A). Statistical analysis revealed 324 DAPs (Fig. 2B) (supplementary Table 1). Pathway enrichment analysis of the upregulated DAPs highlighted pathways such as “metabolism” and “electron transfer activity”, while downregulated DAPs were enriched in pathways such as “muscle contraction” and “cytoskeletal protein binding” (Fig. 2C and D). Specifically, gene regulatory network analysis revealed enrichment of the metabolic regulator PRKAA2 among the upregulated proteins, while the transcription factors SRF and NFE2 were enriched among the downregulated proteins.
Pathway analysis of proteins identified in left atrium from horses with atrial fibrillation. (A) Principal component analysis (PCA) plot of log2-transformed intensities associated with healthy control (CTR) and atrial fibrillation (AF) horse samples. Components 1 and 2 are presented. AF = red, CTR = blue. (B) Volcano plot comparing protein abundance in left atrium (LA) regions from AF horses. Log2 fold change (logFC) difference and non-adjusted p-value are presented in volcano plot. Red = upregulated and blue = downregulated in AF compared to CTR. (C and D) Significance score identified in enrichment analysis of upregulated or downregulated proteins using different databases; GO Biological Process (beige), GO Cellular Component (grey), and GO Molecular Function (blue).
Proteomic profiling of RA in horses with AF revealed changes in extracellular matrix and contractile proteins
Next, we compared the RA region from control horses to horses with AF. Using PCA, we observed two clusters corresponding to the groups along component 4, accounting for 9.2% variance (Fig. 3A). Statistical analysis revealed 268 DAPs (Fig. 3B). Pathway enrichment analysis of the upregulated DAPs highlighted pathways such as “ECM organization” and “carbohydrate metabolism” (Fig. 3C), while downregulated DAPs were associated with pathways such as “muscle contraction” and “carboxylic acid metabolism” (Fig. 3D).
Pathway analysis of proteins identified in right atrium from horses with atrial fibrillation. (A), Principal component analysis (PCA) plot of log2-transformed intensities associated with healthy control (CTR) and atrial fibrillation (AF) horse samples. Components 1 and 4 are presented. AF = red, CTR = blue. (B) Volcano plot comparing protein abundance in right atrium (RA) regions from AF horses. Log2 fold change (logFC) difference and non-adjusted p-value are presented in volcano plot. Red = upregulated and blue = downregulated in AF compared to CTR. (C and D) Significance score identified in enrichment analysis of upregulated or downregulated proteins using different databases; GO Biological Process (beige), GO Cellular Component (grey) and GO Molecular Function (blue).
Proteomic profiling of LV in horses with AF revealed changes in metabolic pathways
Finally, we investigated the protein differences between LV regions from control and AF horses. Using PCA, we observed two separate clusters corresponding to the groups along component 3, accounting for 10% variance (Fig. 4A). Statistical analysis revealed 284 DAPs (Fig. 4B). Pathway enrichment analysis revealed that the upregulated DAPs were enriched in pathways related to “metabolism” and “tRNA aminoacylation”. Analysis of downregulated DAPs also highlighted “metabolism” and “actin” (Fig. 4C and D).
Pathway analysis of proteins identified in left ventricle from horses with atrial fibrillation. (A) principal component analysis (PCA) plot of log2-transformed intensities associated with healthy control (CTR) and atrial fibrillation (AF) horse samples. Components 1 and 3 are presented. AF = red, CTR = blue. (B) Volcano plot comparing protein abundance in left ventricle (LV) regions from AF horses. Log2 fold change (logFC) difference and non-adjusted p-value are presented in volcano plot. Red = upregulated and blue = downregulated in AF compared to CTR. (C and D) Significance score identified in enrichment analysis of upregulated or downregulated proteins using different databases; GO Biological Process (beige), GO Cellular Component (grey) and GO Molecular Function (blue).
Interestingly 21 of the identified DAPS were regulated in the same direction in all three chambers (Supplementary Fig. 2A-C.) comprising a larger overlap between the two atria including upregulation of VCAN and downregulation of MYH6 (Supplementary Fig. 2B). A complete list of all the DAPs across regions are provided in Supplementary Table 1.
Histological and proteomic profiling highlighted extracellular matrix glycoprotein remodeling in early AF
A major hallmark of AF progression is structural remodeling with changes in the atrial tissue structure and fibrosis. To investigate the degree of fibrosis in the three heart regions, we stained tissue with Picro Sirius red and wheat germ agglutinin (WGA). Automated segmentation image analysis of picrosirius red stained tissue revealed an increase in collagen content in the LA samples from horses with AF (p-value = 0.01), but no significant difference in the RA and LV compared to control horses (Fig. 5A). We also identified a significantly higher intensity of WGA positive content, indicative of higher levels of ECM glycoproteins, in the RA of horses with AF (p-value = 0.01) (Fig. 5B).
Fibrosis quantification. (A) Representative images of Picro Sirius red staining and comparison of collagen content in the left atrium (LA), the right atrium (RA) and the left ventricle (LV) between controls (CTR) and horses with atrial fibrillation (AF) (Student’s t-test). (B) representative images from wheat germ agglutinin (WGA) labelling and comparison of WGA intensity in the LA, the RA and the LV between CTR and horses with AF (Student’s t-test), * p < 0.05). Representative examples of Picro Sirius red are shown from control horse 13 (mare, 5 years, 405 kg) and from AF horse 6 (stallion, 3 years, weight unknown) for RA and LV and from AF horse 8 (mare, 3 years, 402 kg) for LA. For WGA, representatives are shown from control horse 14 (gelding, 6 years, 500 kg) for LA and LV and from control horse 10 (gelding, 7 years, 490) for RA and from AF horse 5 (gelding, 2 years, 410 kg).
To gain deeper insight into the specific protein changes associated with ECM remodeling, we enriched our proteomic data for ECM proteins. We identified a total of 324, 268 and 284 ECM-associated proteins in LA, RA, and LV, respectively (Fig. 6A-C; Supplementary Fig. 3). Using PCA based on the ECM-associated proteins only, we observed two clusters corresponding to the groups in all three heart regions (Supplementary Fig. 3). Statistical analysis revealed 23, 21 and 22 differentially expressed ECM proteins in LA, RA, and LV regions, respectively, when comparing AF and control (non-adj. p-value < 0.05; Fig. 6D-F). Interestingly, the majority of differentially regulated glycoproteins were upregulated in the LA and RA but downregulated in the LV of AF horses compared to controls (Fig. 6G-I).
Extracellular matrix profiling of three heart regions from horses with atrial fibrillation. Pie charts representing subgroups of differentially expressed extracellular matrix (ECM)-associated genes/proteins from left atrium (LA) (A), right atrium (RA) (B), and left ventricle (LV) (C) of horses with atrial fibrillation (AF). The annotations were derived from the “matrisome” database55,56. Unsupervised hierarchical clustering of significantly regulated ECM-associated proteins in the three regions; the LA (D), RA (E) and the LV (F). The scale depicts z-scored label-free quantification (LFQ) values. Scatterplots of Log2 fold change (logFC) of all glycoproteins for the the LA (G), RA (H) and the LV (I). Each protein is coloured as ECM proteins = grey, upregulated ECM glycoproteins = red, downregulated ECM glycoproteins = blue.
Based on the particular regulation of ECM glycoproteins observed in the horses with AF, we next aimed to investigate whether similar regulatory patterns were observed in two publicly available datasets from human AF patients20,22. Similar to our equine study, around 6% of the LA DAPs/genes were ECM-associated (Fig. 7A and B, Supplementary Fig. 4). Furthermore, several atrial glycoproteins were upregulated at both proteomic and transcriptomic levels (Fig. 7C and D). Direct cross-species comparison showed that a number of glycoproteins such as POSTN and VCAN were upregulated in both human and horse LA tissue (Fig. 7E and F).
Left atrial extracellular matrix (ECM) glycoprotein expression is increased in both horse and human atrial fibrillation. Pie charts representing subgroups of differentially expressed extracellular matrix (ECM)-associated proteins/genes from previous human proteomic (A) and transcriptomic publications (B). The annotations were based on the “matrisome” database55,56. Scatter plot of log2 fold changes (logFC) for all significantly regulated ECM proteins in selected publications (C and D). Each protein is colored as (ECM proteins = grey, Upregulated ECM glycoproteins = red, Downregulated ECM glycoproteins = blue). E and F, scatter plot of shared ECM proteins using logFC differences from previous publication and horse comparison in our study. Each protein is colored as (shared upregulated ECM proteins = red, shared downregulated ECM proteins = blue, other ECM categories = grey) (E and F). ECM glycoproteins are highlighted as triangles (▲).
Protein changes in the left atrium of horses with AF were comparable to human AF
Next, we investigated the extent to which the AF proteome in horses resembles that of humans by comparing all DAPs from the LA to a curated gene list, containing significantly regulated proteins and genes from the previously mentioned proteomic and transcriptomic studies on AF in humans20,22. Of the 282 DAPs identified in our study, 41 were shared with the proteomic20 (Fig. 8A and B) and 21 were shared with the transcriptomic study22 (Fig. 8C and D). Using scatterplots depicting fold-change differences in our study and the study conducted by Doll et al. (2017), we identified several shared upregulated proteins such as HSPA4, PSMD13 and POSTN along with shared downregulated proteins such as MYH6, TPM2, TNNT2 (Fig. 8B). Additionally, TNNT2 was also a shared downregulated protein when comparing to van den Berg et al. (2021) (Fig. 8D). Pathway enrichment analysis of the overall list of shared proteins indicated that proteins related to “Striated muscle contraction” were downregulated, while proteins from the “Extracellular region” were upregulated (Fig. 8E and F).
Identification of atrial fibrillation induced left atrial protein changes between humans and horses Diagrams showing number of significantly regulated proteins that are shared with two previous studies identifying atrial fibrillation (AF) expression profiles in left atria from humans on either proteomic (A)20 or transcriptomic level (B)22. (C and D) Scatter plot comparing fold change (logFC) differences of shared significantly regulated AF proteins from our study and a previous studies Red = upregulated, blue = downregulated in AF compared to control (CTR). (E and F), significance score (-log10 transformed false-discovery rate (FDR)) identified in enrichment analysis of shared AF proteins using different databases; GO Biological Process (grey), GO Cellular Component (blue), KEGG (purple)59, Reactome (red).
Discussion
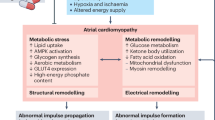
Despite research efforts, comprehending the molecular pathogenesis of AF and mechanisms underlying atrial arrhythmogenic remodeling has proven challenging, impeding effective treatment and prevention strategies. To our knoweldge, this study is first to evaluate the AF-induced proteomic changes in an animal model that spontaneously manifest the disease. Our results revealed substantial overlap in the cardiac proteome between healthy horses and humans, underscoring the translational value of the AF horse model and confirming previous findings23. Our study also highlights atrial metabolic, contractile and structural changes associated with early AF in an otherwise healthy model organism and emphasizes the important role of ECM glycoprotein changes in early disease. Finally, we demonstrated that the AF-induced changes in proteins and their associated signaling pathways were shared between horses with AF and humans with AF. Our proteomic analyses of horses with AF revealed changes in metabolic proteins in all three heart regions. This finding aligns with a human study that investigated samples from AF patients undergoing cardiac surgery20. They found that the most significantly downregulated proteins were enriched for mitochondrial function. Among these, COX7B, IMMT and TIMM8A were > 25-fold less abundant than in healthy tissue. In contrast, we observed an increase in mitochondrial proteins in the LA of horses with AF, as evidenced by an enrichment in oxidative phosphorylation and ATP synthesis pathways. Interestingly, COX7A2 was upregulated in the LA from the AF horses, opposite to what was observed in humans. This apparent discrepancy could be due to species-specific factors, but the different durations of AF between study populations are also likely to be important. In paroxysmal or short-lasting persistent AF, mitochondria are believed to initially compensate for the high atrial rate and boost ATP synthesis. However, over time, continuous metabolic stress may lead to mitochondrial decompensation and a decline in ATP production24. During this process, atrial myocytes can switch their preferred energy substrate from fatty acids to glucose, a phenomenon referred to as a fetal phenotype change25. This metabolic reprogramming has previously been observed in human cardiovascular diseases, including dilated cardiomyopathy, cardiac hypertrophy26 and atrial fibrillation27. This process has been associated with an activation of the energy sensor AMPK, which we identified as a key transcriptional regulator activated in AF in our RA gene regulatory network analysis. In line with this finding, we observed an upregulation of proteins associated with carbohydrate metabolism in the RA, further supporting the role of fetal reprogramming as a component of the atrial metabolic response in equine AF.
We also identified biatrial changes in ECM proteins in AF horses, supported by histological evidence of increased collagen in the LA and increased WGA-staining intensity in the RA. Together, this shows that the ECM remodeling has occurred, even early in the disease process. Structural remodeling is believed to be a leading cause of treatment failure and recurrence in human patients28,29. Our study identified that several key ECM proteins, including MFAP5, VCAN, EFEMP1 and PRELP, were upregulated in both atria. VCAN, an important regulator of cell adhesion, proliferation and migration is produced by cardiac fibroblasts and has previously been associated with an increase in collagen-producing myofibroblast-like cells and ECM gene expression in the LA appendage from humans with AF22. We also observed a downregulation of MXRA5 and TGM2 in both atria of AF horses. MXRA5 has anti-inflammatory and anti-fibrotic properties in the kidneys30 and could potentially have similar effects in the heart. In the LA, the matricellular proteins LGALS1 and POSTN were significantly more abundant in the AF horses, strongly indicative of increased fibrinogenesis as evident from the histological stainings. LGALS1, which facilitates myofibroblast differentiation, has been proposed as a potential target in AF treatment, and is significantly upregulated in humans with AF31. Of note, atrial glycoproteins such as POSTN, MFGE8 and MFAP5 were particularly upregulated, which aligns with previous human data20,22. Together, our findings underscore that substantial remodeling of the atrial ECM proteome occur early in AF pathogenesis, emphasizing the importance of early treatment to prevent structural remodeling.
We uncovered a range of proteins with shared expression profiles between both horses and humans with AF. Specifically, we observed shared upregulation of HSPA4 and PSMD13, both proteins that are involved in monitoring protein homeostasis (“proteostasis”). HSPA4, a member of the heat shock protein family, assist in the folding and refolding of proteins and is believed to support mitochondrion outer membrane assembly32. Interestingly, heat shock proteins are often upregulated in early stages of AF, where they exhibit cardioprotective effects in stressed cytotoxic environments33. PSMD13, on the other hand, is a vital part of the 26 S proteasome, a multi-protein assembly responsible for degrading damaged and ubiquitinated proteins. This complex is crucial for preserving protein balance within cells by degrading misfolded or impaired proteins. While the role of PSMD13 in AF is unknown, it has been shown to inhibit the NF-κB pathway by preventing ubiquitination of the TAK1, a key mediator of pro-inflammatory signalling pathways34. The suppression of TAK1 by PSMD13 could therefore be a protective response in AF. Together, the increased expression of the heat shock proteins and the ubiquitin proteasome-system indicates an activation of the protein quality control system, potentially due to cytoskeletal and sacormeric protein damage triggered by the fast atrial rates35. Derailed proteostasis has been suggested as a driver of AF-induced remodeling36, making these proteins potential upstream targets for AF therapy.
Finally, we identified that contractile proteins were downregulated in AF in both species which included a myosin (MYH6), a tropomyosin (TPM2), and a troponin (TNNT2). Several studies have identified decreased atrial contractile function in AF in both humans and horses37,38. Using echocardiography, we observed a reduced LA reservoir index, an indicator of mechanical dysfunction, in the horses with AF. This finding aligns with a previous study in horses, where atrial contractility was reduced after only three days of induced AF10. Cellular hypocontractility has previously been linked to stroke and thromboembolism in humans, which are significant concerns in AF patients10,39.
While the pathophysiology of AF has previously been studied in horses using tachypacing-induced models10,40,41,42, this study is first to use a preclinical large animal model that spontaneously manifests persistent AF. A recent review on the challenges and solutions in translational AF research specifically emphasized this feature as a limitation of current AF models43. Presently available AF models rely on tachypacing, genetic manipulation or induction of inflammation, autonomic or ventricular dysfunction to create an AF-substrate44. They thereby fail to provide insights into the early AF events in otherwise healthy animals, neglecting factors associated with disease initiation. The horse model stands out not only for its spontaneous disease, but also for its proteomic similarities with humans shown here. Furthermore, horses share pathophysiological characteristics with humans such as pulmonary vein ectopy13,45, and they follow a progressive course of AF similar to humans10. The large size of the atria enables repeated longitudinal transvenous tissue sampling, which could facilitate future studies on temporal substrate changes during AF progression. Overall, the horse holds promise as a valuable translational model for understanding the disease mechanisms underlying AF initiation and early progression, which is a knowledge gap that needs to be filled in order to develop new AF therapies.
The study possesses certain limitations that should be acknowledged. Perfect matching between AF and control horses was not feasible, potentially introducing confounding factors such as breed, sex and age that could influence the DAP identification. Furthermore, two different sampling methods were applied; transvenous and postmortem collection. While this could introduce variability, PCA clustering was primarily driven by disease status and region rather than sample type, age or breed, suggesting that these potential effects were likely minor. Knowing the exact AF duration in these horses would also have been advantageous. However, since AF was not induced experimentally, determining the exact onset time was not possible. Studying AF pathogenesis in humans poses several challenges due to limited atrial tissue availability, and the high prevalence of confounding co-morbidities. While we explored the similarities of AF-induced changes between horses and humans across both RNA and protein levels, we recognize the limitation of directly comparing transcriptomic and proteomic data. Gene expression and protein abundance are governed by different regulatory mechanisms, and rarely correlate perfectly. Yet, our analysis identified shared molecules, supporting the conserved biological response identified in the proteomic comparison. Future studies incorporating equine transcriptomic data will be essential for advancing our comparative understanding of AF. Despite being a different species, studying AF in horse offers clear advantages in capturing isolated spontaneous AF at its earliest stage, without any underlying systemic or local heart disease.
In conclusion, our study provides significant insights into the early AF pathophysiology that may drive arrhythmogenesis in both humans and horses. We identified early metabolic and contractile adaptations, along with signs of structural remodeling in equine hearts, many of which were shared with human AF patients. These findings underscore the importance of early intervention in AF management and provides insights into potential mechanistic targets for mitigating early AF-induced remodeling. Moreover, the successful implementation of non-invasive transvenous heart sampling in live horse patients demonstrates the feasibility of longitudinally studying atrial arrhythmogenic remodeling dynamics. Our study introduces a new horse model with spontaneous disease onset for translational AF research.
Methods
Animals
The study was conducted in compliance with the guidelines from the European Commission Directive 86/609/EEC. Prior to commencing the study, it received ethical approval from the local committee responsible for ethical considerations at the Department of Veterinary Clinical Sciences, University of Copenhagen, and was also reviewed and authorized by the Danish Animal Experiments Inspectorate, granted under license number 2020-15-0201-00425. For client-owned horses, consent was obtained prior to the inclusion of the horses. All methods are reported in accordance with ARRIVE guidelines.
Two of the horses were admitted to the Large Animal Teaching Hospital in Taastrup, Denmark, for cardioversion of AF. Before treatment, informed consent was obtained from the owners to collect transvenous biopsies from the RA. For the remaining six horses, treatment was not pursued by the owners, and the horses were donated for research. From these horses, post-mortem biopsies were collected from the RA, LA and LV. Eight healthy research horses in sinus rhythm served as a control group. For specific details regarding the included horses, please refer to Table 1. Surface 24 h Holter ECG recordings were obtained from all horses prior to inclusion. Routine clinical examination, blood hematology and biochemistry panels were assessed to rule out systemic illness. Echocardiographic evaluations were performed to assess cardiac structure and function, and valvular regurgitations considered either trivial or mild were accepted at inclusion. The postmortem samples were harvested immediately after euthanasia by a lethal dose of pentobarbital (140 mg/kg, Euthasol® vet. (400 mg/mL), Dechra Veterinary Products A/S, Uldum, Denmark). Two of the control horses were sacrificed by exsanguination during a terminal experiment where the horses were under deep general anesthesia. The terminal study was performed in accordance with approved ethical protocols and EU legislation. Further details are available in a previous publication46. The heart was explanted within 10 min, immersed in cold phosphate buffered saline (PBS), and samples were collected and snap-frozen in liquid nitrogen and stored at -80 °C until proteomic analysis. Samples for histology were fixed in 10% formaldehyde for 24 h and then stored in 70% ethanol.
Transvenous biopsy collection
As previously mentioned, transvenous biopsies from the RA were collected from two of the included AF horses. The biopsy sampling was performed as previously described47,48. The horses were sedated intravenously with 0.01 mg/kg body weight (BW) detomidine (Domosedan®, Orion Pharma Animal Health, Copenhagen, Denmark) and 0.01 mg/kg BW butorphanol (Dolorex®, Orion Pharma Animal Health, Copenhagen, Denmark). A shaved area over the mid-region of the right jugular vein was prepared for an aseptic procedure. Before vein puncture, local analgesia with 3 ml of Mepivacaine was administered subcutaneously (Carbocain®, 10 mg/ml mepivacaine; Aspen Pharma Trading Limited, Dublin, Ireland). A 10 Fr sheath was inserted using the modified Seldinger technique, followed by an8 F introducer sheath (Flexor® check-Flo® Introducer Set, 90 cm, COOK Medical) guided to the RA with a guidewire under transthoracic echocardiographic guidance. Once proper placement was confirmed, the bioptome (Argon 190031 Jawz™, Endomyocardial biopsy forceps, 2.4 mm × 105 cm, 7.5 F, Argon Medical) was introduced to collect endomyocardial samples. Following collection, biopsies for proteomics were promptly frozen in liquid nitrogen and stored at -80 °C until subsequent processing. Samples for histology were fixed in 10% formaldehyde for 24 h and then stored in 70% ethanol.
Histology and immunohistochemistry
Paraffin-embedded tissue was sectioned at 4 μm, deparaffinized by a descending series of ethanol (99.9–70%), followed by Tissue-Clear® Xylene substitution, and rehydrated in water. Next, the samples were stained according to the manufacturer’s protocol by a series of steps with Wiegert’s haemoxylin and picrosirius red (Morphisto staining kit Picrosirius red for collagen I + III, Frankfurt, Germany). Images were acquired using the Zeiss Axio Scan.Z1 Slide scanner at a magnification of 10x. Subsequently, collagen quantification was performed using automated segmentation analysis in ZEN Intellesis software, as outlined in a previous study10.
For immunohistochemical staining with WGA, the samples were deparaffinized and blocked by incubation with 2% (w/v) fraction V bovine serum albumin and 2% (w/v) glycine in PBS at room temperature for 1 h. Next, the samples were incubated with WGA diluted in PBS (1:200) (Alexa Flour 594 conjugate, ThermoFisher) for 2 h at room temperature. A final wash in PBS was performed and coverslips were mounted with Prolong Gold Antifade mounting medium (ThermoFisher Scientific). Images were acquired using the Zeiss Axio Scan.Z1 Slide scanner at a magnification of 20x. Area and intensity were quantified using ZEN Intellesis software as previously described for automated segmentation analysis49.
Protein isolation
The frozen samples were homogenized in tissue buffer (5 mM EDTA, 25 mM NaF, 300 mM Sucrose, 30 nM and KH2PO4 in PMSF (final conc: 100µM) with 1 protease inhibitor tablet and 1 phosphatase inhibitor tablet per 10 ml buffer) Lysing 1/8 Matrix Stubes with beads in a Preccellys (CK28) centrifuge with the following program: 5000 rpm for 20 s times 2 with 20 s of cooling between. The homogenate was spun at 4 °C at 15.000 rpm for 5 min and the supernatant was transferred and stored at -80 °C.
Sample Preparation for mass spectrometry analysis
Isolated protein extracts (100 µg) were diluted in digestion buffer (0.5% SDC in 50 mM TEAB) and heat-treated for 5 min at 95 °C. A modified filter-aided sample preparation (FASP) protocol50,51 was performed. Proteins were reduced and alkylated in digestion buffer containing 1:50 (v: v) tris(2-carboxyethyl)phosphine (0.5 M, Sigma) and 1:10 (v: v) 2-chloroacetamide (0.5 M, Sigma) for 30 min at 37 °C. Proteolytic digestion was carried out overnight at 37 °C using Trypsin/LysC mix (Promega) in a 1:100 ratio (µg of enzyme to µg of protein) per sample. The peptide-containing solutions were cleaned up using self-made stage-tips containing a Poly-styrene-divinylbenzene copolymer modified with sulfonic acid groups (SDB-RPS; 3 M) material, as described previously52. After vacuum centrifugation, samples were resuspended in 2% acetonitrile (Sigma) / 0.1% TFA (Sigma).
Data acquisition by mass spectrometry
Peptide mixtures (1 µl direct injections) were analyzed on Bruker timsTOF mass spectrometer (Bruker Daltonics) using positive ion mode with a Captivespray ion source on-line connected to a Dionex Ultimate 3000RSnano chromatography system (Thermo Fisher). Peptides were separated on an Aurora column with captive spray insert (C18 1.6 μm particles, 15 cm, 75 μm inner diameter; IonOptics) at 60 °C. A solvent gradient of buffer A (0.1% formic acid) and buffer B (99.9% acetonitrile/0.1% formic acid) over 28 min, was used at a flow rate of 600 nL/min. The mass spectrometer was operated in DIA PASEF mode with 1.1 s cycle time and TIMS ramp time of 100.0 ms. MS scan range was set to 100–1700 m/z.
Protein identification by mass spectrometry
Bruker raw files (.d) were analyzed with DIA-NN software (v.1.8.0)53 using the UniProt FASTA Equus caballus (UP000002281_9796 and UP000002281_9796_additional) database (downloaded July, 2023). In DIA-NN, the options ‘FASTA digest for library-free search/library generation’ and ‘Deep learning-based spectra, RTs and IMs prediction’ were enabled to generate a library. All other settings were left to default: Digestion protease = Trypsin/P; missed cleavage = 1; max number of variable modifications = 0; N-term M excision = True; C carbamidomethylation = True; Ox(M) methionine oxidation = True; peptide length range = 7–30; precursor charge range = 1–4; precursor m/z range = 300–1800; fragment ion m/z range = 200–1800; Precursor FDR (%) = 1%; Use isotopologues = enabled; MBR = enabled; Neural network classifier = single-pass mode; Protein inference = Genes; Quantification strategy = Any LC (high accuracy); Cross-run normalization = RT-dependent; Library generation = Smart profiling; Speed and RAM usage = Optimal results.
Bioinformatic processing and statistics
The report file generated by DIA-NN was used to acquire the count of unique proteins at a 1% false discovery rate (FDR). Protein quantities were obtained from the unique gene list in DIA-NN (report.unique_genes_matrix.tsv) and implemented in Perseus (v1.6.14.0)54. Data was log2 transformed, group annotated and filtered to include only proteins with at least 70% valid values in each group. Statistical comparisons between groups were made using two-sided Student’s t-test with permutation-based FDR (< 0.05) and 250 randomizations. Given that multiple testing corrections using FDR can be overly restrictive in small n proteomic studies, we used non-adjusted p-values for individual proteins. Volcano plots were generated in GraphPad Prism (v10) by plotting logFC difference and log-transformed p-values from the t-test analysis. Missing values were only imputed for principal component analysis (PCA) (width = 0.4, down shift = 1.8).
For the initial human heart comparison, Supplementary Table 1 from a previous study23 was downloaded. The columns for Gene names and Unique peptides were imported and filtered based on the presence of more than one unique peptide.
Extracellular matrix (ECM) enrichment was achieved by comparing individual regional comparisons with the matrisome homo sapiens database55,56 and selecting overlapping proteins for further analysis. Principal component analysis plots were generated on imputed data using the same parameters as described above. Hierarchical clustering of significantly regulated proteins (p-value < 0.05) was created based on z-scored LFQ values and generated by average linkage, pre-processing with k-means, and Euclidean distance. The z-score normalization was calculated by subtracting the mean intensity from each protein value across all samples followed by division by the standard deviation.
Up and downregulated protein lists were analyzed using STRING app (v2.0.1)57 in Cytoscape (v3.9.1)58. Equus caballus was selected as species in all analyses, except for the last horse/human comparison. Confidence score cutoff was set to 0.4. The STRING enrichment function was used to retrieve functional enrichment with “genome” as the network background. The following ontology reference sets were used; GO Biological Process, GO Cellular Component, GO Molecular Function, KEGG Pathways59, Reactome Pathways. Pathway enrichment data was plotted in GraphPad Prism (v10) using pathway description and -log10(FDR).
We downloaded supplementary data 9 (41467_2017_1747_MOESM11_ESM.xlsx) from a previous study20, and created a new list containing gene names and logFC difference (n = 411 proteins) identified as significant from volcano plot analysis20. For transcriptomic comparison, the supplementary file (1-s2.0-S1547527121019883-mmc1.xls) was downloaded from22. List of genes (IncidentAF_DEG) were loaded into Perseus and filtered based on significant p-value (P.Value_new_onsetAF column; non. adj. p-value < 0.05) and protein-coding genes (by removing NA values in hgnc_symbol column). No filtering was performed at logFC level (logFC_new_onsetAF column) resulting in a list of 1215 genes. In Perseus, the downloaded proteomic and transcriptomic datasets were enriched for ECM proteins by matching gene names with the Matrisome database55,56. Comparison with horse data from our study was made to all proteins identified in the left atria comparison (AF vs. CTR, no p-value filtering).
Data availability
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE60 partner repository with the dataset identifier PXD052818.
Change history
06 October 2025
The original online version of this Article was revised: In the original version of this Article Sarah Dalgas Nissen, Joakim Armstrong Bastrup and Simon Libak Haugaard were omitted as equally contributing authors. The statement has now been added and reads: “Sarah Dalgas Nissen, Joakim Armstrong Bastrup and Simon Libak Haugaard contributed equally to this work.”
References
Iwasaki, Y. K. et al. Atrial fibrillation pathophysiology: implications for management. Circulation 124 (20), 2264–2274 (2011).
Kornej, J. et al. Epidemiology of atrial fibrillation in the 21st century. Circul. Res. 127 (1), 4–20 (2020).
Nath, L. C. et al. Incidence, recurrence, and outcome of postrace atrial fibrillation in thoroughbred horses. J. Vet. Intern. Med. 35 (2), 1111–1120 (2021).
Else, R. W. & Holmes, J. R. Pathological changes in atrial fibrillation in the horse. Equine Vet. J. 3 (2), 56–64 (1971).
Reef, V. B. et al. Recommendations for management of equine athletes with cardiovascular abnormalities. J. Vet. Intern. Med. 28 (3), 749–761 (2014).
Slack, J. et al. Occurrence of cardiac arrhythmias in standardbred racehorses. Equine Vet. J. 47 (4), 398–404 (2015).
Ohmura, H. et al. Risk factors for atrial fibrillation during racing in slow-finishing horses. J. Am. Vet. Med. Assoc. 223 (1), 84–88 (2003).
Decloedt, A. et al. Atrial fibrillation in horses part 1: pathophysiology. Vet. J. 263, 105521 (2020).
van Loon, G. et al. Pacing induced sustained atrial fibrillation in a pony. Can. J. Vet. Res. 64 (4), 254 (2000).
Hesselkilde, E. Z. et al. Longitudinal study of electrical, functional and structural remodelling in an equine model of atrial fibrillation. BMC Cardiovasc. Disord. 19 (1), 228 (2019).
Saljic, A. et al. Increased fibroblast accumulation in the equine heart following persistent atrial fibrillation. IJC Heart Vasculature. 35, 100842 (2021).
Carstensen, H. et al. Long-term training increases atrial fibrillation sustainability in standardbred racehorses. J. Cardiovasc. Transl Res. (2023).
Linz, D. et al. Pulmonary vein firing initiating atrial fibrillation in the horse: oversized dimensions but similar mechanisms. J. Cardiovasc. Electrophys. 31 (5), 1211–1212 (2020).
Lippi, G., Sanchis-Gomar, F. & Cervellin, G. Global epidemiology of atrial fibrillation: an increasing epidemic and public health challenge. Int. J. Stroke. 16 (2), 217–221 (2021).
Ang, Y. S. et al. A new therapeutic framework for atrial fibrillation drug development. Circul. Res. 127 (1), 184–201 (2020).
Kisheva, A. & Yotov, Y. Risk factors for recurrence of atrial fibrillation. Anatol. J. Cardiol. 25 (5), 338–345 (2021).
Buhl, R. et al. Atrial fibrillatory rate as predictor of recurrence of atrial fibrillation in horses treated medically or with electrical cardioversion. Equine Vet. J. (2021).
Nattel, S. Harada, atrial remodeling and atrial fibrillation: recent advances and translational perspectives. J. Am. Coll. Cardiol. 63 (22), 2335–2345 (2014).
Liu, B. et al. Proteomic analysis of atrial appendages revealed the pathophysiological changes of atrial fibrillation. Front. Physiol. 2020, 11 (2020).
Doll, S. et al. Region and cell-type resolved quantitative proteomic map of the human heart. Nat. Commun. 8 (1), 1469 (2017).
Liu, W. et al. Multiple targets related to mitochondrial function unveiled by metabolomics and proteomics profiles of hearts from atrial fibrillation patients. Front. Physiol. 2023, 14 (2023).
van den Berg, N. W. E. et al. PREDICT-AF Investigators. Extracellular matrix remodeling precedes atrial fibrillation: Results of the PREDICT-AF trial. Heart Rhythm, 2021. 18(12):p.2115-2125
Linscheid, N. et al. Quantitative proteome comparison of human hearts with those of model organisms. PLoS Biol. 19 (4), e3001144 (2021).
Muszyński, P. & Bonda, T. A. Mitochondrial dysfunction in atrial fibrillation-mechanisms and Pharmacological interventions. J. Clin. Med. 10, 11 (2021).
Harada, M. et al. Metabolic considerations in atrial fibrillation- mechanistic insights and therapeutic opportunities. Circ. J. 81 (12), 1749–1757 (2017).
Ritterhoff, J. & Tian, R. Metabolic mechanisms in physiological and pathological cardiac hypertrophy: new paradigms and challenges. Nat. Rev. Cardiol. 20 (12), 812–829 (2023).
Barth, A. S. et al. Reprogramming of the human atrial transcriptome in permanent atrial fibrillation. Circul. Res. 96 (9), 1022–1029 (2005).
Verheule, S. et al. Loss of continuity in the thin epicardial layer because of endomysial fibrosis increases the complexity of atrial fibrillatory conduction. Circulation: Arrhythmia Electrophysiol. 6 (1), 202–211 (2013).
Nattel, S., Burstein, B. & Dobrev, D. Atrial remodeling and atrial fibrillation. Circulation: Arrhythmia Electrophysiol. 1 (1), 62–73 (2008).
Poveda, J. et al. MXRA5 is a TGF-β1-regulated human protein with anti-inflammatory and anti-fibrotic properties. J. Cell. Mol. Med. 21 (1), 154–164 (2017).
Liu, L. et al. Potential target genes in the development of atrial fibrillation: a comprehensive bioinformatics analysis. Med. Sci. Monit. 27, e928366 (2021).
Hartl, F. U. Molecular chaperones in cellular protein folding. Nature 381 (6583), 571–580 (1996).
Liu, D. et al. Role of heat shock proteins in atrial fibrillation: from molecular mechanisms to diagnostic and therapeutic opportunities. Cells 12, 1 (2022).
Li, Q. et al. PSMD13 inhibits NF-κB pathway by targeting TAK1 for K63-linked ubiquitination in Miiuy croaker (Miichthys Miiuy). Fish Shellfish Immunol. 138, 108857 (2023).
Brundel, B. J. J. M. et al. Atrial fibrillation. Nat. Rev. Dis. Prim.. 8 (1), 21 (2022).
Kishore, P., Collinet, A. C. T. & Brundel, B. J. J. M. Prevention of atrial fibrillation: putting proteostasis derailment back on track. J. Clin. Med. 12 (13), 4352 (2023).
Leistad, E., Christensen, G. & Ilebekk, A. Atrial contractile performance after cessation of atrial fibrillation. Am. J. Physiol. Heart Circ. Physiol. 264 (1), H104–H109 (1993).
Schwarzwald, C. C., Schober, K. E. & Bonagura, J. D. Echocardiographic evidence of left atrial mechanical dysfunction after conversion of atrial fibrillation to sinus rhythm in 5 horses. J. Vet. Intern. Med. 21 (4), 820–827 (2007).
Thomas, L. et al. Abnormalities of left atrial function after cardioversion: an atrial strain rate study. Heart 93 (1), 89–95 (2007).
Carstensen, H. et al. Time-dependent antiarrhythmic effects of flecainide on induced atrial fibrillation in horses. J. Vet. Intern. Med. 32 (5), 1708–1717 (2018).
Fenner, M. F. et al. Effect of selective ik,ach Inhibition by XAF-1407 in an equine model of tachypacing-induced persistent atrial fibrillation. Br. J. Pharmacol. 177 (16), 3778–3794 (2020).
De Clercq, D. et al. Atrial and ventricular electrical and contractile remodeling and reverse remodeling owing to short-term pacing-induced atrial fibrillation in horses. J. Vet. Intern. Med. 22 (6), 1353–1359 (2008).
Nattel, S. et al. Why translation from basic discoveries to clinical applications is so difficult for atrial fibrillation and possible approaches to improving it. Cardiovasc. Res. 117 (7), 1616–1631 (2021).
Schüttler, D. et al. Animal models of atrial fibrillation. Circul. Res. 127 (1), 91–110 (2020).
Haïssaguerre, M. et al. Spontaneous initiation of atrial fibrillation by ectopic beats originating in the pulmonary veins. N. Engl. J. Med. 339 (10), 659–666 (1998).
Carstensen, H. et al. Long-term training increases atrial fibrillation sustainability in standardbred racehorses. J. Cardiovasc. Transl. Res. 16 (5), 1205–1219 (2023).
Decloedt, A. et al. Right atrial and right ventricular ultrasound-guided biopsy technique in standing horses. Equine Vet. J. 48 (3), 346–351 (2016).
Hesselkilde, E. et al. First catheter-based high-density endocardial 3D electroanatomical mapping of the right atrium in standing horses. Equine Vet. J. 53 (1), 186–193 (2021).
Nissen, S. D. et al. Muscarinic acetylcholine receptors M2 are upregulated in the atrioventricular nodal tract in horses with a high burden of second-degree atrioventricular block. Front. Cardiovasc. Med. 2023, 10 (2023).
Wiśniewski, J. R. et al. Universal sample Preparation method for proteome analysis. Nat. Methods. 6 (5), 359–362 (2009).
Bastrup, J. A., Aalkjær, C. & Jepps, T. A. Identification of novel proteins and mechanistic pathways associated with early-onset hypertension by deep proteomic mapping of resistance arteries. J. Biol. Chem. 298 (1), 101512 (2022).
Bastrup, J. A. & Jepps, T. A. Proteomic mapping reveals dysregulated angiogenesis in the cerebral arteries of rats with early-onset hypertension. J. Biol. Chem. 299 (10), 105221 (2023).
Demichev, V. et al. DIA-NN: neural networks and interference correction enable deep proteome coverage in high throughput. Nat. Methods. 17 (1), 41–44 (2020).
Tyanova, S. et al. The perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods. 13 (9), 731–740 (2016).
Naba, A. et al. The extracellular matrix: tools and insights for the omics era. Matrix Biol. 49, 10–24 (2016).
Naba, A. et al. The matrisome: in Silico definition and in vivo characterization by proteomics of normal and tumor extracellular matrices *. Mol. Cell. Proteom. 11, 4 (2012).
Perez-Riverol, Y. et al. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 47 (D1), D442–D450 (2018).
Shannon, P. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13 (11), 2498–2504 (2003).
Kanehisa, M. & Goto, S. Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28 (1), 27–30 (2000).
Perez-Riverol, Y. et al. The PRIDE database resources in 2022: a hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res. 50 (D1), D543–d552 (2022).
Acknowledgements
We would like to acknowledge the horse owners for generously allowing us to include material from their horses in this study. Furthermore, we extend sincere gratitude to Susanne Nørskov Sørensen for her technical assistance in sample preparation.
Funding
SDN was funded by the Danish Independent Research Foundation (grant number 1032-00053B). JAB was funded by the Lundbeck Foundation (grant number R400-2022-1213). SLH was funded by a research grant from the Danish Cardiovascular Academy, which is funded by the Novo Nordisk Foundation (grant number NNF20SA0067242) and the Danish Heart Foundation. MJS was funded by a research grant from the Danish Cardiovascular Academy, which is funded by the Novo Nordisk Foundation (grant number PhD 2022002-HF) and the Danish Heart Foundation. SN is funded by the Canadian Institutes of Health Research and the Heart and Stroke Foundation of Canada.
Author information
Authors and Affiliations
Contributions
All authors discussed the results and contributed to the final manuscript. SDN conceived and designed the study, executed the collection of samples, stainings, sample preparation, data analysis, and interpretation, drafted the main manuscript, and prepared the figures. JAB contributed to the study conception, executed sample preparation, conducted formal data analysis and interpretation, prepared the figures and drafted the main manuscript. SLH contributed to the study conception, executed the collection of samples, performed data analysis and interpretation, and drafted the main manuscript. RMK was responsible for histopathological stainings and analysis. MS and STK executed the collection of samples and contributed to data interpretation. HC, CHI, and SN contributed to the study conception and data interpretation. TAJ and RB conceived and designed the study, contributed to data interpretation, and provided supervision and project administration.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Nissen, S.D., Bastrup, J.A., Haugaard, S.L. et al. Horse model of spontaneous atrial fibrillation share proteomic changes with humans. Sci Rep 15, 31694 (2025). https://doi.org/10.1038/s41598-025-16885-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-025-16885-7