Abstract
Early-life stress sensitizes individuals to subsequent stressors to increase lifetime risk for psychiatric disorders. Within the ventral tegmental area (VTA), a mesolimbic brain region implicated in stress response and mental health, early-life stress causes long-lasting changes in gene expression and chromatin modifications that in turn cause latent physiological and behavioral sensitivity to stress. These molecular consequences of early-life stress are indicative of epigenetic priming, a form of molecular memory in which developmental or environmental cues open chromatin at enhancers to facilitate transcriptional response to stimuli. However, the long term impact of early-life stress on chromatin architecture in the VTA was not yet known. Using a combination of activity-dependent cellular tagging and ATAC-sequencing, we find that early-life stress opens chromatin specifically in stress-activated cells of the VTA, that this remodeling persists into adulthood, and that opening chromatin at cis-regulatory elements including enhancers augments transcriptional response to adult stress. Together, this data supports enhancer priming within ELS-responsive cells in the VTA as a biological mechanism for lifelong stress sensitivity.
Similar content being viewed by others
Introduction
Millions of children worldwide experience early-life stress (ELS), which increases lifetime risk for developing mood, anxiety, and substance use disorders1,2,3,4. ELS broadly encompasses many adverse experiences including maltreatment (neglect, and/or physical, sexual, and/or emotional abuse), death or loss of a caregiver, poverty, systemic racism, exposure to community violence, or experience of a natural disaster5,6,7,8,9. Studies in humans and preclinical animal models have shown that ELS sensitizes individuals to future stressors, which may be at the root of increased psychiatric disease risk10,11,12,13,14. However, the mechanisms through which childhood experiences impart continued stress sensitivity across the lifespan are poorly understood.
ELS exerts prominent and long-lasting effects on the function and regulation of the ventral tegmental area (VTA), a dopaminergic brain region involved in motivation, reward processing, learning, and emotion regulation15,16. Dopamine neurons within the VTA have protracted development and continue to mature through adolescence which may leave them vulnerable to stress and other early environmental insults17. ELS blunts VTA development and functional connectivity in children18,19 and blunts reward processing, which in turn predict depressive symptoms20,21. Rodent models show that ELS increases excitability of dopaminergic neurons in VTA, increases baseline dopamine levels released from VTA into nucleus accumbens (NAc)22,23,24,25, blunts physiological response to rewarding stimuli, and alters dopaminergic response to stressors26,27,28,29.
These functional consequences of ELS are likely a result of altered molecular development12,30,31. ELS induces both long-lasting changes to baseline gene expression in VTA12, and latent gene expression changes revealed by a second hit of stress in adulthood30, pointing to epigenetic regulatory mechanisms. Indeed, we and others have observed broad changes in post-translational histone modifications and chromatin-modifying enzymes in VTA which functionally regulate excitability of dopamine and GABA neurons in VTA and behavioral sensitivity to stress32,33,34. These previous findings support the hypothesis that ELS epigenetically primes chromatin to be more reactive to stress and other stimuli. Epigenetic priming —a form of molecular memory in which developmental or environmental cues open chromatin at enhancers— facilitates a faster, stronger, and/or sensitized transcriptional response to recurring stimuli54,55,56,57. However, the long term impact of ELS on chromatin architecture in the VTA was not yet known.
Here, we sought to determine how ELS alters chromatin architecture in the VTA. We specifically hypothesized that ELS would open chromatin at enhancers, and that cells initially activated by ELS would have the strongest molecular changes. To test this, we performed activity-dependent ATAC-sequencing (assay for transposase accessible chromatin) in the VTA of adult male and female mice exposed to ELS or standard rearing conditions, and predicted gene expression response to subsequent stress. Our results provide novel insights into how the epigenome encodes long-lasting stress sensitivity.
Results
Tagging and capturing stress-responsive and non-responsive cells of the VTA
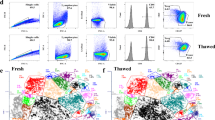
To test the hypothesis that cells initially activated by ELS would have the greatest chromatin changes, we examined chromatin profiles in ELS-activated and non-activated cell populations separately. To permanently tag and isolate these separate cell populations, we utilized double transgenic ArcCreERT2 x Sun1-sfGFP-Myc mice (Fig. 1a) in which transgene expression is both neuronal activity- and ligand-dependent35,36. ELS occurred in a stress sensitive window from postnatal day P10-17 as previously described12,37. Genetic recombination was induced on P17 by administration of 4-OHT either at the beginning of a 3-hour period of maternal separation in a clean cage (ELS) or exploration of a novel enrichment object (standard-reared pups, Std) (Fig. 1b). We previously validated that this approach tags similar numbers of cells in ELS and Std mice so as not to bias downstream analyses, and that experience-dependent neuronal tagging is specific and engages distinct ensembles of cells36. Permanent expression of nuclear membrane-localized protein SUN1-sfGFPMyc enabled us to affinity-purify tagged nuclei later in adulthood (> P60) by the INTACT method (isolation of nuclei tagged in specific cell types)38,39(Fig. 1c, d). Both male and female samples were collected from each group.
We performed ATAC-sequencing in both tagged and untagged cell populations to characterize and compare open chromatin regions across groups and cell populations (Fig. 1c). Initial investigation of ATAC-seq tracks found that experience-activated cells have open chromatin peaks at the Arc gene promoter used to drive transgene expression in both Std and ELS samples, even > 40 days after tagging, potentially suggesting these cells remain highly active (Fig. 1e). For both Std and ELS groups, approximately 25% of nuclei in the VTA were tagged and captured (Fig. 1f). To investigate the types of cells activated and tagged in Std and ELS mice, we visualized chromatin accessibility at promoters of cell-type marker genes, and found opening at Th, Slc6a3, and Slc17a6 at relatively similar levels across groups, indicating that both conditions activated dopamine, GABA, and glutamate neurons, respectively (Supplemental Fig. 1).
Tagging and capturing stress-responsive and non-responsive cells of the VTA. (a) Breeding scheme of ArcCreERT2 x Sun1-sfGFP-Myc mice. (b) Experimental timeline for ELS from P10-17, 4-OHT -induced recombination at P17 in standard-reared (Std) and ELS pups, weaned at P21 and tissue collected at > P60 in adulthood. (c) Schematic of 4-OHT- and activity-dependent recombination and tagging of VTA neurons, followed by nuclear isolation, affinity-purification of tagged cells, and ATAC-sequencing and analysis in both tagged and untagged cells. (d) Example images of purified nuclei expressing SUN1GFP in the nuclear membrane (scale bar 150 μm). (e) Example browser tracks of ATAC peaks at the Arc promotor from experience-activated samples. (f) Average counts of total and affinity-purified (GFP) nuclei from standard-reared (gray) and ELS (red) samples. Error bars are mean ± SEM.
ELS opens chromatin in experience-activated cells of the VTA
Among the experienced-activated population of VTA cells, we identified 53,689 total accessible regions of chromatin. In order to determine whether there are persistent changes in chromatin in early experience-tagged cells that last into adulthood, we calculated differences in chromatin accessibility by DESeq2 using a likelihood ratio test modeling both group and sex. We found 178 differentially accessible regions [DARs, log2(fold change) > 0.5, p < 0.01], all of which were opened by ELS (Fig. 2a). While this group difference was striking, no single peak reached adjusted p-value significance. Therefore, we sought to further test this broad pattern of chromatin opening in two additional ways. First, we shuffled the ELS and Std sample labels through all possible permutations and compared the log2fold-change (LFC) of the top 100 most significant regions (100 lowest p-value within shuffled labeled matrices) to the matrix with the correctly labeled samples (Fig. 2b). We found the greatest average fold change between ELS and Std-reared with the actual correct labels (one-sample z-test, p = 0.02; Fig. 2b), indicating that the opening of chromatin by ELS is unlikely to be artificial. Second, as an alternative to significance testing, we performed parameter estimation using the R package apeglm to shrink the LFC estimates in such a manner that higher-variance observations are given adjusted LFCs near zero40. We find that many of the DARs retain substantial effect sizes even after LFC shrinkage, again indicating that the opening effect we see following ELS is not the result of noise (Fig. 2c). Browser tracks of top regions of opened chromatin confirm consistent opening across all ELS compared to Std samples (Fig. 2d). These findings are in striking contrast to those in un-tagged cells of the VTA, which had only 13 DARs (four opened and nine closed by ELS; Fig. 2e) and which was not different than shuffled values (Supplemental Fig. 2). It should be noted, however, that non-tagged cells are likely to be more heterogeneous than the experience-activated population. These results indicate that ELS induces persistent opening of chromatin in experience-responsive cells, but this effect is absent in the broader VTA cell population.
ELS opens chromatin in experience-activated cells of the VTA. (a) Volcano plot of differentially accessible regions (DARs) within experience-activated cells. (DESeq2, p-value < 0.01 and |logFC|>1). DARs opened by ELS shown in red. (b) Average log2(FoldChange) for actual and shuffled sample labels of the 100 lowest p-value regions comparing ELS and Std with each set of labels. Actual labels z-score = 1.96 vs. shuffled means (p = 0.02). (c) Volcano plot with log2(foldchange) shrinkage showing opening (red, +) and closing (blue, -) regions in activated population. (d) Example browser tracks of ATAC peaks opened by ELS in experience-activated cells, RPM-normalized (chr12:54877473–54877631–left, chr4:80149575–80150001–center, chr11:38084665–38084873–right). (e) Volcano plot of DARs within non-activated cells. (DESeq2, p-value < 0.01 and |logFC|>1). DARs opened by ELS shown in orange; closed by ELS shown in dark grey. (f) Enrichment or depletion (log10 of observed over expected for each genomic feature in the mm39 genome) of DARs mapped to each genomic feature for DARs. Inset shows proportion of DARs found in putative cis-regulatory elements (> 200bb from the nearest TSS (red, 88%), within proximity to the TSS (light red, 9%), or at the TSS (white, 3%). (g) Top transcription factor motifs found in DARs.
We next sought to characterize these activated-cell DARs to better understand how ELS alters gene regulatory functions and biological processes. Overall, 88% of DARs were in putative cis-regulatory elements (CREs), defined as > 200 bp from the nearest transcription start site, as enhancers may also be found within intronic, exonic, and intergenic regions (Fig. 2f, pie chart inset)41. Further characterization of genomic features observed within our DAR set compared to expected proportions of the mouse genome revealed that ELS-opened regions were enriched for all genomic features besides those classified as strictly intergenic, with the greatest enrichment of enhancers (Fig. 2f; Ensembl-defined enhancers).
To determine whether experience-activated DARs were enriched for functional elements of transcription factor binding sites, we performed motif analysis42(Fig. 2g). Genomic loci that gained ATAC peaks after ELS were significantly enriched for binding sites for three motifs: SOX3, CTCF, and NFATC2, indicating that ELS impacts regions involved in nervous system development, gene regulation by modulation of chromatin looping, and neural response to activity43,44,45,46(Fig. 2g). Together, these results support the notion that long-lasting opening of chromatin after ELS may alter epigenetic development of the VTA and prime the epigenetic landscape to respond to future stress and other stimuli.
Chromatin opening after ELS predicts gene expression response to adult stress
An important question is whether the observed changes in chromatin accessibility after ELS are relevant to gene expression. Given an enrichment of DARs within CREs (Fig. 2f) and that chromatin changes may govern transcriptional response to future stress32, we first determined which genes are putatively regulated by CRE-DARs. As enhancers and CREs do not necessarily regulate their nearest gene neighbor, we used publicly available long-range interaction mapping to determine which genes are likely regulated by each CRE-DAR (T-Gene47). We then used previously published RNA-seq data from adult VTA30 to infer expression changes of the putatively-regulated genes after different stress experiences, relative to each group’s own baseline: adult stress (AS; social defeat for males, subthreshold variable stress for females) alone vs. Std-reared control, or ELS + adult stress (ELS + AS) vs. ELS-control (Fig. 3a). We found that these putatively-regulated genes were more highly expressed after adult stress among mice with a history of ELS [unpaired two-tailed t-test: t(1,206) = 2.638, p = 0.0090]. Gene Ontology (GO) revealed an enrichment of genes related to development and chromatin organization48 (Fig. 3b). RNA-expression data for selected example DAR-regulated genes Ampd2 and Slco4a1 show an increased gene expression response to adult stress after ELS (relative to ELS baseline expression) compared to the response to adult stress in Std-reared mice (Fig. 3c, d; two-tailed Mann-Whitney Ampd2 U = 16.50, p = 0.0014; Slco4a1 U = 17.0, p = 0.0017). AMPD2 is involved in purine metabolism and protein translation49. Slco4a1 encodes OATP-E, a transporter for thyroid hormones and other organic anions. Interestingly, thyroid hormone regulation is known to be affected by both ELS and adult stress, and we recently showed that rescuing developmental deficits in thyroid hormone availability can rescue sensitivity to later stress50,51,52,53 (Fig. 3d). These results support the interpretation that long-lasting chromatin changes following ELS may prime transcriptional responses to adult stress later in life.
Chromatin opening after ELS predicts gene expression in response to adult stress. (a) log2(fold change) of genes putatively contacted by CRE-DARs after adult stress alone (Std-AS) compared to Std-control or ELS and adult stress together (ELS-AS) compared to ELS-controls. (b) Gene Ontology (GO) terms for gene list putatively contacted by ELS-opened DARs for biological process. (c,d) Example gene expression in Std-AS (vs. Std-Ctl) and ELS-AS (vs. ELS-Ctl) for Ampd2 (c) and Slco4a1 (d). Left: normalized gene expression for both males (circles) and females (triangles); right: browser tracks showing each CRE-DAR and putative gene contact (yellow highlights).
Discussion
ELS sensitizes, or primes, individuals to subsequent stressors to increase lifetime risk of psychiatric disorders. At a molecular level, ELS programs both long-lasting baseline gene expression changes in the VTA and other efferent brain regions, primes gene expression response to a second hit of stress, and alters post-translational histone modifications12,30,32. Here, we tested the hypothesis that ELS encodes these persistent changes within the VTA at an epigenetic level by priming cis-regulatory regions of the genome through open chromatin. Using activity-dependent cellular tagging and ATAC-sequencing in both tagged and untagged populations, we show that ELS exclusively opens chromatin in ELS-responsive cells, that this remodeling persists into adulthood, and that opening chromatin at these CREs may serve to augment transcriptional response to adult stress.
Epigenetic priming is associated with open chromatin and the presence of the post-translational histone modification H3K4me1, and facilitates transcriptional response to stimuli54,55,56,57. This phenomenon is known to occur in botany, immunology, embryological development, and cancer56,58,59,60,61. Elegant work also demonstrated that experience can prime enhancers in the adult hippocampus through changes in 3D enhancer architecture, that priming is stable, and that memory recall activates enhancers62. Our results reveal that ELS also leads to stable epigenetic priming. In the aftermath of ELS, we find that ELS largely opens chromatin (Fig. 2a-d), and that differentially accessible chromatin is enriched for enhancers and other CREs (Fig. 2f). Through long-range interaction mapping to putatively contacted genes, we also show that ELS-opened regions predict the transcriptional response to a second hit of stress (Fig. 3a, c,d). This is also consistent with recent work showing that broad postnatal deposition of H3K4me1 in the VTA increases dopamine neuron excitability and behavioral response to adult stress32, demonstrating that epigenetic priming is functionally relevant and sufficient to recapitulate the effect of ELS at a behavioral level. Together, these data support an emerging role for enhancer priming as a biological mechanism for ELS-induced lifelong stress sensitivity.
The persistence of this open chromatin state in ELS-activated cells into adulthood is consistent with stable epigenetic remodeling during development. Chromatin remodeling in early embryological development is an essential component of cell fate specification and differentiation; maintenance of remodeled chromatin states maintains cellular identity and prevents aberrant gene expression which could lead to disease63,64,65. Maturation of the epigenome extends beyond fetal development into the postnatal period. Recent research has demonstrated that the epigenetic landscape within cortical interneurons continues to mature between mouse postnatal weeks 1 and 3, after which it crystalizes and is highly stable into adulthood66. This window of postnatal epigenetic maturation overlaps with the postnatal sensitive period for stress exposure in mice12,30,37,67,68, suggesting that ELS may interfere with chromatin development. This is supported by our finding that chromatin is aberrantly open in ELS-activated cells of the VTA (Fig. 2a-c), and by an enrichment of motifs for transcription factors essential to brain development and three-dimensional chromatin structure (Fig. 2g). However, this now begs the question of whether ELS opens chromatin de novo, potentially through recruitment of pioneer factors, or whether ELS blunts closing of regions that should normally close during development, leaving chromatin in a more open, immature state.
Several caveats of this study should be considered. First, because cell tagging was specifically done in distinct contexts (novelty/enrichment vs. ELS), the cells tagged in each condition may belong to distinct pathways. However, recent work has demonstrated that individual dopamine neurons can respond to multiple distinct stimuli69, refuting a labeled-line hypothesis, and we did not find that cells tagged in each of these contexts were reactivated by adult stress at different overall rates36. Nevertheless, future studies may determine the extent to which other developmental experiences and exposures shape chromatin remodeling. Second, analysis used to infer genes contacted by putative enhancers relies on 3D chromatin data from the brain that isn’t specific to VTA, and much data now indicates that chromatin profiles are cell-type-specific. Future studies generating VTA-specific contact data may improve gene response prediction. Similarly, predicted gene expression changes are from bulk-sequencing rather than tagged cells. Yet given these caveats, we still see that genes predicted to be primed by ELS-opened enhancers have greater transcriptional response to adult stress. Third, we point out that the DARs are defined by uncorrected p-values. This is likely due to a low number of samples with successful ELS-trapped cells and ATAC-seq. Despite this, the pattern of chromatin opening in ELS samples is clear and supported by label-shuffled analysis, and example regions of opened and closed chromatin clearly demonstrate a robust effect of ELS across all samples and sexes.
In sum, these data provide novel evidence that ELS primes transcriptional sensitivity to future stress though developmental changes in chromatin, akin to an epigenetic memory of stress within the VTA. While facilitated stress response may be adaptive in some contexts, constant stress hypersensitivity and vigilance are thought to precipitate mood, anxiety, and other psychiatric disorders70. These findings provide insight into how early experience programs lifelong hypersensitivity to stress, with translational implications.
Methods
Reported in accordance with ARRIVE guidelines.
Mice
All experiments were conducted in accordance with relevant guidelines and protocols approved by the Institutional Animal Care and Use Committee at Princeton University. Mice were housed in a temperature- and humidity-controlled vivarium and maintained on a 12-h light/dark cycle (lights on at 4a.m.) and were group-housed in sets of three to five with ad libitum access to food and water. Mice were considered to be “male” or “female” based on external genitalia (adult) and/ or anogenital distance (pups). For all experiments, mice were bred in-house in trios, males were removed after 5–7 days, and females were separated into individual cages 1–3 days prior to parturition. All offspring regardless of early life experience were weaned at postnatal day P21, with males and females weaned into separate cages, keeping littermates together and only combining pups from different litters of the same experimental condition in order to maintain at least three mice/cage. Males and females were used in all experiments.
In order to tag and isolate experience-responsive cells for ATAC-seq experiments, we generated Arc-CreERT2 X Sun1-GFP double-transgenic mice on a C57BL/6J background as previously described36. Briefly, heterozygous Arc-CreERT2/+ mice35 (stock #022357, The Jackson Laboratory) were bred in-house with R26-CAG-LSL-Sun1-sfGFP-Myc39 knock-in mice (“Sun1-GFP”; stock #030952, The Jackson Laboratory). Genotyping of double-transgenic mice was performed as previously described36.
Early-life stress
Early-life stress (ELS): Litters for ATAC-seq were randomly assigned at birth to standard (Std)-rearing or ELS conditions. The ELS paradigm used was a combination of maternal separation and limited nesting resources during a stress sensitive period from postnatal day P10-17, based on established protocols12. Briefly, pups in ELS-assigned litters were separated from their dams into a clean cage containing a tablespoon of bedding from an adult male aggressor mouse for 3–4 h/day from P10-17. During the week of daily separations, home cage nesting material was also reduced to 1/2 of a nesting puck, which was replaced with a full nesting puck after the last day of stress. Std-reared litters were left with their dams undisturbed except for saline and 4-OHT injections for transgenic animals, and a novel object (running wheel) given on the last day of early life experience for 3 h, in order to induce recombination related to a specific experience as previously validated36. Pups were observed to explore the wheel as a novel object but not to run on it.
Administration of 4-hydroxytamoxifen
To induce experience-dependent recombination, 4-hydroxytamoxifen (4-OHT) was administered as described36. 4-OHT (catalog #H7904, SigmaAldrich) was first prepared as 25 mg/ml stock solutions in ethanol and frozen at -20 °C. For injections, 4-OHT was prepared from stock to 5 mg/ml, dissolved in ethanol and corn oil (1:4).
Double-transgenic Arc-CreERT2 X Sun1-GFP pups were first habituated to injections of saline (60 µL i.p.) from P14-16. To drive recombination on P17, (the last day of the stress for ELS pups), ELS and Std-reared transgenic pups were injected with 4-OHT (50 mg/kg body weight, approximately 60 µL/pup, i.p.) at the beginning of maternal separation. Standard-reared pups were returned to their home cage and given a novel enrichment object (running wheel: pups were observed to explore the novel object but not necessarily run on it) in order to tag a specific positive-valence experience. The mice were then left undisturbed for at least three days (P17-P21) to allow time for expression of the transgenes to develop before weaning. Animals were allowed to mature until adulthood, at which time tissue was collected and nuclei isolated. We previously validated the use of these mice for experience specific tagging, and showed that we were able to tag similar numbers of neurons in ELS and Std VTA so as not to artificially bias downstream analyses36.
Nuclear isolation and ATAC-sequencing
Double-transgenic Arc-CreERT2 X Sun1-GFP mice allowed for affinity purification of nuclei expressing the SUN1-GFP-MYC fusion protein in the nuclear membrane by the INTACT approach38,39. Experience-activated samples included 2 Std and 3 ELS samples (ELS: 2 male and 1 female samples, Std: 1 male and 1 female samples). Non-activated samples included 2 Std and 3 ELS samples (ELS: 2 male and 1 female samples, Std: 2 female samples). All samples were from individual mice and distinct litters. Brains were harvested from mice after cervical dislocation and rapid decapitation into ice-cold homogenization buffer in adulthood (P60-90). 16-gauge bilateral punches of the VTA were taken from 1 mm slices and flash-frozen until use. To isolate nuclei, VTA punches were thawed in ice-cold homogenization buffer and dounce homogenized in 1.5mL of 1X Unstable homogenization buffer71. Homogenate was transferred to 2mL lo-bind tubes then spun down at 500 x g for 5 min (at 4 C). Nuclei were collected by iodixanol gradient and counts were collected using a hemocytometer. Nuclei from tagged cells were separated from non-tagged nuclei by incubation with anti-GFP (Sigma; cat#SAB4301138-100UL) and anti-myc (Sigma; cat#: SAB4300318-100UG) antibodies that bound to magnetic Protein G Dynabeads (ThermoFisher; cat#10004D). Total yield of nuclei for all samples was ~ 90k nuclei, and previous calculations of nuclear yield in the striatum estimates activated nuclei made up ~ 10% of the total yield. Both populations of cells were kept and immediately used for ATAC-sequencing. All reactions were performed on ice.
Nuclei from tagged and non-tagged cells were transposed using Illumina’s transposition kit (Illumina; cat#20034198) for 30–35 min for non-activated and 35–40 min for activated nuclei at 37 °C at 1000RPM. Fragmented DNA was recovered with Qiagen MinElute kits (Qiagen; cat# 28604). Samples were amplified and purified according to Corces et al. 201771 using Illumina UD indexes (Illumina; cat#20027214 A or B) and AMPure XP beads (ThermoFisher; cat#NC9959336). Resulting yield of DNA fragment sizes was determined by Bioanalyzer dsDNA High Sensitivity protocol and normalized by concentration for sequencing. All samples were sequenced within the Genomics Core Facility of the Lewis-Sigler Institute at Princeton University on a NovaSeq using SP 100nt chemistry to yield ~ 325–400 M read depth. FASTQ quality controls were used to show quality of reads on average (> 30 Phred) for all samples, and only samples with a Phred score > 30 were used.
Sequencing and statistical analysis
Adapters were trimmed from ATAC-seq reads using fastp (v0.23.4), with base correction enabled, and aligned to mm39 using bowtie2 (v2.4.5) (“--no-unal --no-mixed --no-discordant”). Using samtools (v1.13), reads with MAPQ > 30 were retained, followed by samtools collate, fixmate, sort, and markdup to identify but not remove PCR duplicates in the absence of UMIs. MACS2 (v2.2.9.1) was used to call narrowpeaks for each sample (which ignores duplicate reads by default).
Using R, a consensus peak set was generated using GenomicRanges (v1.58.0) “reduce” command. ATAC reads were imported using BRGenomics (v1.17.1) “import_bam” with “field = NULL” and unique reads in peaks were counted using “getCountsByRegions”. We selected only peaks that were reproducibly discovered in either Std or ELS-reared samples based on the Irreproducibility Discovery Rate (IDR) (Li et al., 2011) via R package idr (v1.3) “est.IDR” (mu = 0.1, sigma = 1, rho = 0.2, p = 0.5), keeping any peaks with idr < 0.1 (n = 53689). Differential accessibility was assessed using DESeq2 (v1.46.0) using a likelihood ratio test comparing a full model “~sex + condition” (with condition being ELS or Std) to a reduced model of sex alone. Log2(Fold-Change) Shrinkage analysis used apeglm. Related plots were regenerated using ggplot2 (v3.5.1). We identified differentially accessible regions (DARs) as peaks with |log2FoldChange|>0.5 and an uncorrected p-value < 0.01. For the shuffling analysis, sex was ignored, and all possible shuffled assignments of ELS or Std condition were applied to the samples. In each case, the average log2FoldChange was calculated for the peaks with the lowest 100 unadjusted p-values. We used a one-sample z-test to compare the mean log-fold change (LFC) from the actual labels to the mean of the distribution of mean LFCs obtained from shuffled-label permutations. The resulting z-score was converted to a two-tailed p-value using the standard normal distribution.
RPM/CPM-normalized bigWig files were also generated in R using GenomicRanges “coverage” via BRGenomics “getStrandedCoverage” with strand information removed.
Bioinformatic analysis: genomic features, motif analysis, contacted gene prediction, looping visualizations, and gene ontology analysis
Track visualization of example open and closed differentially accessible regions (DARs) was done by loading the generated RPM-normalized bigWig files into the Integrated Genome Viewer (IGV; igv.org/app/); mm39 genome build).
ChIPseeker (v.1.36.0) was used to define broad genomic feature classes (TSS, near TSS, putative enhancers) overlapping differentially accessible regions (DARs). GenomicDistributions (v.1.8.0) and GenomicDistributionsData (v.1.8.0) were used for higher resolution genomic feature mapping. Since the mm39 genome was not added to the GenomicDistributionsData default package, we manually added the genome. We also manually added ENCODE cCREs (enhancers) (obtained via ENSEMBL, https://oct2024.archive.ensembl.org/) to the list of features.
The complete list of DARs in each condition (activated and non-activated) were supplied to geneontology.org (PANTHER) to perform gene ontology (GO) for genes within or potentially contacted by DARs.
Motif Analysis was carried out using XTREME, of the MEME suite (https://meme-suite.org/meme/tools/xstreme). Significantly upregulated and downregulated regions were supplied to the motif discovery pipeline, using the HOCOMOCO Mouse v11 CORE parameter for known motifs. All other parameters were kept at default; an E value of ≤ 0.05 was used as a significance threshold.
In order to identify genes putatively contacted and regulated by DARs, and run gene ontology (GO) analysis on the resulting list, we used the T-Gene tool within MEMEsuite. We first lifted-over our ATACseq peaks from mm39 to mm9 using a UCSC genome browser chain file (https://hgdownload.soe.ucsc.edu/goldenPath/mm39/liftOver/mm39ToMm9.over.chain.gz) and the re-implementation of UCSC liftOver found in the R/Bioconductor package rtracklayer (v1.66.0). During liftOver, peaks that failed to lift over, or lifted over into multiple non-contiguous peaks, were dropped from this analysis (n = 2 for BB_openRegions_rmBg.bed). Regions that survived the lift over were then used in T-Gene. Output of T-Gene is limited to 1000 putative gene contacts. We applied a cutoff of the correlation and distance (CnD) p-value < 0.01 to identify the strongest contact predictions.
We used these putatively-contacted genes to predict the impact of changes in chromatin accessibility on gene expression response to adult stress. We mapped these genes to previously published bulk RNA-seq from adult VTA among mice exposed to ELS and/or adult stress (GEO GSE89692). Four groups included: Std-Control, ELS-Control, Std-Adult Stress, and ELS-Adult Stress. ELS in these experiments were identical to that in the current study. For male and female samples, adult stress consisted of chronic social defeat stress and sub-threshold variable stress, respectively12,30. Genes with expression < 10 CPM and outliers (based on Grubb’s test and standard-deviation > 2) were excluded. Log2Fold-Change (L2FC) was previously calculated using DESeq2. One-way ANOVA comparisons were made on L2FC values for ELS-AdultStress vs. ELS-Ctl, and Std-AdultStress vs. Std-Ctl male samples. Next, we calculated individual sample fold-change values for all male and female subjects exposed to adult stress relative to average expression of their own rearing group [i.e.: ELS-AdultStress vs. ELS-Ctl (n = 6 M, 5 F) and Std-AdultStress vs. Std-Ctl (n = 6 M, 6 F)] to determine how expression changes after adult stress within each rearing group. Two-tailed Mann-Whitney was used to evaluate effects.
The complete list of potential gene contacts were supplied to STRING(v.12) to perform gene ontology (GO) of biological processes, only including terms that passed an adjusted significance p-value < 0.05.
Data availability
All sequencing data is deposited in NCBI’s Gene Expression Omnibus, accession number GSE268468, or by contact with corresponding author (CJP). Sequencing data was analyzed via standardized pipelines as described.
References
Green, J. G. et al. Childhood adversities and adult psychiatric disorders in the National comorbidity survey replication I: associations with first onset of DSM-IV disorders. Arch. Gen. Psychiatry. 67, 113–123 (2010).
Kessler, R. C. et al. Childhood adversities and adult psychopathology in the WHO world mental health surveys. Br. J. Psychiatry J. Ment Sci. 197, 378–385 (2010).
Kendler, K. S. et al. The impact of environmental experiences on symptoms of anxiety and depression across the life span. Psychol. Sci. 22, 1343–1352 (2011).
Scott, K. M., McLaughlin, K. A., Smith, D. A. R. & Ellis, P. M. Childhood maltreatment and DSM-IV adult mental disorders: comparison of prospective and retrospective findings. Br. J. Psychiatry. 200, 469–475 (2012).
Hackman, D. A., Farah, M. J. & Meaney, M. J. Socioeconomic status and the brain: mechanistic insights from human and animal research. Nat. Rev. Neurosci. 11, 651–659 (2010).
UNICEF. Child Displacement. UNICEF. (2018).
Giano, Z., Wheeler, D. L. & Hubach, R. D. The frequencies and disparities of adverse childhood experiences in the U.S. BMC Public. Health. 20, 1327 (2020).
Webb, E. K., Carter, S. E., Ressler, K. J. & Fani, N. Harnett, N. G. The neurophysiological consequences of racism-related stressors in black Americans. Neurosci. Biobehav Rev. 161, 105638 (2024).
Brieant, A., Sisk, L. M., Keding, T. J., Cohodes, E. M. & Gee, D. G. Leveraging multivariate approaches to advance the science of early-life adversity. Child. Abuse Negl. 106754 https://doi.org/10.1016/j.chiabu.2024.106754 (2024).
McLaughlin, K. A., Conron, K. J., Koenen, K. C. & Gilman, S. E. Childhood adversity, adult stressful life events, and risk of past-year psychiatric disorder: a test of the stress sensitization hypothesis in a population-based sample of adults. Psychol. Med. 40, 1647–1658 (2010).
Zhang, Z. Y. et al. Early adversity contributes to chronic stress induced depression-like behavior in adolescent male rhesus monkeys. Behav. Brain Res. 306, 154–159 (2016).
Peña, C. J. et al. Early life stress confers lifelong stress susceptibility in mice via ventral tegmental area OTX2. Science 356, 1185–1188 (2017).
Saxton, K. & Chyu, L. Early life adversity increases the salience of later life stress: an investigation of interactive effects in the PSID. J. Dev. Orig Health Dis. 147, 1–12 (2019).
Sidamon-Eristoff, A. E., Cohodes, E. M., Gee, D. G. & Peña, C. J. Trauma exposure and mental health outcomes among central American and Mexican children held in immigration detention at the united States–Mexico border. Dev. Psychobiol. 64, e22227 (2022).
Peña, C. J., DeBerardine, M. & Sullivan, K. E. Molecular heterogeneity and development of the ventral tegmental area. Curr. Opin. Behav. Sci. 61, 101478 (2025).
Hanson, J. L., Williams, A. V., Bangasser, D. A. & Peña, C. J. Impact of early life stress on reward circuit function and regulation. Front. Psychiatry. 12, 1799 (2021).
Hoops, D. & Flores, C. Making dopamine connections in adolescence. Trends Neurosci. 40, 709–719 (2017).
Marusak, H. A., Hatfield, J. R. B., Thomason, M. E. & Rabinak, C. A. Reduced ventral tegmental Area-Hippocampal connectivity in children and adolescents exposed to early threat. Biol. Psychiatry Cogn. Neurosci. Neuroimaging. 2, 130–137 (2017).
Park, A. T. et al. Early childhood stress is associated with blunted development of ventral tegmental area functional connectivity. Dev. Cogn. Neurosci. 47, 100909 (2021).
Hanson, J. L. et al. Cumulative stress in childhood is associated with blunted reward-related brain activity in adulthood. Soc. Cogn. Affect. Neurosci. 11, 405–412 (2016).
Hanson, J. L., Hariri, A. R. & Williamson, D. E. Blunted ventral striatum development in adolescence reflects emotional neglect and predicts depressive symptoms. BPS 78, 598–605 (2015).
Spyrka, J. et al. Early life stress-induced alterations in the activity and morphology of ventral tegmental area neurons in female rats. Neurobiol. Stress. 13, 100250 (2020).
Matthews, K., Dalley, J. W., Matthews, C., Tsai, T. H. & Robbins, T. W. Periodic maternal separation of neonatal rats produces region- and gender-specific effects on biogenic amine content in postmortem adult brain. Synap N Y N. 40, 1–10 (2001).
Arborelius, L. & Eklund, M. Both long and brief maternal separation produces persistent changes in tissue levels of brain monoamines in middle-aged female rats. Neuroscience 145, 738–750 (2007).
Afonso, V. M., King, S. J., Novakov, M., Burton, C. L. & Fleming, A. S. Accumbal dopamine function in postpartum rats that were Raised without their mothers. Horm. Behav. 60, 632–643 (2011).
Cabib, S., Puglisi-Allegra, S. & D’amato, F. R. Effects of postnatal stress on dopamine mesolimbic system responses to aversive experiences in adult life. Brain Res. 604, 232–239 (1993).
Brake, W. G., Zhang, T. Y., Diorio, J., Meaney, M. J. & Gratton, A. Influence of early postnatal rearing conditions on mesocorticolimbic dopamine and behavioural responses to psychostimulants and stressors in adult rats. Eur. J. Neurosci. 19, 1863–1874 (2004).
Matthews, K., Wilkinson, L. S. & Robbins, T. W. Repeated maternal separation of preweanling rats attenuates behavioral responses to primary and conditioned incentives in adulthood. Physiol. Behav. 59, 99–107 (1996).
Jahng, J. W. et al. Mesolimbic dopaminergic activity responding to acute stress is blunted in adolescent rats that experienced neonatal maternal separation. Neuroscience 171, 144–152 (2010).
Peña, C. J. et al. Early life stress alters transcriptomic patterning across reward circuitry in male and female mice. Nat. Commun. 10, 5098 (2019).
Vosberg, D. E., Leyton, M. & Flores, C. The Netrin-1/DCC guidance system: dopamine pathway maturation and psychiatric disorders emerging in adolescence. Mol. Psychiatry. 25, 297–307 (2020).
Geiger, L. T. et al. Early-life stress alters chromatin modifications in VTA to prime stress sensitivity. https://doi.org/10.1101/2024.03.14.584631 (2025).
Authement, M. E. et al. Histone deacetylase Inhibition rescues maternal Deprivation-Induced GABAergic metaplasticity through restoration of AKAP signaling. Neuron 86, 1240–1252 (2015).
Shepard, R. D., Langlois, L. D., Authement, M. E. & Nugent, F. S. Histone deacetylase Inhibition reduces ventral tegmental area dopamine neuronal hyperexcitability involving AKAP150 signaling following maternal deprivation in juvenile male rats. J. Neurosci. Res. 98, 1457–1467 (2020).
Denny, C. A. et al. Hippocampal memory traces are differentially modulated by experience, time, and adult neurogenesis. Neuron 83, 189–201 (2014).
Balouek, J. A. et al. Reactivation of Early-Life stress-Sensitive neuronal ensembles contributes to lifelong stress hypersensitivity. J. Neurosci. 43, 5996–6009 (2023).
Peña, C. J., Nestler, E. J. & Bagot, R. C. Environmental programming of susceptibility and resilience to stress in adulthood in male mice. Front. Behav. Neurosci. 13, 272 (2019).
Deal, R. B. & Henikoff, S. A simple method for gene expression and chromatin profiling of individual cell types within a tissue. Dev. Cell. 18, 1030–1040 (2010).
Mo, A. et al. Epigenomic signatures of neuronal diversity in the mammalian brain. Neuron 86, 1369–1384 (2015).
Zhu, A., Ibrahim, J. G. & Love, M. I. Heavy-tailed prior distributions for sequence count data: removing the noise and preserving large differences. Bioinformatics 35, 2084–2092 (2019).
Moore, J. E. et al. Expanded encyclopaedias of DNA elements in the human and mouse genomes. Nature 583, 699–710 (2020).
Grant, C. E., Bailey, T. L. & XSTREME Comprehensive motif analysis of biological sequence datasets. https://doi.org/10.1101/2021.09.02.458722 (2021).
Schepers, G. E., Teasdale, R. D. & Koopman, P. Twenty pairs of Sox: extent, homology, and nomenclature of the mouse and human Sox transcription factor gene families. Dev. Cell. 3, 167–170 (2002).
Ong, C. T. & Corces, V. G. CTCF: an architectural protein bridging genome topology and function. Nat. Rev. Genet. 15, 234–246 (2014).
Ranger, A. M. et al. The nuclear factor of activated T cells (NFAT) transcription factor NFATp (NFATc2) is a repressor of chondrogenesis. J. Exp. Med. 191, 9–22 (2000).
Vihma, H., Luhakooder, M., Pruunsild, P. & Timmusk, T. Regulation of different human NFAT isoforms by neuronal activity. J. Neurochem. 137, 394–408 (2016).
O’Connor, T., Grant, C. E., Bodén, M. & Bailey, T. L. T-Gene: improved target gene prediction. Bioinformatics 36, 3902–3904 (2020).
Thomas, P. D. et al. Making genome-scale phylogenetics accessible to all. Protein Sci. 31. PANTHER, 8–22 (2022).
Akizu, N. et al. AMPD2 regulates GTP synthesis and is mutated in a potentially treatable neurodegenerative brainstem disorder. Cell 154, 505–517 (2013).
Murray, M. & Zhou, F. Trafficking and other regulatory mechanisms for organic anion transporting polypeptides and organic anion transporters that modulate cellular drug and xenobiotic influx and that are dysregulated in disease. Br. J. Pharmacol. 174, 1908–1924 (2017).
Fujiwara, K. et al. Identification of thyroid hormone transporters in humans: different molecules are involved in a Tissue-Specific Manner*†. Endocrinology 142, 2005–2012 (2001).
Privitera, M. et al. Noradrenaline release from the locus coeruleus shapes stress-induced hippocampal gene expression. eLife 12, RP88559 (2024).
Bennett, S. N., Chang, A. B., Rogers, F. D., Jones, P. & Peña, C. J. Thyroid hormones mediate the impact of early-life stress on ventral tegmental area gene expression and behavior. Horm. Behav. 159, 105472 (2024).
Calo, E. & Wysocka, J. Modification of enhancer chromatin: What, How, and why? Mol. Cell. 49, 825–837 (2013).
D’Urso, A. & Brickner, J. H. Epigenetic transcriptional memory. Curr. Genet. 63, 435–439 (2017).
Lämke, J. & Bäurle, I. Epigenetic and chromatin-based mechanisms in environmental stress adaptation and stress memory in plants. Genome Biol. 18, 124 (2017).
Nord, A. S. & West, A. E. Neurobiological functions of transcriptional enhancers. Nat. Neurosci. 23, 5–14 (2020).
Cui, K. et al. Chromatin signatures in multipotent human hematopoietic stem cells indicate the fate of bivalent genes during differentiation. Cell. Stem Cell. 4, 80–93 (2009).
Bevington, S. L. et al. Inducible chromatin priming is associated with the establishment of immunological memory in T cells. EMBO J. 35, 515–535 (2016).
Vicente-Dueñas, C., Hauer, J., Cobaleda, C. & Borkhardt, A. Sánchez-García, I. Epigenetic priming in cancer initiation. Trends Cancer. 4, 408–417 (2018).
Martin, E. W. et al. Chromatin accessibility maps provide evidence of multilineage gene priming in hematopoietic stem cells. Epigenetics Chromatin. 14, 2 (2021).
Marco, A. et al. Mapping the epigenomic and transcriptomic interplay during memory formation and recall in the hippocampal engram ensemble. Nat. Neurosci. 23, 1606–1617 (2020).
Ziffra, R. S. et al. Single-cell epigenomics reveals mechanisms of human cortical development. Nature 598, 205–213 (2021).
Su-Feher, L. et al. Single cell enhancer activity distinguishes GABAergic and cholinergic lineages in embryonic mouse basal ganglia. Proc. Natl. Acad. Sci. 119, e2108760119 (2022).
Mirabella, A. C., Foster, B. M. & Bartke, T. Chromatin deregulation in disease. Chromosoma 125, 75–93 (2016).
Stroud, H. et al. An Activity-Mediated transition in transcription in early postnatal neurons. Neuron https://doi.org/10.1016/j.neuron.2020.06.008 (2020).
Moriceau, S., Roth, T. L. & Sullivan, R. M. Rodent model of infant attachment learning and stress. Dev. Psychobiol. 52, 651–660 (2010).
Rincón-Cortés, M. & Sullivan, R. M. Early life trauma and attachment: immediate and enduring effects on neurobehavioral and stress axis development. Front. Endocrinol. 5, 33 (2014).
Willmore, L. et al. Overlapping representations of food and social stimuli in mouse VTA dopamine neurons. Neuron https://doi.org/10.1016/j.neuron.2023.08.003 (2023).
McEwen, B. S. Stress, Adaptation, and disease: allostasis and allostatic load. Ann. N Y Acad. Sci. 840, 33–44 (1998).
Corces, M. R. et al. An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nat. Methods. 14, 959–962 (2017).
Acknowledgements
We would like to thank Wei Wang and the Genomics Core Facility in the Lewis-Sigler Institute for Integrative Genomics at Princeton (LSI) where ATAC-seq was performed. Parts of Fig. 1 were generated with BioRender.com.
Funding
This research was funded by NIH R00MH115096 (CJP); NIH R01MH129643 (CJP); NIH F31MH131351 (RR); and the New York Stem Cell Foundation (CJP). CJP is a New York Stem Cell Foundation Robertson Investigator. This manuscript is the result of funding in whole or in part by the National Institutes of Health (NIH). It is subject to the NIH Public Access Policy. Through acceptance of this federal funding, NIH has been given a right to make this manuscript publicly available in PubMed Central upon the Official Date of Publication, as defined by NIH.
Author information
Authors and Affiliations
Contributions
These studies were designed by CJP and RLR. ATAC-seq data were collected by RLR and analyzed by RLR, MD, SD, and CJP; mouse work was performed by RLR and SNB. The manuscript was written by RLR, CJP, and MD with input from all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Rashford, R.L., DeBerardine, M., Dan, S. et al. Persistent open chromatin state in early-life stress-activated cells of the VTA. Sci Rep 15, 36118 (2025). https://doi.org/10.1038/s41598-025-21157-5
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-21157-5