Abstract
Ubiquitin conjugating enzyme UBE2S is an important enzyme in ubiquitin mediated proteolysis. The mechanisms of UBE2S in the colorectal cancer (CRC) proliferation need further exploration for new ubiquitination-related drugs exploitation. Firstly, we performed immunohistochemistry of UBE2S protein on CRC and adjacent tissues and found that UBE2S protein expressed higher in adjacent tissues than in tumor. TCGA and GTEx databases revealed a positive correlation between UBE2S expression and overall survival rate in CRC (P < 0.05). Cell proliferation experiments on HCT116 and RKO cell lines invalidated that UBE2S overexpression inhibited CRC cell proliferation and progression from G0/1 phase to G2 phase while knocking down UBE2S enhanced it. Mechanistically, combined with enrichment analysis based on RNA sequencing, it was identified molecularly that UBE2S simultaneously inhibited the activation of MAPK/ERK and PI3K/AKT pathways through promoting the ubiquitin-mediated degradation of TRAF6 protein, an upstream molecule to activate the both pathways, which can be blocked by inhibitor MG132. Animal experiments demonstrated that knocking down UBE2S resulted in larger transplanted tumor volumes than the control group, while overexpressing UBE2S resulted in the smaller ones in vivo. Summarily, we firstly identified that the UBE2S inhibited CRC proliferation by negatively regulating MAPK/ERK and PI3K/AKT/mTOR pathways through ubiquitinating TRAF6 protein.
Similar content being viewed by others
Colorectal cancer (CRC), as one of the most frequent malignancies worldwide, ranks third in incidence rate and second in mortality rate1. Therefore, there are increasing researches on the mechanisms of CRC progression. As a common post-translational modification process, the ubiquitination exerted a significant effect on various tumors including CRC, gastric cancer, lung cancer and so on. Increasing evidence identified that ubiquitination can cause target protein degradation in eukaryotic cells to regulate intracellular protein levels and maintaining intracellular homeostasis2,3which played important roles in tumor metastasis, cell cycle, and proliferation4,5,6. As an important member of the conjugating enzyme E2, UBE2S have been reported to take part in the progression of multiple tumors. UBE2S is a K11 chain specific E2 enzyme associated with human APC/C, which plays an important role in both mitosis and meiosis, thereby regulating the cell cycle and proliferation7,8. Though UBE2S was reported to play a promoting role in some tumors, there was still controversy over the regulation of UBE2S in the occurrence and development of CRC. In view of this, we further explore the potential mechanisms of UBE2S on the CRC proliferation.
During recent years, researchers revealed the abnormal expression of UBE2S in a variety of tumors, including breast cancer, ovarian cancer, lung cancer, colorectal cancer and so on9. UBE2S can affect biological processes such as cell cycle, DNA damage repair, tumor progression, and sensitivity to anti-tumor drugs by regulating the level of protein ubiquitination modification. Gong et al. found that UBE2S targeted RPL26 for ubiquitination and degradation, thereby promoting the progression of non-small cell lung cancer by regulating c-Myc10. It was also reported that UBE2S promoted ovarian cancer development by regulating the PI3K/AKT/mTOR signaling pathway to regulate the cell cycle and apoptosis8. Zhang et al. reported that UBE2S inhibited autophagy by activating the PI3K/AKT/mTOR signaling axis, affecting the resistance of ovarian cancer to cisplatin11. Meanwhile, studies have reported that UBE2S can induce K11 specific ubiquitination and degradation of several substrate proteins, thereby participating in various cellular processes and catalyzing the conversion of ubiquitin to substrates for protein degradation through E3 dependent or E3 independent mechanisms12. However, the molecular mechanisms regulated by UBE2S remain poorly defined. Further fundamental research is needed to elucidate its signaling regulatory axis, which is crucial for understanding its therapeutic potential and advancing clinical translation..
The activation of the PI3K/Akt pathway regulates the expression of downstream substrates, including apoptosis related proteins Bad and Caspase 9, thereby regulating various cell phenotypes, including proliferation, differentiation, apoptosis, and migration13. The PI3K/Akt pathway can also regulate mammalian rapamycin targets (mTOR), and the mTOR pathway is an intracellular signaling pathway activated by extracellular signals, promoting various biological processes such as angiogenesis, cell growth, cell proliferation, and cell metabolism via downstream transcription factors FoxO, c-Myc, and HIF1 α14. The transcription factor FOXM1 up-regulated UBE2S expression in liver cancer cells for ubiquitination of PTEN at Lys327 and Lys60 sites and AKT phosphorylation, thereby enhancing chemotherapy resistance15. The malignant phenotype of hepatocellular carcinoma may be promoted by the FOXM1/UBE2S/PTEN/p-AKT axis, indicating that this disease can be treated by targeting UBE2S. Multiple studies have reported that activation of the MAPK pathway can promote the progression of various cancers, including gastrointestinal cancer, hepatocellular carcinoma, cancer and so on16,17,18. Extracellular Signal Regulated Kinase (ERK) is an important molecule in the MAPK pathway. ERK is highly activated and plays a role in various malignant tumors through phosphorylation, promoting cell proliferation, inhibiting apoptosis, and regulating the cell cycle, hence participating in the occurrence and development of tumors. For instance, the STAT5B/ARRB2 pathway mediated by PRL promoted the progression of prostate cancer by activating MAPK signaling19.
TRAF6 is a pivotal signal transduction protein that participates in regulating various biological processes. In tumors, TRAF6 can regulate multiple signaling pathways, such as MAPK/ERK and PI3K/AKT, and therefore affect the proliferation, differentiation, apoptosis, and function of immune cells, and participate in regulating immune responses20,21. It was reported that TRAF6 was upregulated in colon cancer and promoted the proliferation of colon cancer cells22. In the PI3K/Akt signaling pathway, TRAF6 can interact with PI3K (phosphatidylinositol 3-kinase), activate AKT protein kinase, and promote prostate cancer metastasis23.
In this study, we will conduct RNA sequencing, animal experiments, immunohistochemistry, Co-IP experiments, mass spectrum, and ubiquitination inhibition experiments to explore the mechanisms of UBE2S in CRC proliferation in vivo and in vitro, which may provide new potential therapeutic targets for the treatment of CRC.
Materials and methods
Cell culture and reagents
CRC cells HCT116 and RKO used in this investigation were sourced from The Cell Bank of the Chinese Academy of Sciences and stored at the Shanghai Institute of Digestive Surgery, cultured in RPMI-1640 medium supplemented with 10% fetal bovine serum (Gibco, USA) and 5 µg/ml penicillin-streptomycin in a Humidified incubator at 37 °C with 5% CO2. Blasticidin (5mM, SELLECK, USA), MG132 (10mM, SELLECK, USA), U0126 (2.5mM, SELLECK, USA), cycloheximide (CHX, 2 mg/ml, SELLECK, USA), Rapamycin (10mM, MCE, USA) and Ipataserib (5mM, SELLECK, USA) were stored in a −20℃ refrigerator. The final concentrations used in the experiments were as follows: 10 µg/ml CHX (cycloheximide), 5 µM U0126 (MAPK inhibitor), 5 µM MG132 (proteasome inhibitor), 5 µM Ipatasertib (AKT inhibitor), 10 µM Rapamycin (mTOR inhibitor) and 10 µM Blasticidin (antibiotic).
Patients and samples
The samples of CRC and adjacent tissues used in the investigations were obtained from the patients of the Shanghai Ruijin Hospital, Shanghai Jiao Tong University school of medicine. Histopathological diagnosis was consistent with World Health Organization criteria by hematoxylin and eosin staining. All samples were obtained with informed consent. We obtained ethical approval from the Ruijin Hospital, Shanghai Jiao Tong University school of medicine. All the methods were performed in accordance with the relevant guidelines and regulations.
Lentiviral constructs and cell transfection
To knock down UBE2S, the shRNA sequences were subcloned into H_UBE2S-shRNA596 (PGMLV-mScarlet-Blasticidin) (Genomeditech, Shanghai, China). The shRNA of UBE2S targeted the following sequence: 5′- ATGCTGGCGAGCGCGATAAGA − 3′. Human full-length cDNA of UBE2S were subcloned into PGMLV-CMV-H_UBE2S-3×Flag-EF1-mScarlet-T2A-Blasticidin (Genomeditech, Shanghai, China) for UBE2S overexpression. Then 15 µl SH-UBE2S or OE-UBE2S and corresponding control lentiviruses containing blasticidin resistance were added to culture dishes of HCT116 and RKO adherent cells. After cultured in the RPMI-1640 medium with 10% inactivated FBS for 48 h, 10µM of blasticidin was added. Then we continued to incubate for 48 h and washing it with PBS to remove the non-transfected cells. Under a fluorescence microscope, HCT116 and RKO cell lines were observed to stably express red fluorescence protein. Finally, we constructed HCT116 and RKO cells as group UBE2S-NC, UBE2S-SH, UBE2S-Vector, and UBE2S-OE. After further screening with and blasticidin (10µM), amplification was then carried out for subsequent experiments.
RNA sequencing and data analysis
RKO cells with UBE2S knockdown and overexpressing were used to perform RNA sequencing. Proper RKO cells were planted in 10 cm dish with RPMI-1640 medium and 10% FBS. Three repeated dishes were set for each group. After incubation for 48 h, cells were collected into 1.5 ml tube with 1 ml Trizol (Invitrogen, USA) for RNA extraction. RNA purification was checked by NanoPhotometer® spectrophotometer (IMPLEN, CA, USA), and RNA integrity was evaluated by Bioanalyzer 2100 system (Agilents, CA, USA). After the RNA quality control, sequencing libraries were generated using NEBNext® UltraTM RNA Library Kit for Illumina® (NEB, USA). RNA sequencing was performed on Illumina Novaseq6000 platform (Jiayin Biomedical Technology Co., Ltd., Shanghai, China). Indexes clustering was performed on cBot Cluster System using TruSeq PE Cluster Kit v3-cBot-HS (Illumia). HTSeq v0.6.0 was used to count the reads mapped to each gene, and then the FPKM (Fragments Per Kilobase per Millions base pairs) of each gene was calculated based on the length of each gene and reads counts. The detailed procedures of raw data acquirement were as described previously24.Then, the DESeq2 algorithm was used to filter the expression of differentially expressed genes (DEGs). Enrichment analysis of DEGs (at the standard |log2 (fold change)| >1 and P<0.05) between UBE2S-SH and UBE2S -NC or UBE2S-OE and UBE2S -Vector groups were performed, including signaling pathway analysis.
Co-immunoprecipitation (Co-IP) assay, Immunoprecipitation (IP) assay and mass spectrometry (MS)
Co-IP assay with the Kit (#88804, Thermo Fisher Scientific) was used to verify the physical interactions between UBE2S and TRAF6, and the IP assay for TRAF6 protein was used to detect the ubiquitination level of TRAF6. Cell lysates were incubated with the primary antibodies of TRAF6 at 4 °C overnight. After the immune complex was incubated with A/G magnetic beads for 6 h, the magnetic beads were washed twice with IP buffer, followed by washing the antigen/antibody complex once with pure water. Finally, the products of IP were boiled for 15 min. The protein from IP was analyzed by western blotting and the protein from Co-IP was analyzed and mass spectrometry (Thermo Fisher Scientific, MA, USA).
Quantitative real-time PCR (qRT-PCR)
TRIzol solution (Invitrogen, USA) was used to extract total RNA according to the manufacturer’s protocols and concentration were measured by multi-wavelength measurement system (Infinite 200pro, Tecan). Proper RNA was reversely transcribed to cDNA using HiScript RT Mix (Vazyme, Jiangsu, China). And then, qRT-PCR was conducted using the SYBR Premix Ex Taq Kit (TaKaRa Biotechnology, Dalian, China). 10 µl reaction mixture was used and analyzed on HT 7900 (Applied Biosystems, Foster City, USA). The detailed procedures were as described previously24. All reactions were run in triplicates and normalized to GAPDH by 2−ΔΔ CT method. The relevant primers for qRT-PCR are shown in Table S1.
Western blotting
Cell lysates were separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), electroblotted onto polyvinylidene fluoride (PVDF) membranes. The membranes were incubated with respective specific primary antibodies overnight at 4 °C and then corresponding secondary antibodies at room temperature for 2 h. The main primary antibodies included UBE2S (1:1000, 11415-1-AP, Proteintech, China), TRAF6 (1:1000, ab137452, abcam, UK), ERK1/2 (1:1000, 4695, CST, USA), Phospho-ERK1/2 (1:1000, 4370, CST), AKT(1:1000, 4691, CST), Phospho-AKT(Thr308, 1:1000, 13038, CST), Phospho-AKT(Ser473, 1:1000, 4060, CST), mTOR(1:1000, 2983, CST), Phospho-mTOR(Ser2448, p-mTORC1, 1:1000, 5536 and 2976, CST), Phospho-mTOR(Ser2481, p-mTORC2, 1:1000, 2974, CST), Phospho-AP-1(p-c-Jun, 1:1000, 9261, CST), AP-1(c-Jun, 1:1000, 9165, CST), Smad2(1:1000, 5339, CST), Phospho-Smad2(1:1000, 3108, CST), TAK1(1:1000, 4505, CST), Phospho-TAK1(1:1000, 9339, CST), HRP-conjugated GAPDH (1:1000, 3683, CST), K63-linkage Specific Polyubiquitin(1:1000, 5621, CST) and Ubiquitin (1:1000, 58395, CST). The secondary antibodies were respectively anti-mouse and anti-rabbit HRP-conjugated ones.
CCK-8 assay
To examine cell proliferation, HCT116 and RKO cells (3 × 103/well) were seeded into 96-well plate and continued to incubate for 24 h, 48 h and 72 h respectively to measure. In the rescue experiments, we changed corresponding mediums with inhibitors on the second day after cell seeding. The 100-ml solution of 10% CCK8 was added for 2 h, and OD values at 450 nm were measured by a spectrophotometry (BioTek, Vermont, USA). The detailed procedures were as described previously24.
Colony formation assay
In the colony formation assays, HCT116 and RKO cells (2 × 103/well) were seeded to six-well plates, and incubated with 3 ml mediums with 10% FBS for 7 days. And then, the mediums were changed and U0126 (5µM), Ipataserib (5µM), Rapamycin (10µM) or MG132 (5µM) were added respectively. The cells were incubated for another 7 days. On the 14th day, the colony formation formation units were fixed and dyed with 1% crystalline purple diluted in methanol for 20 min. After the six-well plates were washed gently with clean water, the triplicates of colony formation units were photographed and counted. The detailed procedures were as described previously24.
Tumor growth model on nude mice
To observe the CRC growth and related protein expression, five-week-old female nude mice (BALB/C nude, Beijing Vitolihua Experimental Animal Technology Co., Ltd.) were selected and used in all the in vivo experiments. HCT116 cells (5 × 106/mouse) with UBE2S knockdown and overexpression were injected into the right flank of female nude mice. After 24 days, all mice were anaesthetized and sacrificed with CO2 and the xenografts were dissected. The tumors were measured and calculated using the formula (volume = length ×width ×width/2). The tumors were fixed with 4% paraformaldehyde to make 4 μm-thick slices for further observations. All experiments complied with the ARRIVE. All animal experiments in this study were approved by the Guide for the Care and Use Laboratory Animals of the Ruijin Hospital, Shanghai Jiao Tong University School of Medicine.
Immunohistochemistry and Immunofluorescence
The 4 μm thick slices were used to perform IHC staining by streptavidin-peroxidase method as described in previous research25. Rabbit monoclonal antibody anti-UBE2S, anti-TRAF6, anti-p-ERK1/2, anti-AKT and anti-mTOR was HRP-labeled goat anti-rabbit antibody (1: 200, Servicebio, GB23305, China) were used. All photographs were taken under the Nikon Digital Sight DS-U2 (Nikon, Tokyo, Japan) at 400× field. For the immunofluorescence, 4 μm thick slices were co-incubated with UBE2S (1:200, 11415-1-AP, Proteintech, China) and TRAF6 (1:200, ab137452, abcam, UK) overnight, followed by secondary antibodies. Detailed procedures were as described previously26. Slides were stained with DAPI/mounting medium (ThermoFisher, USA) and examined by fluorescence microscopy.
SiRNA transfection
The siRNA for TRAF6 was transfected into HCT116 and RKO cells using Lipo 2000 reagent at the concentration of 100 nmol/L according to the manufacturers’ protocol. After cells were transfected with siRNA for 6 h, mediums were changed. In 48 h, transfected cells were collected for verification by qPCR and western blot and used for relevant experiments. The sequences of siRNAs for TRAF6 are shown in Table S2.
Cell cycle analysis by flow cytometry
The collected cells were fixed in 75% ethanol overnight, washed for three times with PBS, and then stained with PI/RNase Staining Buffer (BD Biosciences) for 30 min. The detailed procedures were as described previously25. Finally, the cell cycle was analyzed by flow cytometry using the FACSCalibur system (BD Biosciences, United States).
Statistical analysis
All data were performed using GraphPad Prism 8.0 (Inc., La Jolla, CA, USA) and shown as the mean ± standard deviation (SD). Student’s t-test and Mann-Whitney U test were used to analyze the data. Two-tailed p values smaller than 0.05 were considered statistically significant.
Results
Low expression of UBE2S in colorectal cancer enhances its proliferation
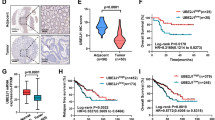
We firstly used the online database TCGA and analysis website GEPIA (http://gepia.cancer-pku.cn). By conducting survival analysis of clinical information corresponding to transcriptome, we found that high expression of UBE2S was associated with a higher overall survival rate in colorectal cancer patients, showing a protective effect (Fig. 1A, P < 0.05). To explore the expression of UBE2S in colorectal cancer, we performed immunohistochemistry on 6 pairs of colorectal cancer patients and adjacent tissues. The experimental results showed that the expression of UBE2S protein was lower in colorectal cancer than in adjacent tissues (Figs. 1B and 6.7 ± 1.1 vs. 2.7 ± 0.8, P < 0.01). To explore the potential biological effects of UBE2S on colorectal cancer, we successfully constructed stable expression cell lines of HCT116 and RKO with UBE2S knockdown and overexpression using lentiviruses, and validated them using qPCR and Western blotting techniques (Fig. 1C and D). In CCK8 cell proliferation experiments on cell lines HCT116 and RKO, knocking down UBE2S enhanced cell proliferation ability (Fig. 1E, 72 h, HCT116 0.93 ± 0.03 vs. 1.61 ± 0.05, P < 0.01; RKO 1.01 ± 0.05 1.81 ± 0.06, P < 0.001), while overexpression of UBE2S weakened cell proliferation ability (Fig. 1E, 72 h, HCT116 1.16 ± 0.01 vs. 0.74 ± 0.01, P < 0.001; RKO 1.26 ± 0.04 0.82 ± 0.02, P < 0.001); Cloning experiments were conducted in cell lines HCT116 and RKO. Knocking down UBE2S increased the number of colony formation units (Fig. 1F, HCT116 374.0 ± 11.5 vs. 767.3 ± 18.0, P < 0.01; RKO 162.0 ± 11.0 vs. 275.3 ± 13.3, P < 0.01), while overexpression of UBE2S reduced the number of colony formation units (Fig. 1F, HCT116 399.3 ± 13.6 vs. 237.0 ± 6.5, P < 0.01; RKO 182.3 ± 8.7 vs. 85.3 ± 6.5, P < 0.05). Flow cytometry experiments were conducted on cell lines HCT116 and RKO, and overall, knocking down UBE2S resulted in a decrease in the proportion of G0/1 cells and an increase in the proportion of S and G2 phase cells. However, overexpression of UBE2S resulted in an increase in the proportion of G0/1 cells and a decrease in the proportion of S and G2 phase cells (Fig. 1G, P < 0.05).
Low expression of UBE2S in colorectal cancer enhances its proliferation. (A) High expression of UBE2S is positively correlated with overall survival rate in CRC patients; (B) UBE2S protein expression is lower in CRC tissues than in adjacent tissues (n = 6, 400×); (C) and (D) qPCR and protein analysis of stable cell line HCT116 and RKO with UBE2S knockdown or overexpression; (E) CCK8 experiments on HCT116 and RKO cell lines (n = 3); (F) Colony formation assays on HCT116 and RKO cell lines (n = 3); (G) Flow cytometry experiments are conducted on HCT116 and RKO cell lines (n = 3). *P < 0.05, **P < 0.01, ***P < 0.001, ‘ns’ represents no statistical significance.
UBE2S affects the activity of MAPK and PI3K/AKT pathways by regulating TRAF6 protein
Based on the above functional experimental results, to further explore the molecular mechanism of UBE2S in the proliferation of colorectal cancer, we knocked down UBE2S on RKO cells and performed RNA sequencing and differential gene enrichment analysis. It was found that UBE2S can significantly affect the activity of both MAPK and PI3K/AKT pathways simultaneously (Fig. 2A). Next, we performed protein analysis on stable cell lines HCT116 and RKO with UBE2S knockdown and overexpression. The results showed that after UBE2S knockdown, TRAF6, p-AKT, and p-ERK proteins were all increased, while overexpression of UBE2S resulted in a decrease in TRAF6, p-AKT, and p-ERK proteins (Fig. 2B and C, P < 0.05); To further explore the effects of UBE2S on TRAF6 at RNA level, qRT-PCR experiments were conducted. Results indicated that there was no difference after both UBE2S knockdown or overexpression (Fig. 2D). Based on the results obtained from the cell model, we performed immunohistochemical validation on the cancer and adjacent tissues of colorectal cancer patients. The results showed that the expression of UBE2S protein in tumor decreased compared to adjacent tissues, while TRAF6, p-AKT(at Thr308, instead of Ser473), and p-ERK proteins increased in tumor (Fig. 2E, Figure S1A and S1B, UBE2S 6.0 ± 0.3 vs. 2.7 ± 0.4, P < 0.001; TRAF6 2.1 ± 0.3 vs. 4.9 ± 0.4, P < 0.05; p-AKT 3.9 ± 0.2 vs. 7.6 ± 0.2, P < 0.01; p-ERK 3.5 ± 0.3 vs. 6.6 ± 0.2, P < 0.01). Besides, some other downstream proteins of TRAF6 were affected by UBE2S such as TAK1, Smad2 and AP1(Figure S1C). In the colony formation assays, knocking down UBE2S resulted in an increase in the number of colony formation units (Fig. 2G, HCT116 274.0 ± 14.7 vs. 650.0 ± 9.9, P < 0.0001; RKO 190.0 ± 13.5 vs. 317.0 ± 17.0, P < 0.01), while the number decreased after using MAPK inhibitor U0126 and PI3K/AKT inhibitor Ipataserib respectively (Fig. 2E, U0126 650.0 ± 9.9 vs. 369.7 ± 10.3, P < 0.0001; 317.0 ± 17.0 vs. 250.7 ± 9.7, P < 0.05; Ipataserib 650.0 ± 9.9 vs. 391.7 ± 14.1, P < 0.01; 317.0 ± 17.0 vs. 223.7 ± 10.0, P < 0.05), and the number of colony formation units was lower after using both inhibitors simultaneously (Fig. 2E, HCT116 650.0 ± 9.9 vs. 215.0 ± 15.6, P < 0.0001; 317.0 ± 17.0 vs. 152.0 ± 8.6, P < 0.05). Similarly, in our CCK8 cell proliferation experiment, knocking down UBE2S can enhance cell proliferation ability. The use of MAPK inhibitor U0126 and PI3K/AKT inhibitor Ipataserib, respectively, weakened cell proliferation ability, while the use of both inhibitors resulted in weaker cell proliferation ability (Fig. 2F, P < 0.05).
UBE2S affects the expression of TRAF6 protein and the activity of MAPK and PI3K/AKT pathways. (A) RNA sequencing and differentially expression gene enrichment analysis on RKO cells with UBE2S knockdown; (B) Protein analysis with UBE2S knockdown or overexpression; (C) Statistical analysis of UBE2S, TRAF6, p-AKT, and p-ERK protein expression in colorectal cancer and adjacent tissues (n = 3); (D) Statistical analysis of UBE2S and TRAF6 mRNA expression (n = 3); (E) Immunohistochemical quantification scores for colorectal cancer and adjacent tissues (n = 3, 400×); (F) CCK8 proliferation experiment with UBE2S knockdown, adding MAPK inhibitor U0126, PI3K/AKT inhibitor Ipataserib or both inhibitors together (n = 3); (G) Colony formation assays with UBE2S knockdown, adding MAPK inhibitor U0126, PI3K/AKT inhibitor Ipataserib or both inhibitors together (n = 3). ‘*’ represents comparison with adjacent cancer tissues or ‘NC’ group, and ‘#’ represents comparison with the ‘SH’ group. * P < 0.05, **P < 0.01, ***P < 0.001;#P < 0.05, ##P < 0.01, ###P < 0.001, #### P < 0.0001.
UBE2S affects transplanted tumor growth via the TRAF6/MAPK and TRAF6/PI3K/AKT pathways in vivo
To further explore the effects of UBE2S in vivo, we conducted subcutaneous tumor transplantation animal experiments using HCT116 stable cell lines. The experimental results showed that the volume of transplanted tumors in nude mice in the UBE2S knockdown group was larger than that in the control group (Fig. 3A, the control group was 0.77 ± 0.29 cm3 vs. 1.40 ± 0.33 cm3, P < 0.001), while the volume of transplanted tumors in nude mice in the UBE2S overexpression group was smaller than that in the control group (Fig. 3B, the control group was 0.95 ± 0.19 cm3 vs. 0.17 ± 0.08 cm3, P < 0.001). The growth records of transplanted tumors demonstrated that there were significant differences between the knockdown and control group or overexpression and control group (Fig. 3C, P < 0.01). We further analyzed the RNA of transplanted tumor tissue and found that UBE2S knockdown or overexpression had no effect on TRAF6 at the transcriptional level (Fig. 3D). However, we found that the expression of TRAF6 protein significantly increased after UBE2S knockdown, and decreased after overexpression of UBE2S in the transplanted tumor tissue protein analysis (Fig. 3E, P < 0.01). After immunofluorescence analysis of the transplanted tumor tissue samples, it was found that UBE2S regulated the expression of TRAF6 at protein level (Fig. 3F and G, P < 0.01), which were consistent with the previous results of in vitro cell experiments. In immunohistochemistry of nude mouse transplanted tumors, knocking down UBE2S increased the expression of TRAF6, p-AKT and p-ERK proteins (Fig. 3H, P < 0.05), while overexpression of UBE2S reduced the expression of TRAF6, p-AKT, and p-ERK proteins (Fig. 3I, P < 0.05).
The effect of UBE2S on the expression level of TRAF6 and the activity of MAPK and PI3K/AKT pathways in vivo. (A) Animal experiments demonstrate larger volumes of the transplanted tumor with UBE2S knockdown (n = 5); (B) Overexpression of UBE2S reduces the volumes of the transplanted tumor (n = 5); (C) Volume measurement in the growth of transplanted tumors in nude mice(n = 5); (D) RNA analysis of transplanted tumor tissue reveals that both UBE2S knockdown and overexpression have no effects on the transcription level of TRAF6 (n = 5); (E) Protein analysis of transplanted tumor tissues reveals an impact of UBE2S on TRAF6 protein (n = 5); (F) Immunofluorescence of transplanted tumors for the UBE2S and TRAF6 protein expression ((n = 5, 400×); (G) Statistical analysis of immunohistochemistry of transplanted tumors for UBE2S and TRAF6 proteins with UBE2S knockdown and overexpression; (H) In immunohistochemistry of nude mouse transplanted tumors, UBE2S knockdown results in an increase in the expression of TRAF6, p-AKT, and p-ERK proteins (n = 5, 400×); (I) In immunohistochemistry of nude mouse transplanted tumors, UBE2S overexpression results in a decrease in the expression of TRAF6, p-AKT, and p-ERK proteins(n = 5, 400×). *P < 0.05, **P < 0.01, ***P < 0.001, ‘ns’ represents no statistical significance.
UBE2S regulates the activation of mTOR through the PI3K/AKT pathway
To further investigate the molecular mechanism by which UBE2S affects the proliferation of colorectal cancer, we also performed RNA sequencing and differential gene enrichment analysis after overexpressing UBE2S on RKO cells. In addition to the top MAPK and PI3K/AKT pathways, UBE2S also affects the activity of the mTOR pathway in the enriched pathways (Fig. 4A). Next, we overlapped the 240 pathways enriched by overexpressing UBE2S (counts ≥ 5) with the 12 pathways enriched by knocking down UBE2S, and thus got three pathways: MAPK, PI3K/AKT, and mTOR (Fig. 4B). Next, through immunohistochemical analysis of cancer and adjacent tissue sections in colorectal cancer patients, we firstly found that the expression of UBE2S protein decreased compared to adjacent tissues (Fig. 4C, P < 0.001), while the expression of p-mTOR protein increased (Fig. 4C, p-mTOR: adjacent tissues 2.4 ± 0.2 vs. cancer tissues 5.6 ± 0.3, P < 0.05). In HCT116 and RKO cell lines, knocking down UBE2S resulted in an increase in p-mTORC1 protein (but not p-mTORC2, and later abbreviated as p-mTOR), while overexpression of UBE2S resulted in a decrease in p-mTOR protein (Fig. 4D and Figure S1B, P < 0.01). In the colony formation assays, knocking down UBE2S resulted in an increase in the number of colony formation units (Fig. 4E, control group compared to knocking down group, HCT116 125.0 ± 8.6 vs. 222.7 ± 11.1, P < 0.05; RKO 100.7 ± 10.5 vs. 265.3 ± 10.7, P < 0.01), but it could be reversed by the mTORC1 inhibitor rapamycin (Fig. 4E, knockdown group compared to rapamycin group, HCT116 222.7 ± 11.1 vs. 164.7 ± 5.8, P < 0.05; RKO 265.3 ± 10.7 vs. 184.0 ± 7.9, P < 0.01). In the CCK8 proliferation experiment, the enhanced cell proliferation ability after knocking down UBE2S (Fig. 4F, control group vs. knocking down group, HCT116 0.78 ± 0.03 vs. 1.56 ± 0.04, P < 0.001; RKO 0.87 ± 0.02 vs. 1.97 ± 0,01, P < 0.001) was reversed by the mTORC1 inhibitor rapamycin (Fig. 4F, knocking down group vs. rapamycin group, HCT116 1.56 ± 0.04 vs. 1.08 ± 0.04, P < 0.01; RKO 1.97 ± 0.01 vs. 1.02 ± 0.01, P < 0.01). In protein detection, knocking down UBE2S resulted in an increase in both p-AKT and p-mTOR proteins. Further using the PI3K/AKT inhibitor Ipataserib resulted in a consistent decrease in p-AKT and p-mTOR protein (Fig. 4G).
UBE2S affects mTOR expression through PI3K/AKT pathway. (A) RNA sequencing and DEGs enrichment analysis on RKO cells with UBE2S overexpression; (B) Overlapped pathways (counts ≥ 5) between overexpression and knockdown of UBE2S are MAPK, PI3K/AKT, and mTOR signaling pathways; (C) Immunohistochemistry of colorectal cancer and adjacent tissues for UBE2S and p-mTOR protein (n = 3, 400×); (D) In HCT116 and RKO cell lines, knocking down UBE2S results in an increase in p-mTOR protein, while overexpressing UBE2S results in a decrease in mTOR protein (n = 3); (E) Colony formation assays with UBE2S knockdown and further mTOR inhibitor rapamycin (n = 3); (F) CCK8 proliferation experiments with UBE2S knockdown and further mTOR inhibitor rapamycin (n = 3); (G) Knocking down UBE2S results in an increase in both p-AKT and mTOR proteins, while Ipataserib results in a decrease in both p-AKT and p-mTOR proteins. ‘*’ represents comparison with adjacent cancer tissues or the ‘NC’ group, and ‘#’ represents comparison with the “SH” group. *P < 0.05, **P < 0.01, ***P < 0.001;##P < 0.01.
Inhibition on colorectal cancer proliferation by overexpression of UBE2S can be reversed by MG132
All the above data suggested that UBE2S may regulate TRAF6 expression at post-transcription level. To test this hypothesis, we performed a cycloheximide (CHX) chase assay to determine TRAF6 half-life in CRC cells of UBE2S overexpression and control groups. As shown in Fig. 5A, UBE2S overexpression could significantly shortened TRAF6 half-life, which suggested that UBE2S regulated TRAF6 stability in CRC cells. As known to us, ubiquitination is an important and prevalent protein modification that regulates protein levels. Therefore, the ubiquitination of TRAF6 CRC cells was examined. The results of IP experiments demonstrated that overexpression of UBE2S enhanced the ubiquitination of TRAF6 protein (Fig. 5B), which was much more obvious in the presence of the proteasome inhibitor MG132 (Fig. 5B). TRAF6 polyubiquitination enhanced by UBE2S overexpression suggested that TRAF6 proteins may be degraded by the ubiquitin/proteosome pathway in CRC cells. It’s reported that UBE2S can induce K11 specific ubiquitination and degradation of several substrate proteins through E3 dependent or E3 independent mechanisms. Our experiments verified it (Figure S1D and S1E). Therefore, Co-IP and MS assays were performed to validate the interaction between UBE2S and TRAF6 proteins, and the results indicated that there indeed existed an interaction between the both proteins (Table S3). Besides, UBE2S overexpressed in HCT116 and RKO cells resulted in a decrease in TRAF6 protein expression and a corresponding decrease in p-AKT and p-ERK proteins (Fig. 5C). Further use of the ubiquitination inhibitor MG132 resulted in an increase in TRAF6, and a corresponding increase in p-AKT and p-ERK proteins (Fig. 5C). In the CCK8 proliferation experiment, overexpression of UBE2S inhibited cell proliferation (Fig. 5D, 72 h, control group vs. overexpression group, HCT116 2.33 ± 0.04 vs. 1.02 ± 0.03, P < 0.0001; RKO 1.91 ± 0.04 vs. 1.04 ± 0,02, P < 0.01), which could be reversed by MG132 (Fig. 5D, overexpression group vs. MG132 group, HCT116 1.02 ± 0.03 vs. 1.47 ± 0.01, P < 0.01; RKO 1.04 ± 0,02 vs. 1.36 ± 0.02, P < 0.01). In the colony formation assays, overexpression of UBE2S increased the number of colony formation units (Fig. 5E, control group compared to overexpression group, HCT116 438.3 ± 10.3 vs. 235.7 ± 8.2, P < 0.001; RKO 314.0 ± 8.0 vs. 159.7 ± 10.7, P < 0.01), but could be reversed by MG132 (Fig. 5E, overexpression group compared to MG132 group, HCT116 235.7 ± 8.2 vs. 332.3 ± 11.0, P < 0.01; RKO 159.7 ± 10.7 vs. 223.7 ± 13.7, P < 0.05). In immunohistochemistry of transplanted tumors, both p-AKT and mTOR proteins were elevated in tissues with UBE2S knockdown, while p-AKT and mTOR proteins were reduced in tissues with UBE2S overexpression (Fig. 5F, P < 0.05).
Overexpression of UBE2S inhibits CRC proliferation and can be restored by the ubiquitination inhibitor MG132. (A) Cycloheximide (CHX) chase assay to determine TRAF6 half-life in CRC cells of UBE2S overexpression and control groups (n = 3); (B) The results of IP experiments for the detection of TRAF6 protein ubiquitination. (C) After overexpression of UBE2S, TRAF6 protein decreases, and the corresponding p-ERK and p-AKT proteins decreases. Further use of MG132 results in an increase in TRAF6, p-AKT, and mTOR proteins; (D) CCK8 proliferation experiments with UBE2S overexpression and further ubiquitination inhibitor MG132 (n = 3); (E) Colony formation assays with UBE2S overexpression and further ubiquitination inhibitor MG132 (n = 3); (F) Immunohistochemistry of transplanted tumors for UBE2S, p-AKT and mTOR proteins (n = 3). ‘*’ represents comparison with the ‘NC’ group, and ‘#’ represents comparison with the ‘SH’ group. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.001;##P < 0.01.
UBE2S regulates PI3K/AKT pathway activity through TRAF6 regulation
To further verify that UBE2S regulates the PI3K/AKT pathway activity through TRAF6, we first designed three interfering sequences of TRAF6 to synthesize siRNA, and then screened the most effective siRNA to knock down TRAF6 for subsequent experiments. After transfection on wild-type HCT116 and RKO cell lines, qPCR validation revealed that si-3 had the best knockdown effect (Fig. 6A). Further protein detection was performed on the transfected cells, and the results showed that si-3 had the strongest knockdown effect on TRAF6 (Fig. 6B). Next, we used si-3 to knock down TRAF6, and then p-mTOR protein expression accordingly decreased in wild-type HCT116 and RKO cells (Fig. 6C, P < 0.01). To explore the effect of UBE2S combined with TRAF6 on the downstream PI3K/AKT pathway, we first knocked down UBE2S, resulting in an increase in TRAF6 expression, as well as an increase in p-AKT and p-mTOR proteins (Fig. 6D, P < 0.05). However, further TRAF6 knockdown resulted in a decrease in p-AKT and p-mTOR proteins (Fig. 6D). In the CCK8 proliferation experiment, knocking down UBE2S enhanced cell proliferation ability (Fig. 6E, 72 h, HCT116, P < 0.001; 72 h, RKO, P < 0.0001), which could be reversed by siRNA of TRAF6 (Fig. 6E, knocking down group compared to siRNA group, HCT116 1.96 ± 0.04 vs. 0.96 ± 0.04, P < 0.001; RKO 2.18 ± 0.01 vs. 1.23 ± 0.01, P < 0.0001). In the colony formation experiments, knocking down UBE2S resulted in an increase in the number of colony formation units (Fig. 6F, HCT116 P < 0.01; RKO P < 0.001), which could also be reversed by siRNA of TRAF6 (Fig. 6F, knockdown group compared to siRNA group, HCT116 248.0 ± 10.2 vs. 179.0 ± 5.7, P < 0.05; RKO 289.0 ± 5.9 vs. 154.0 ± 9.6, P < 0.001).
UBE2S regulates the activity of the PI3K/AKT pathway through TRAF6, thereby affecting mTOR expression. (A) Screening the most effective siRNA on TRAF6 expression at the RNA level; (B) Screening the most effective siRNA on TRAF6 expression at the protein level; (C) TRAF6 knockdown with siRNA reduces p-mTOR expression in wild-type HCT116 and RKO cells (n = 3); (D) TRAF6, p-AKT and p-mTOR proteins expression with UBE2S knockdown and further TRAF6 knockdown (n = 3); (E) CCK8 proliferation experiments with UBE2S knockdown and further TRAF6 knockdown (n = 3); (F) Colony formation assays with UBE2S knockdown and further TRAF6 knockdown (n = 3). ‘*’ represents comparison with the ‘NC’ group, and ‘#’ represents comparison with the ‘SH’ group. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.001;###P < 0.001, ####P < 0.0001.
Discussion
During recent years, increasing evidence has verified the significant roles of ubiquitination in tumor progression. With more detailed researches on mechanisms of the ubiquitin proteasome system in tumors, more and more anti-cancer drugs targeting relevant molecules in the ubiquitination process have been developed, some of which have entered the clinical trial stage and even been approved for clinical treatment. For example, the Bortezomib was the first approved ubiquitination inhibitor for the treatment of multiple myeloma27, which inhibited the proteasome, thereby blocking the degradation of ubiquitinated proteins and ultimately induced tumor cell death.. Pitinib (MLN4924) impeded the functions of various ubiquitination enzymes to treat various types of cancer and immune system diseases, including lupus, solid tumors, and hematological tumors28,29,30. These drugs exert an essential role in clinical treatment by intervening in the ubiquitination process to affect protein expression to achieve therapeutic effects. However, the existing drugs mainly have an inhibitory effect for the ubiquitination process currently, and development of drugs to enhance ubiquitination at specific sites is still in the early stages and needs further exploring.
UBE2S, as a conjugating enzyme E2, was reported to regulate tumor progression through multiple mechanisms, including proliferation, cell cycle and metastasis7,8,9. In this study, UBE2S inhibited proliferation of CRC through enhancing ubiquitination of substrate TRAF6 protein and degrading it. Previous studies demonstrated that UBE2S promoted the development of ovarian cancer by promoting the PI3K/AKT/mTOR signaling pathway to regulate cell cycle and apoptosis11. However, in this study, overexpression of UBE2S in CRC cells weakened their proliferation and prevented cell cycle from G0/G1 phase to G2 phase. It is common for the same gene to play a dual role in different tumors, and even in different stages of the same tumor. Some may play a completely opposite role, which reflects the complexity of gene function. For instance, activation of p53 in hepatocytes paradoxically promoted non-cell autonomous liver carcinogenesis31. ID4 acted as a tumor suppressor in prostate cancer32,33, but a proto-oncogene in bladder cancer8,34.
In our study, we found that UBE2S had a significant effect on the MAPK and PI3K/AKT pathways, and the results of experiments showed that UBE2S can simultaneously inhibit the activation of both pathways, thereby impeding CRC proliferation. Knocking down UBE2S increased the CRC proliferation, which can be reversed by MAPK pathway inhibitor U0126 or PI3K/AKT pathway inhibitor Ipataserib respectively. As reported before, MAPK pathway inhibitors and PI3K/AKT pathway inhibitors have been used as chemotherapy drugs in clinical practice. For example, Periposine served as an oral AKT inhibitor for the treatment of various cancers, including colorectal cancer, lung cancer, and renal cell carcinoma35,36,37. Ipatasertib and Capivasertib were currently in clinical trials and were expected to be used to treat solid tumors, such as ER positive breast cancer and CRC38,39. Clinical trials have shown that drugs targeting the PI3K/AKT pathway exhibit good anti-tumor activity in certain types of tumors. The PI3K/AKT/mTOR (PAM) pathway is a highly conserved signal transduction network in eukaryotic cells, which can promote cell survival, growth, and cell cycle progression. It’s reported that PI3K/AKT promoted the activation of mTOR by directly phosphorylating another inhibitory protein PRAS40 of mTOR, intensifying the activation of mTOR and participating in the regulation of key processes such as cell metabolism, protein synthesis, and cell proliferation40. As for therapy, mTORC1 inhibitors such as rapamycin and everolimus have been widely used in the treatment of various tumors. They achieve the effect of inhibiting tumor growth by weakening the activation of the mTOR signaling pathway, blocking the proliferation, metastasis, and invasion of tumor cells. These drugs have achieved certain success in the treatment of renal cell carcinoma, breast cancer, prostate cancer, lung cancer, CRC and other tumor types. AKT and mTOR inhibitors were used as monotherapies and combined with other anti-tumor drugs in clinical trials as strategies to overcome therapeutic resistance. The combination of mTOR inhibitors and 5-FU can enhance anti-tumor effects in the treatment of colorectal cancer, potentially becoming a new field of treatment for colorectal cancer41. Clinical trials identified that KU-0063794 exhibited an efficacy in breast cancer, lung cancer and other tumor types, providing a new direction for tumor therapy42. Overall, the mTOR signaling pathway plays an important role in CRC. Drug targeting mTOR has been applied clinically, bringing new treatment options for certain cancer patients. In this study, it was confirmed that UBE2S can inhibit the PI3K/AKT pathway activity by reducing the TRFA6 protein, and then mTOR, as a downstream of the PI3K/AKT pathway, was hence blocked. After inhibition of the PI3K/AKT pathway by inhibitors, the expression of p-mTOR decreased, and the proliferation of CRC was weakened. At the same time, the enhanced proliferation of CRC caused by TRAF6 mediated activation of the PI3K/AKT/mTOR pathway can be reversed by rapamycin.
TRAF6, as a pivotal signal transduction protein, can activate multiple downstream signaling pathways to influence physiological functions and pathological processes, including MAPK and PI3K/AKT signaling pathways. For example, KDM4B promoted cancer growth and glucose metabolism in CRC through TRAF6 mediated AKT activation20. In addition, TRAF6 can activate ERK in CD40 signaling along the Ras independent pathway21. In our study, UBE2S knockdown promoted the CRC proliferation by increasing TRAF6 protein, which was reversed by MAPK inhibitor U0126, PI3K/AKT inhibitor Ipataserib or mTORC1 inhibitor rapamycin, respectively. Sun et al. reported that TRAF6 was upregulated in colon cancer and promoted the proliferation of colon cancer cells22. In gastric cancer progression, POU5F1 promoted cancer cell proliferation by reducing the ubiquitination level of TRAF643. In our study, the expression of UBE2S was lower in CRC than in adjacent tissues while the expression of TRAF6 protein was increased, which led to the CRC growth. Increased expression of TRAF6 protein can promote metastasis of colorectal cancer.
In CRC cells, we found that TRAF6 also plays an important role in their proliferation. As a hub gene for activating the MAPK and PI3K/AKT pathways, TRAF6 protein was negatively correlated with UBE2S protein expression. Interfering TRAF6 expression in wild-type HCT116 and RKO cells showed that MAPK/ERK and PI3K/AKT pathways were significantly inhibited with decreased p-AKT and mTOR proteins, and thus CRC cell proliferation was inhibited. A novel inhibitor targeting TRAF6 for hepatocellular carcinoma treatment has also been identified, and further clinical trials are needed44. This study identified that UBE2S inhibited CRC proliferation through reducing TRAF6 protein mediated activation of MAPK/ERK and PI3K/AKT/mTOR pathways. Due to the lack of effective ubiquitination agonist widely applied in clinical practice, this study only used inhibitors for experiments. In the future, further experiments can be conducted to detect the role of ubiquitination agonists in the protective function of UBE2S in CRC. In addition, significant progress has been made in reactivating ubiquitinated systems through the utilization of advanced techniques and a deeper understanding of ubiquitin biology, such as protein-targeted chimeric molecules (PROTACs) and hydrophobic tags (HYTs), to generate novel therapeutic strategies for various human diseases, including cancer45,46. Consequently, the advancement of technology stands poised to greatly facilitate the unraveling of the enigma surrounding UBE2S. Future studies should focus on UBE2S and its downstream pathways as potential anticancer drug targets. This includes developing UBE2S-specific chemical probes and evaluating their efficacy in clinical trials, which may provide new supplementary therapeutics for CRC.
Conclusion
This study reveals that UBE2S expression is lower in colorectal cancer tissues than in adjacent tissues, and is positively correlated with the overall survival rate of CRC patients. We firstly identify that the UBE2S/TRAF6/MAPK and PI3K/AKT/mTOR axes are novel pathways mediating CRC proliferation. Drugs targeting UBE2S and MAPK and PI3K/AKT/mTOR pathways may have potential therapeutic value for colorectal cancer.
Data availability
Mass spectrometry data of Co-immunoprecipitation protein was submitted as a supplementary file ‘Table S3’. The datasets generated and/or analysed during the current study are available in the GEO repository (GSE301888). Further queries can be directed to the Corresponding Author.
References
Siegel, R. L., Miller, K. D., Wagle, N. S. & Jemal, A. Cancer statistics, 2023. Cancer J. Clin. 73, 17–48. https://doi.org/10.3322/caac.21763 (2023).
Swatek, K. N. & Komander, D. Ubiquitin modifications. Cell Res. 26, 399–422. https://doi.org/10.1038/cr.2016.39 (2016).
Senft, D., Qi, J. & Ronai, Z. A. Ubiquitin ligases in oncogenic transformation and cancer therapy. Nat. Rev. Cancer. 18, 69–88. https://doi.org/10.1038/nrc.2017.105 (2018).
Bui, Q. T., Hong, J. H., Kwak, M., Lee, J. Y. & Lee, P. C. Ubiquitin-conjugating enzymes in cancer. Cells 10. https://doi.org/10.3390/cells10061383 (2021).
Popovic, D., Vucic, D. & Dikic, I. Ubiquitination in disease pathogenesis and treatment. Nat. Med. 20, 1242–1253. https://doi.org/10.1038/nm.3739 (2014).
Oh, E., Akopian, D. & Rape, M. Principles of Ubiquitin-Dependent signaling. Annu. Rev. Cell Dev. Biol. 34, 137–162. https://doi.org/10.1146/annurev-cellbio-100617-062802 (2018).
Zhang, R. Y. et al. UBE2S interacting with TRIM28 in the nucleus accelerates cell cycle by ubiquitination of p27 to promote hepatocellular carcinoma development. Signal. Transduct. Target. Therapy. 6, 64. https://doi.org/10.1038/s41392-020-00432-z (2021).
Zhang, M. et al. UBE2S promotes the development of ovarian cancer by promoting PI3K/AKT/mTOR signaling pathway to regulate cell cycle and apoptosis. Mol. Med. (Cambridge Mass). 28, 62. https://doi.org/10.1186/s10020-022-00489-2 (2022).
Zhang, M. et al. Diverse roles of UBE2S in cancer and therapy resistance: biological functions and mechanisms. Heliyon 10, e24465. https://doi.org/10.1016/j.heliyon.2024.e24465 (2024).
Gong, D. et al. UBE2S targets RPL26 for ubiquitination and degradation to promote non-small cell lung cancer progression via regulating c-Myc. Am. J. Cancer Res. 13, 3705–3720 (2023).
Zhang, M., Wang, J., Guo, Y., Yue, H. & Zhang, L. Activation of PI3K/AKT/mTOR signaling axis by UBE2S inhibits autophagy leading to cisplatin resistance in ovarian cancer. J. Ovarian Res. 16, 240. https://doi.org/10.1186/s13048-023-01314-y (2023).
Liu, Z., Haas, A. L., Diaz, L. A., Conrad, C. A. & Gíudice, G. J. Characterization of a novel keratinocyte ubiquitin carrier protein. J. Biol. Chem. 271, 2817–2822. https://doi.org/10.1074/jbc.271.5.2817 (1996).
Hu, L. et al. UBE2S, a novel substrate of Akt1, associates with Ku70 and regulates DNA repair and glioblastoma multiforme resistance to chemotherapy. Oncogene 36, 1145–1156. https://doi.org/10.1038/onc.2016.281 (2017).
Bai, D., Ueno, L. & Vogt, P. K. Akt-mediated regulation of NFkappaB and the essentialness of NFkappaB for the oncogenicity of PI3K and Akt. Int. J. Cancer. 125, 2863–2870. https://doi.org/10.1002/ijc.24748 (2009).
Gui, L. et al. UBE2S promotes cell chemoresistance through PTEN-AKT signaling in hepatocellular carcinoma. Cell. Death Discovery. 7. https://doi.org/10.1038/s41420-021-00750-3 (2021).
Sun, H. et al. USP11 promotes growth and metastasis of colorectal cancer via PPP1CA-mediated activation of ERK/MAPK signaling pathway. EBioMedicine 48, 236–247 (2019).
Xiang, Z. et al. A positive feedback between IDO1 metabolite and COL12A1 via MAPK pathway to promote gastric cancer metastasis. J. Experimental Clin. Cancer Research: CR. 38, 314. https://doi.org/10.1186/s13046-019-1318-5 (2019).
Xie, M. et al. FGF19/FGFR4-mediated elevation of ETV4 facilitates hepatocellular carcinoma metastasis by upregulating PD-L1 and CCL2. J. Hepatol. 79, 109–125. https://doi.org/10.1016/j.jhep.2023.02.036 (2023).
Yang, T. et al. PRL-mediated STAT5B/ARRB2 pathway promotes the progression of prostate cancer through the activation of MAPK signaling. Cell Death Dis. 15, 128. https://doi.org/10.1038/s41419-023-06362-2 (2024).
Li, H. et al. KDM4B facilitates colorectal cancer growth and glucose metabolism by stimulating TRAF6-mediated AKT activation. J. Exp. Clin. cancer Res.: CR 39, https://doi.org/10.1186/s13046-020-1522-3 (2020).
Kashiwada, M. et al. Tumor necrosis factor receptor-associated factor 6 (TRAF6) stimulates extracellular signal-regulated kinase (ERK) activity in CD40 signaling along a ras-independent pathway. J. Exp. Med. 187, 237–244. https://doi.org/10.1084/jem.187.2.237 (1998).
Sun, H. et al. TRAF6 is upregulated in colon cancer and promotes proliferation of colon cancer cells. Int. J. Biochem. Cell Biol. 53, 195–201. https://doi.org/10.1016/j.biocel.2014.04.010 (2014).
Hamidi, A. et al. TGF-β promotes PI3K-AKT signaling and prostate cancer cell migration through the TRAF6-mediated ubiquitylation of p85α. Sci. Signal. 10. https://doi.org/10.1126/scisignal.aal4186 (2017).
Li, J. et al. Genetic profiles affect the biological effects of Serine on gastric cancer cells. Front. Pharmacol. 11, 1183. https://doi.org/10.3389/fphar.2020.01183 (2020).
Qu, Y., Li, J., Cai, Q. & Liu, B. Hec1/Ndc80 is overexpressed in human gastric cancer and regulates cell growth. J. Gastroenterol. 49, 408–418. https://doi.org/10.1007/s00535-013-0809-y (2014).
Pan, J. et al. CD36 mediates palmitate acid-induced metastasis of gastric cancer via AKT/GSK-3β/β-catenin pathway. J. Experimental Clin. Cancer Research: CR. 38, 52. https://doi.org/10.1186/s13046-019-1049-7 (2019).
Sonneveld, P. et al. Daratumumab, bortezomib, lenalidomide, and dexamethasone for multiple myeloma. N. Engl. J. Med. 390, 301–313. https://doi.org/10.1056/NEJMoa2312054 (2024).
Zhang, Y. et al. Neddylation is a novel therapeutic target for lupus by regulating double negative T cell homeostasis. Signal. Transduct. Target. Therapy. 9. https://doi.org/10.1038/s41392-023-01709-9 (2024).
Pomella, S. et al. MYOD-SKP2 axis boosts tumorigenesis in fusion negative rhabdomyosarcoma by preventing differentiation through p57(Kip2) targeting. Nat. Commun. 14, 8373. https://doi.org/10.1038/s41467-023-44130-0 (2023).
He, Z. X. et al. Targeting Cullin neddylation for cancer and fibrotic diseases. Theranostics 13, 5017–5056. https://doi.org/10.7150/thno.78876 (2023).
Makino, Y. et al. Constitutive activation of the tumor suppressor p53 in hepatocytes paradoxically promotes Non-Cell autonomous liver carcinogenesis. Cancer Res. 82, 2860–2873. https://doi.org/10.1158/0008-5472.Can-21-4390 (2022).
Tian, L. Y., Smit, D. J. & Jücker, M. The role of PI3K/AKT/mTOR signaling in hepatocellular carcinoma metabolism. Int. J. Mol. Sci. 24. https://doi.org/10.3390/ijms24032652 (2023).
Tewari, D., Patni, P., Bishayee, A., Sah, A. N. & Bishayee, A. Natural products targeting the PI3K-Akt-mTOR signaling pathway in cancer: A novel therapeutic strategy. Sem. Cancer Biol. 80, 1–17. https://doi.org/10.1016/j.semcancer.2019.12.008 (2022).
Yu, L., Wei, J. & Liu, P. Attacking the PI3K/Akt/mTOR signaling pathway for targeted therapeutic treatment in human cancer. Sem. Cancer Biol. 85, 69–94. https://doi.org/10.1016/j.semcancer.2021.06.019 (2022).
Bendell, J. C. et al. Randomized placebo-controlled phase II trial of perifosine plus capecitabine as second- or third-line therapy in patients with metastatic colorectal cancer. J. Clin. Oncology: Official J. Am. Soc. Clin. Oncol. 29, 4394–4400. https://doi.org/10.1200/jco.2011.36.1980 (2011).
Wu, D. W. et al. FHIT loss confers cisplatin resistance in lung cancer via the AKT/NF-κB/Slug-mediated PUMA reduction. Oncogene 34, 2505–2515. https://doi.org/10.1038/onc.2014.184 (2015).
Srivastava, N. & Cho, D. C. Perifosine in renal cell carcinoma. Expert Opin. Investig. Drugs. 22, 285–291. https://doi.org/10.1517/13543784.2013.754422 (2013).
Shi, Z. et al. Functional mapping of AKT signaling and biomarkers of response from the FAIRLANE trial of neoadjuvant Ipatasertib plus Paclitaxel for Triple-Negative breast cancer. Clin. Cancer Research: Official J. Am. Association Cancer Res. 28, 993–1003. https://doi.org/10.1158/1078-0432.Ccr-21-2498 (2022).
Sun, L. et al. Ipatasertib, a novel Akt inhibitor, induces transcription factor FoxO3a and NF-κB directly regulates PUMA-dependent apoptosis. Cell Death Dis. 9, 911. https://doi.org/10.1038/s41419-018-0943-9 (2018).
Wang, H. et al. Proline-rich Akt substrate of 40 kda (PRAS40): a novel downstream target of PI3k/Akt signaling pathway. Cell. Signal. 24, 17–24. https://doi.org/10.1016/j.cellsig.2011.08.010 (2012).
Chen, J., Shao, R., Li, L., Xu, Z. P. & Gu, W. Effective Inhibition of colon cancer cell growth with MgAl-layered double hydroxide (LDH) loaded 5-FU and PI3K/mTOR dual inhibitor BEZ-235 through apoptotic pathways. Int. J. Nanomed. 9, 3403–3411. https://doi.org/10.2147/ijn.S61633 (2014).
Jeon, Y. W. et al. Potentiation of the anticancer effects by combining docetaxel with Ku-0063794 against Triple-Negative breast cancer cells. Cancer Res. Treat. 54, 157–173. https://doi.org/10.4143/crt.2020.1063 (2022).
Yang, W. et al. POU5F1 promotes the proliferation, migration, and invasion of gastric cancer cells by reducing the ubiquitination level of TRAF6. Cell Death Dis. 14, 802. https://doi.org/10.1038/s41419-023-06332-8 (2023).
Li, N. et al. Identification of a new TRAF6 inhibitor for the treatment of hepatocellular carcinoma. Int. J. Biol. Macromol. 182, 910–920. https://doi.org/10.1016/j.ijbiomac.2021.04.081 (2021).
Xie, S. et al. Discovery of Norbornene as a novel hydrophobic Tag applied in protein degradation. Angew. Chem. Int. Ed. Engl. 62, e202217246. https://doi.org/10.1002/anie.202217246 (2023).
Dai, X. J. et al. Degraders in epigenetic therapy: protacs and beyond. Theranostics 14, 1464–1499. https://doi.org/10.7150/thno.92526 (2024).
Acknowledgements
We thank the National Natural Science Foundation of China (82271766) for financial support.
Author information
Authors and Affiliations
Contributions
J.L.: Designed methods, conducted experiments, analyzed data, drafted the manuscript. H.H. & H.F.: Assisted with methods, experiments, collected samples and data analysis. X.C.: Helped with methods and analysis, reviewed and edited the draft. T.Z., X.J. & K.L.: Provided supervision. R.Z.: Oversaw the project, reviewed the draft, contributed to the concept, and coordinated efforts. All authors have read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval and consent to participate
Our study was approved by Medical Ethics Committee of the Shanghai Ruijin Hospital (No.028-2021), Shanghai Jiao Tong University school of medicine, Shanghai, China. Treatment of experimental animals followed the internationally recognized 3R principle and welfare ethics in accordance with the requirements of the guidelines for ethical review of experimental animal welfare in China.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Li, J., Feng, H., Liu, K. et al. UBE2S inhibits colorectal cancer proliferation by regulating the PI3K/AKT and MAPK/ERK pathways via TRAF6. Sci Rep 15, 35610 (2025). https://doi.org/10.1038/s41598-025-18637-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-025-18637-z