Abstract
Hepatocellular carcinoma (HCC) is one of the most common malignant tumors with high morbidity and mortality worldwide. Angiogenesis is essential for HCC progression and metastasis. Some angiogenesis-related genes promote this process, whereas other antiangiogenic genes inhibit HCC growth and metastasis. Therefore, finding new potential biomarkers for HCC prognosis prediction and treatment is essential. Public RNAseq and clinical data from TCGA and GEO database, download angiogenesis-related genes from the GeneCards, MSigDB database, through the single factor analysis of Cox, LASSO build risk score—Cox regression analysis model and external validation verified from the GEO. Cox regression analysis, Kaplan Meier (KM) curve, ROC curve, and decision-curve analysis will be used to evaluate and examine the risk score prediction effect of the model. GSVA analysis was used to assess the variation of gene sets between groups, and ClBERSOFT, ESTIMATE, and TIMER databases were used to analyze the immune infiltration in the single-cell level analysis of gene expression differences between cells. Finally, in the three pairs of HCC tissues and tissue adjacent to carcinoma by real-time fluorescent quantitative PCR (qRT_PCR) and western blotting (WB) to evaluate angiogenesis-related genes (ATP2A3 AEBP1 PNMA1, PLAT) expression level in HCC, and AEBP1 was knocked out in HCCLM3 cells, which is to study AEBP1 biological function in HCC. We established a prognostic risk assessment model based on 13 significant genes associated with HCC prognosis by Cox analysis and LASSO-Cox regression analysis. The median was used to divide these patients into high-risk and low-risk groups, and the prognosis of the high-risk group was worse than that of the low-risk group. Through the multivariate Cox regression analysis, it was found that the risk score was an independent predictor of overall survival (OS). The GSVA analysis suggested that the predicted high-risk population showed higher activity in the purine, pyrimidine, and riboflavin metabolic pathways. Compared with the low-risk group, the tumor microenvironment in the high-risk group showed a reduction in the number of cells promoting anti-tumor immunity and an increase in the number of cells inhibiting anti-tumor immunity, as well as a reduction in overall immune infiltration and matrix components. On the single-cell level, it was confirmed that the key genes (AEBP1, ATP2A3, PLAT, and PNMA1) expressed differently between liver cancer and adjacent tissue cell groups. Finally, qRT_PCR and WB results showed that ATP2A3, AEBP1, PNMA1, and PLAT were highly expressed in liver cancer tissue compared to adjacent tissue, and the proliferation, migration, and invasion of HCCLM3 cells were inhibited after knocking out AEBP1. We constructed novel risk score models as prognostic biomarkers for HCC, which has the potential to guide the development of more personalized treatment strategies for HCC patients. In addition, AEBP1 is a potential therapeutic target for HCC.
Similar content being viewed by others
Introduction
Liver cancer is the third leading cause of cancer-related deaths and ranks sixth in the incidence new cancer cases1. The incidence of liver cancer has been increasing in recent years, and it is estimated that by 2025, more than 1 million patients will be diagnosed with liver cancer each year2. Hepatocellular carcinoma (HCC) is the most common primary liver malignant tumor, accounting for more than 90% of primary liver malignant tumors3. The risk factors for HCC mainly include viral hepatitis (hepatitis B and C), non-alcoholic steatohepatitis, alcohol-related liver disease, and exposure to dietary toxins such as aflatoxin and aristolochic acid4. Although HCC has made great progress in surgical resection, liver transplantation, radiofrequency ablation, transcatheter arterial chemoembolization, targeted therapy, and immunotherapy, the efficacy of current treatment strategies for HCC patients is still limited5. The median survival time of patients with advanced liver cancer is 1–1.5 years, and the 5-year survival rate of overall liver cancer patients is less than 20%6. Thus, there is a critical need to identify new biomarkers that can enhance the prediction of HCC prognosis and inform more effective treatment approaches.
Angiogenesis is a critical process in tumorigenesis, Thus, there is a critical need to identify new biomarkers that can enhance the prediction of HCC prognosis and inform more effective treatment approaches. Additionally, angiogenesis continuously provides oxygen and nutrients to the tumor to nourish the rapid growth of the tumor and can promote immune escape by changing the immunosuppressive microenvironment, thus leading to tumor progression and metastasis7,8. Angiogenesis is a crucial factor affecting the progression of HCC which is a typical hypervascular tumor. Due to the rich vascular characteristics of HCC, anti-angiogenic therapy has shown great potential in the treatment of HCC. For example, sorafenib is the first angiogenesis inhibitor approved by the FDA for the first-line treatment of advanced HCC9. Subsequently, other angiogenesis inhibitors (tyrosine kinase inhibitors, TKI) such as imatinib and regorafenib were developed to overcome drug resistance and improve the therapeutic effect10. However, due to the large heterogeneity among HCC patients, the clinical benefits of anti-angiogenesis therapy are not significant7,11. HIF1A, TMPRSS4 and SDF are related to angiogenesis, and studies have found that these genes promote HCC progression and metastasis by positively regulating angiogenesis in HCC12,13. Nevertheless, the specific prognostic value of angiogenesis-related genes in HCC remains inadequately understood.
In the study, we systematically analyzed angiogenesis, and the expression of related genes, and discussed the angiogenesis-related gene in patients with HCC prognosis prediction of potential value. A previous study14 showed that angiogenesis-related gene signatures can be used to predict the prognosis of HCC. We constructed a new risk assessment model based on 13 angiogenesis-related genes from the TCGA dataset. This model was subsequently validated using the GSE116174 cohort. Additionally, we conducted in vitro experiments to investigate the functional role of AEBP1 in HCC. Our findings indicate that risk models based on angiogenesis-related genes could serve as effective biomarkers for predicting HCC prognosis, thereby assisting clinicians in tailoring personalized treatment strategies. Furthermore, our results suggest that AEBP1 represents a promising therapeutic target in the treatment of HCC.
Methods and materials
Data download
Eight samples of LIHC scRNA-seq data were obtained from the GEO Database with the accession number GSE14022. With the help of the website https://tisch.compgenomics.org/, we performed an analysis on liver cancer at the single-cell level. We obtained LIHC bulk RNA-seq data from TCGA (361 tumor and 50 normal samples) and GSE116174 (64 tumor samples) databases15. Then, 36 angiogenesis-related genes were mined from the GnneCards database (http://www.genecards.org/) and Molecular Signatures Database (MSiDB, http://www.broad. mit.edu/gsea/msigdb).
WGCNA analysis
We collected 36 genes associated with angiogenesis and scored each patient using the R package ssGSEA. According to the score, we use WGCNA analysis. The R package ‘WGCNA’ was utilized to create gene coexpression networks in TCGA-LIHC, which included 361 tumor samples. This network formation involved several key steps: Firstly, a similarity matrix was defined. Next, the ideal gentle threshold of 6 was utilized to convert this similarity matrix into an adjacency matrix. Additionally, the adjacency matrix underwent additional modifications to become a topological overlap matrix (TOM). The next step involved organizing the dissTOM using Tom Cluster to produce a hierarchical clustering tree. Module identification within this tree was achieved by employing dynamic techniques for tree cutting. Subsequently, module eigengenes (MEs) for each module were computed, serving as indicators of the module’s collective expression profile. The MEs of various modules were examined, and the average distance metric between them was determined by utilizing the reciprocal of the Pearson correlation coefficient. Finally, an average linkage hierarchical clustering approach was employed to cluster the MEs of all modules, setting the minimum genome value at 5016. This process culminated in the merging of highly similar modules, resulting in a comprehensive coexpression network.
Development of the predictive angiogenesis scoring model
Through the use of WGCNA (Weighted Gene Co-expression Network Analysis), we were able to pinpoint genes associated with angiogenesis. The angiogenesis related genes that overlapped were integrated into LASSO Cox regression analysis using the glmnet package in R. We conducted univariate Cox regression analysis for overall survival (OS) employing the survival package in R to identify HAIRGs with prognostic significance. The selection of the penalty parameter (λ) was guided by the minimum criterion, utilizing tenfold cross-validation. The risk score for each patient was calculated by using the formula Risk Score = Σ (gene expression × coefficient), which takes into account the gene expression levels and their corresponding regression coefficients. Afterward, individuals diagnosed with liver carcinoma were divided into high or low-risk categories according to the median of these computed risk scores.
Examining the clinical significance of the high and low-risk categories
By combining clinical information from liver cancer patients in TCGA with a prognostic model focused on angiogenesis, we classified them into two groups based on their risk levels: high and low. Subsequently, we analyzed the correlation of these risk groups with clinical parameters such as age, gender, tumor-node-metastasis (TNM) classifications, and cancer stage, concerning the survival status of the patients. Distinct clinical profiles were observed among patients in various risk categories.
The construction of nomogram and calibration curves
Using the “rms” package in R, we created nomograms and calibration plots. The nomogram served to assess the alignment between observed outcomes and predicted probabilities. In contrast, the calibration graph was utilized to predict the likelihood of survival after 1, 3, and 5 years17. Following that, the training set underwent univariate and multivariate Cox regression analyses to assess whether the risk score served as an autonomous prognostic marker for overall survival (OS) in LIHC patients. Covariates included age, stage, gender, and risk score. T-tests were used to analyze the correlation between risk scores and clinicopathological characteristics. Adhering to scientific research standards, statistical significance was determined by establishing a p-value threshold below 0.05 (p < 0.05).
GSVA analysis
After categorizing patients into high and low-risk groups using the previously developed angiogenesis-based prognostic model, we performed pathway analysis utilizing Gene Set Variation Analysis (GSVA) in our study. Our initial step involved the selection of pertinent gene sets that represent biological pathways, with a primary emphasis on those linked to angiogenesis and the advancement of tumors. These gene sets were chosen from well-established databases (c2.cp.kegg.v7.5.1.symbols.gmt). After categorizing our gene expression data into high and low-risk groups, we proceeded to perform GSVA using the corresponding R package. This involved the mapping of genes to their respective pathways and computing enrichment scores for each pathway in each sample group. The enrichment scores obtained from GSVA provided a quantification of how actively involved each pathway was in the high versus low-risk groups. The scores played a crucial role in evaluating the disparities in biological processes and pathways among the two risk categories.
Analysis of immune cell infiltration in LIHC samples
Through our research, we utilized the prognostic model we created based on angiogenesis to categorize liver cancer patients into groups with high and low risks. CIBERSORT was employed for the prediction of scores for 22 immune cell types, and the Wilcoxon test was utilized to identify variations in immune cell scores among these risk groups. Subsequently, the ESTIMATE algorithm was applied to gauge the degree of immune infiltration, revealing that the low-risk group exhibited higher immune infiltration. Furthermore, we investigated the expression of immune checkpoint genes in both the high-risk and low-risk groups. In the end, we utilized the TIDE online tool to forecast the TIDE (Tumor Immune Dysfunction and Exclusion) scores for the samples. A higher TIDE score suggests a higher probability of immune evasion, offering a valuable understanding of the immunological environment of liver cancer in terms of patient risk classification.
IMvigor210 immunotherapy cohort
Utilizing the IMvigor210 immunotherapy cohort, we reconstructed the risk model using 13 key genes. This was done in conjunction with clinical information from the immunotherapy cohort, such as cancer staging and the degree of response to immunotherapy. Afterwards, we performed a comparative examination, incorporating these clinical variables, to assess the differences between the high and low-risk groups. By employing this method, a deeper comprehension of the significance of the risk model within the framework of immunotherapy results was made possible.
Single-cell analysis of angiogenesis-related genes
Using our approach, we analyzed the genes selected for the prognostic model-related to angiogenesis through single-cell analysis on the web-based platform http://www.tisch.comp-genomics.org/. This enabled a detailed examination of their expression profiles at the single-cell level, providing insights into the cellular heterogeneity within the context of angiogenesis.
Collection of clinical samples
In this study, 3 pairs of fresh HCC tissues and adjacent normal tissues were obtained from 3 patients with primary HCC who underwent surgical resection in the Affiliated Hospital of Guizhou Medical University. None of the patients received radiotherapy, chemotherapy or immunotherapy before surgery. This study was approved by the Ethics Committee of the Affiliated Hospital of Guizhou Medical University (Ethics No. 2022375). All participants provided written informed consent.
Cell culture
HCCLM3 cells were donated from the biomolecular Biology Laboratory of Guizhou Medical University. All HCCLM3 cell cultures containing 100 IU/ml penicillin, streptomycin 100 IU/ml, and 10% FBS DMEM medium, put at 37 °C, and CO2 content is 5% of the cultivation in the box. Cell growth status was observed and recorded daily.
RNA extraction and quantitative real-time PCR
Using TRIzol reagent (Takara Biotechnology Co, LTD, Dalian, China), following the instructions from the 3 fresh HCC tissue and total RNA extracted tissue adjacent to carcinoma. Using the PrimeScript ™ RT kit (Takara, Dalian, China) will be a total RNA reverse transcription of cDNA. Using SYBR ® Premix Ex Taq ™ II kit (Takara) for RT–qPCR detection of gene expression. b-actin was used as a reference gene, and the relative gene expression levels were calculated using the 2-DDCt method.
Western blot analysis
HCC tissue and tissue adjacent to carcinoma in RIPA cracking buffer and PMSF ice slurry. Proteins were extracted and quantified with a BCA Protein Assay kit (Beyotime, Shanghai, China). Twenty µg of protein were separated by SDS–polyacrylamide gel electrophoresis and transferred to a polyvinylidene fluoride membrane. After blocking with 5% skim milk at room temperature for 2 h, the cells were treated with primary antibodies (anti-ADH1B, anti-ALDOB, anti-ADH1a, anti-FBP1, anti-ADH6, anti-b-actin; Cloning Technology Co, Wuhan, China) were placed at 4 °C overnight. Subsequently, the enzyme league second incubation membrane resistance and ECL chemiluminescence reagent kit (Beyotime) for testing. Using ImageJ software will protein to a to b-actin quantitatively.
Cell counting kit-8
The well-grown cells were seeded in 96-well plates, the cell concentration was adjusted to 1.5 × 104/ml, 200 µl of cell suspension was added to each well, and each group of cells was repeated three times and cultured in an incubator. After cell adhesion to the wall, 100 µl of CCK-8 reagent was added to each well, measurements of 0 h were recorded, and cells were placed in the incubator for 2.5 h. Finally, at the indicated time points (0, 24, 48, and 72 h), the absorbance values at 450 nm wavelength were measured with a microplate reader (Varioskan Flash, version: 4.00.53), and the cell viability curves were plotted based on the absorbance values.
Transwell invasion
On early – 20 °C under the precooling of the cell plate, 50 ml per hole to join the matrix evenly spread out, the next day and a half hour early put holes in serum-free medium for hydration. Will grow well cells after digestion, centrifugal, adjust the cell concentration and the 2 × 104/ml, every hole to join 200 (including l cell suspension, after 48 h sample, use 20 min 4% paraformaldehyde-fixed cells, crystal violet staining 20 min 0.1%, PBS scrub after 3 times to dry. Five fields were taken under an inverted microscope with a 200 × field of view and counted by Image J software.
Statistic analysis
All statistical analyses were performed with the use of R software, version 4.2.2. A two-sided t-test was used to compare gene expression in HCC tissues and adjacent non-tumor tissues. Kaplan–Meier analysis and log-rank test were used to compare the differences in OS between groups. Univariate and multivariate Cox regression analysis and LASSO Cox regression analysis were used to determine the independent predictors of OS, and the Mann–Whitney test was used to compare the SsGSEA scores of immune pathways or cells between the two groups. If not specifically required, P values < 0.05 were considered statistically significant, and all P values were two-tailed.
Results
Obtaining angiogenesis-related genes through co-expression networks
To construct a gene set related to angiogenesis, we first conducted a comprehensive search in the GeneCards database using keywords related to angiogenesis. We also integrated and screened the gene sets related to angiogenesis in the MSigDB database. Based on the existing research literature, after careful alignment, deduplication and preliminary evaluation of gene function, we finally selected 36 genes closely related to angiogenesis (as shown in Supplementary Table 1), which laid the foundation for subsequent analysis of angiogenesis-related genes.
For our research, we performed a Weighted Gene Co-expression Network Analysis (WGCNA) by utilizing the ‘WGCNA’ software in R. Initially, we evaluated liver cancer patients in the TCGA database using an angiogenesis-related gene set and the “ssGSEA” software. The creation of a weighted gene co-expression network was made easier by this scoring system. A soft-thresholding power of 6 was chosen to transform the expression matrix into an adjacency matrix. This adjacency matrix was then converted into a topological overlap matrix (TOM). Genes were clustered and modules were identified using the dynamic tree-cut method, with a minimum module size of 50 genes (Fig. 1A), through average linkage hierarchical clustering based on TOM. Modules were further clustered, merging closely related modules into new ones (mergeCutHeight = 0.15, maxBlockSize = 22,000, minModuleSize = 50), resulting in a total of nine distinct modules (Fig. 1D). Of these, the green and brown modules showed the highest correlation with angiogenesis scores (Fig. 1B,C). The brown modules contains 909 genes, the green modules contains 625 genes. We extracted genes from these two modules and further refined our gene set by selecting those with angiogenesis scores greater than 0.4 and gene significance greater than 0.7. These genes were earmarked for subsequent analyses, providing a focused gene set pertinent to angiogenesis in liver cancer.
WGCNA results: (A) Differential gene cluster map based on topological overlap and given color to the modules. (B) A scatter-plot of gene significance (GS) of weight versus number of module members (MM) in the brown module, where GS and MM have a highly significant correlation. (C) The gene significance (GS) scatter plot of the weight and the number of module members (MM) in the green module, where GS and MM have a highly significant correlation. (D) Correlation maps of co-expression modules with clinical features.
Construction of prognostic signature within the brown and green module
Afterward, we conducted a univariate Cox regression analysis to discover genes that are significantly linked to the prognosis of individuals with liver cancer18. This analysis yielded 30 genes with prognostic relevance, as listed in Supplementary Table 2. To further refine this gene set, we applied LASSO Cox regression analysis to these 30 genes. We then calculated risk scores for each patient using a formula that incorporated the expression levels of 13 selected genes and their corresponding coefficients derived from the LASSO analysis (Fig. 2A). The risk score was calculated using the following formula: Risk score = (expression level of AEBP1 × − 0.044) + (expression level of ATP2A3 × − 0.207) + (expression level of C15orf39 × 0.1) + (expression level of CD3E × 0.003) + (expression level of CD4 × − 0.129) + (expression level of EFEMP1 × − 0.088) + (expression level of GMIP × 0.522) + (expression level of HLA-DPB1 × − 0.078) + (expression level of HLA-DRA × − 0.1406) + (expression level of LAMB1 × 0.220) + (expression level of LPCAT4 × 0.070) + (expression level of PLAT × − 0.147) + (expression level of PNMA1 × 0.065). Afterward, the patients in the training cohort, TCGA data set were divided into a low-risk group and a high-risk group according to the median value of their calculated risk scores. The risk plot showed that individuals belonging to the high-risk category had a less favorable outlook in comparison to those in the low-risk category and the mortality rate in the training cohort increased with the increasing risk scores (left column of Fig. 2B), This risk stratification offers a novel approach to predicting patient outcomes in liver cancer. Kaplan–Meier curves showed that patients in the low-risk group had a higher probability of OS than those in the high-risk group (P < 0.0001) (left column of Fig. 2C). Then this risk scoring model was evaluated by time-dependent ROC analysis. In the training set from the TCGA database, our prognostic model demonstrated Area Under the Curve (AUC) values of 0.73, 0.67, and 0.76 for 1-year, 3-year, and 5-year survival predictions, respectively (left column of Fig. 2D). These results indicated that the risk scoring model had good predictive accuracy for prognosis of HCC in the TCGA cohort.
LASSO regression analysis in the TCGA dataset and construction and validation of the scoring model. (A) LASSO coefficient distribution of ARPG in TCGA data set (left) and selection of optimal parameters in LASSO model (right). (B) Distribution of ARPG survival status, score, and prognostic gene expression in HCC patients in the TCGA (left) and GSE116174 cohorts (right). (C) Kaplan–Meier curves of OS differences between middle- and low-risk groups in the TCGA (left) and GSE116174 cohorts (right). (D) Time-dependent ROC curves for risk scores in the TCGA (left) and GSE116174 cohorts (right).
To demonstrate the predictive value of the risk scoring model, the GSE116174 dataset was employed as the validation cohort. The risk plot presented a clear separation of survival status between the two groups and the red dots represented deceased patients and blue dots alive patients (right column of Fig. 2B). The validation affirmed that individuals categorized as high-risk experienced poorer prognoses in comparison to those in the low-risk category (right column of Fig. 2C). In this external cohort, the model achieved AUC values of 0.79, 0.76, and 0.92 for 1-year, 3-year, and 5-year survival predictions, respectively (right column of Fig. 2D), indicating a robust predictive performance of the model across different datasets.
Analysis of groups classified as high and low risk based on clinical factors
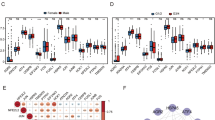
By combining clinical data of liver cancer patients obtained from the TCGA repository with our prognostic model, we categorized patients into high and low-risk groups according to their risk scores19. Our analysis revealed that different age and sex of patients, there were no significant differences in risk scores indicating that both prognostic risks without additional contribution to the model (Fig. 3A,B). In the tumor TNM stage, there was no significant difference in the risk scores for tumor M stage (Fig. 3D), N stage (Fig. 3E), and in the T stage, higher risk scores in patients with T1-T3 stage were generally associated with larger tumor size (Fig. 3C). Likewise, we noticed a connection between elevated risk scores and the progression of stage categories in cancer stages 1 through 3(Fig. 3F). Notably, individuals with elevated risk scores often experienced worse survival outcomes, frequently leaning towards death (Fig. 3G). The importance of the risk score in assessing the progression of liver cancer and the survival of patients is highlighted by this relationship.
Relationship between characteristics and clinical features of HCC patients. There was no difference in risk scores for patients of different genders (A) and ages (B). T grade (C), M grade (D), N grade (E), tumor stage (F) and patient outcomes (G). In the T1–T3 grade and tumor stage 1–3, the risk increased with the increase of the score. As the survival situation (G) score increased, the survival situation became worse. The p-value is displayed as: ns: not significant; *P < 0.05; **P < 0.01; ***P < 0.001.
Independent prognostic indicator of the prognostic risk mode
To determine if the risk score is a vital factor in evaluating the outlook of patients with liver cancer, we performed univariate analyses on the training set specifically for liver cancer. This analysis included clinical factors such as age, gender, stage, and risk score20. The examination revealed that the risk score was highly significant in forecasting the outcome of individuals with liver cancer. Univariate analysis showed that the training set (HR 2.718, 95% CI 2.105 3.511, P < 0.001), the risk score in predicting the prognosis of patients with liver cancer has a significant correlation (Fig. 4A). In addition, additional multivariate analyses were performed, and after adjusting for other confounding factors, the risk score model remained an independent predictor of prognosis in the training set (HR 2.64, 95% CI 1.990 to 3.52, P < 0.001) (Fig. 4B), further supporting the original findings, and the risk score model remained an independent predictor of prognosis in the training set (HR 2.64, 95% CI 1.990 to 3.52, P < 0.001). And stressed the risk score as the importance of the key factors in assessing the prognosis of liver cancer. In addition, stage was also an independent prognostic factor (HR 1.882, 95% CI 1.338–2.464, P < 0.001) (Fig. 4B).
Univariate and multivariate Cox regression analysis in the training set and construction of the nomogram of OS prediction in HCC. (A) Univariate Cox regression analysis in the training set. (B) Multivariate Cox regression analysis in the training set. (C) Columns used to predict the probability of survival in the training set, (right column) columns used to predict the prognosis of HCC patients in the training set, and (left column) calibration charts used to predict 1-year, 3-year, and 5-year survival in the TCGA dataset.
In order to offer healthcare professionals an improved quantitative approach to forecasting the prognosis of patients with LIHC, we created a nomogram that integrates various factors, such as cancer stage and risk score (right column of Fig. 4C). The effectiveness of this nomogram as a prognostic assessment tool was demonstrated through its utilization in estimating the probabilities of survival at 1-year, 3-year, and 5-year intervals. Furthermore, calibration graphs were built, demonstrating a robust agreement between the projected and real survival percentages at 1, 3, and 5 years. This further confirms the accuracy and usefulness of the nomogram in the clinical prognosis of LIHC. The length of each line represents the degree of influence on prognosis, with different values corresponding to different results. In particular, the risk score corresponds to the longest line, indicating its strongest predictive power for prognosis. The column chart as a prognostic assessment tool is effective through its estimated 1 year, 3 years, and 5 years survival probability interval is confirmed. In addition, calibration plots were created that showed strong agreement between predicted and actual survival rates at 1, 3, and 5 years (left column of Fig. 4C). This further confirms the accuracy and utility of the nomogram in the clinical prognosis of LIHC.
Conduct GSVA analysis on groups with high and low-risk
We performed a pathway analysis using the GSVA R package (Gene Set Variation Analysis) for patients classified into high and low-risk groups. The results of our study showed that people in the high-risk category displayed increased activity in metabolic pathways, specifically in the areas of purine metabolism, pyrimidine metabolism, and riboflavin metabolism (Fig. 5). This elevated expression in metabolic pathways suggests a potential link between angiogenesis and altered metabolic processes in liver cancer patients.
The correlation between increased blood vessel formation and alteration of metabolic processes in the high-risk category may suggest a more hostile tumor characteristic, potentially leading to a worse prognosis. This insight not only underscores the importance of angiogenesis in liver cancer pathology but also opens avenues for exploring metabolic pathways as potential therapeutic targets in high-risk liver cancer patients.
Immune infiltration analysis of angiogenesis-related models
We examined the association between our angiogenesis-related model and the infiltration of immune cells by utilizing the CIBERSORT algorithm to estimate the scores of 22 different types of immune cells21. It was noted that individuals classified as high risk exhibited reduced levels of specific immune cell infiltration, such as CD8 T cells, gamma delta T cells, M1 macrophages, M2 macrophages, resting dendritic cells, and resting mast cells (Fig. 6A). Additional examination utilizing the ESTIMATE algorithm indicated that the high-risk group had diminished immune and stromal scores, suggesting a decrease in overall immune infiltration (Fig. 6B). In addition, we analyzed the expression of immune checkpoint genes and observed lower levels of CD200 and CD27 in the high-risk group (Fig. 6C), indicating a possible immune evasion mechanism. Finally, we utilized the TIDE (Tumor Immune Dysfunction and Exclusion) online tool to forecast the TIDE scores for the samples. Despite higher TIDE scores usually suggesting a higher probability of immune evasion, our analysis revealed no notable disparity in TIDE scores between the high and low-risk groups (Fig. 6D). This outcome suggests that while our angiogenesis-related model is strongly associated with varying immune profiles in liver cancer, it does not necessarily correlate with the predicted immune escape potential as assessed by TIDE22. The results emphasize the intricacy of the tumor immune microenvironment in liver cancer and the necessity of employing diverse strategies to comprehend and potentially control these interactions for therapeutic advantage.
Analysis of immune status based on the risk score associated with angiogenesis. (A) The degree of infiltration of 22 types of immune cells between the high and low risk groups in the TCGA dataset. (B) Matrix scores, immune scores and ESTIMATE scores for high- and low-risk groups in the TCGA dataset. (C) Expression of immune checkpoint genes between high- and low-risk groups in the TCGA dataset. (D) TIDE scores for high- and low-risk groups in the TCGA dataset. The p-value is displayed as: ns: not significant; *P < 0.05; **P < 0.01; ****P < 0.001.
Angiogenesis-related models and immunotherapy databases
By employing the IMvigor210 immunotherapy cohort, we developed a risk assessment model using 13 crucial genes. This model unveiled that individuals categorized as high-risk experienced unfavorable prognoses. Surprisingly, in the group of patients undergoing immunotherapy, there was no notable disparity in the response to this treatment between the high-risk and low-risk groups (Left column of Fig. 7A). However, a notable finding was the varying proportions of high and low-risk patients across different cancer stages. In particular, individuals in the high-risk category were found to have a higher occurrence in stages 1 and 3 during the advancement of cancer (Right column of Fig. 7A). This indicates that although the gene expression-based risk model can predict the overall prognosis, its association with the response to immunotherapy is not as evident (Fig. 7B). The effectiveness of immunotherapy is determined by various factors, including tumor microenvironment, immune cell infiltration, and other aspects of the complex tumor-immune interaction, rather than solely relying on gene expression. The prevalence of high-risk patients in early and more advanced stages of cancer highlights the potential utility of this model in early detection and in anticipating aggressive disease progression, which could inform treatment decisions, including the consideration of immunotherapy.
Analysis of angiogenic genes at the single-cell level
In our investigation, single-cell liver cancer dataset GSE14022 was analyzed using the online platform TISCH. Following dimensionality reduction and clustering, cells were categorized into 12 distinct subgroups (Fig. 8A,B). We focused on the expression patterns of four key genes from our prognostic model: AEBP1, ATP2A3, PLAT, and PNMA1. Our analysis revealed that AEBP1 was predominantly expressed in innate lymphoid cells and mast cells (Fig. 8C). ATP2A3 and PNMA1 exhibited widespread expression across various cell subgroups (Fig. 8D,F). Interestingly, PLAT expression was found exclusively in mast cells (Fig. 8E). The differential expression of these genes across cell subgroups highlights their potential roles in tumor biology. The significant expression of AEBP1 in innate lymphoid and mast cells suggests a role in modulating immune responses within the tumor microenvironment. Mast cells, known for their role in inflammation and tissue remodeling, may contribute to tumor progression and angiogenesis, potentially influenced by the expression of PLAT. The broad expression of ATP2A3 and PNMA1 across cell subgroups underscores their ubiquitous importance in liver cancer pathology. These findings provide valuable insights into the cellular heterogeneity of liver cancer and the specific roles of these key genes in different cell types, offering new avenues for targeted therapeutic strategies.
Single-cell analysis reveals major cell types and prognostic gene expression. (A) Uniform manifold approximation and projection (UMAP) maps of different cell clusters. (B) UMAP mapping of major cell types based on specific marker genes. (C) AEBP1 gene expression in each cell type. (D) ATP2A3 gene expression in all cell types. (E) PLAT gene expression in all cell types. (F) PNMA1 gene expression in various cell types.
AEBP1 inhibits the proliferation, migration and invasion of HCCLM3 cells in vitro
In 3 pairs of HCC tissues and corresponding adjacent tissues, RT_qPCR was used to detect the expression levels of 4 prognostic related genes (ATP2A3, AEBP1, PNMA1, PLAT). The level of gene expression of the West_Blot analysis was used to detect liver cancer and adjacent tissues of prognosis of four genes related protein expression level, found that compared with the tissue adjacent to carcinoma, genetic ATP2A3, AEBP1, PNMA1, PLAT were upregulated in liver cancer tissues compared to adjacent tissues (Fig. 9A), The corresponding related proteins are highly expressed in HCC (Fig. 9B). To determine the role of these four genes in HCC, we selected one of them, A1BP1, for cell-level validation. In HCCLM3 cells, knockdown of AEBP1 mediated by siRNA, and CCK8 results showed that knockdown of AEBP1 slowed down the proliferation rate of HCCLM3 cells (Fig. 9C), while Transwell migration and invasion assays confirmed that AEBP1 knockdown inhibited the migration and invasion of HCCLM3 cells (Fig. 9D).
Experimental verification of the expression of four prognostic genes and the functional role of AEBP1. (A) The mRNA expression levels of ATP2A3, PNMA1, AEBP1, and PLAT mRNA in liver cancer tissues and adjacent tissues. (B) Western blot analysis of ATP2A3, PNMA1, AEBP1, and PLAT protein expression levels in liver versus adjacent non-cancerous tissue. (C) CCK8 assayed cell viability after SiRNA-mediated target gene knockdown compared with control and negative control (NC) siRNA. (D) Transwell assays demonstrating the migration and invasion capabilities of HCCLM3 cells, with comparisons made between control, SiRNA-negative, and SiRNA-mediated target gene knockdown groups. Representative images of the invasion and invasion assays are shown on the left, while quantification of migrating and invading cells is displayed on the right.The p-value is displayed as: ns: not significant; *P < 0.05; **P < 0.01; ***P < 0.001.
These findings indicated that ATP2A3, AEBP1, PNMA1, and PLAT were highly expressed in HCC, which was consistent with the bioinformatics findings (Fig. 2B). Among them, after knocking down AEBP1 in HCCLM3 cells, the proliferation, migration and invasion abilities of HCCLM3 cells were decreased.
Discussion
HCC is a leading cause of cancer-related death in many areas of the world. Although the measures of prevention, surveillance, early detection, diagnosis and treatment have been widely implemented, the incidence and mortality of HCC continue to increase in many countries. Angiogenesis is the process of generating new capillaries regulated by angiogenic and anti-angiogenic factors. Angiogenesis allows the tumor cells to provide sufficient oxygen and nutrition, which promotes tumorigenesis, metastasis, and deterioration7,23. As a highly vascularized malignant tumor, HCC produces angiogenic and anti-angiogenic factors under the co-regulation of related genes, thereby regulating the angiogenic process24. High expression of angiogenesis-promoting genes leads to abnormal vasculature and enhances HCC metastasis, while antiangiogenic genes slow HCC progression25. Therefore, it is of great significance to provide prognostic prediction for HCC patients by integrating angiogenesis-related genes as effective and reliable biomarkers for HCC.
In our study, we systematically analyzed the transcript levels of 30 prognostic angiogenesis-related genes in TCGA dataset. LASSO Cox regression analysis was used to screen out 13 prognostic genes related to angiogenesis, and a risk score model was established based on these genes. According to the score, HCC patients were divided into high-risk and low-risk groups. Survival analysis showed that higher scores were significantly associated with poor prognosis of HCC, and the OS of patients in the low-risk group was significantly higher than that in the high-risk group. The predictive power of the risk score model was determined by the time-dependent ROC curve, and the AUC was 0.73, 0.67, and 0.76 in the prediction of 1, 3, and 5 years of generation, respectively, showing relatively robust diagnostic value. In addition, the scoring model was further demonstrated in the validation cohort GSE116174 dataset. By combining the clinical data of liver cancer patients in TCGA database with our prognostic model, we found that the size and stage of HCC were associated with higher scores, and individuals with higher risk scores tended to have poor survival status and tended to die. In addition, principal component analysis further verified that the risk score model could be used as a good predictor for the prognosis of HCC patients, and the risk score model could be used as an independent prognostic factor for clinical outcome. Therefore, this risk score model can be used as a prognostic biomarker for HCC patients.
GSVA analysis showed that the high risk score patients had active purine, pyrimidine, and riboflavin metabolic pathways, which may influence angiogenesis26. Mullen et al. pointed out that enhanced purine metabolic activity promotes the occurrence of hepatocellular carcinoma. Among the hallmarks of cancer, the most important feature of cancer cells is the induction of uncontrolled proliferation, which in turn depends heavily on purine and pyrimidine metabolism27. In HCC patients with poor prognosis, purine metabolism and purine biosynthesis pathway activity are significantly activated28. Yang et al. found that the use of nucleotide reverse transcription inhibitors can inhibit angiogenesis in vivo and in vitro by affecting the proliferation and migration of vascular endothelial cells29. The correlation between increased angiogenesis and altered metabolic processes may indicate that the tumor has more aggressive features that may lead to a worse prognosis. This finding not only highlights the importance of angiogenesis in HCC pathology, but also opens the way to explore metabolic pathways as potential therapeutic targets in high-risk HCC patients.
The Tumor immune microenvironment (TIME) of HCC is a complex mixture of hepatic non-parenchymal resident cells, tumor cells, immune cells and tumor-associated fibroblasts30. The interaction between tumor cells and the TIME plays an important role in the occurrence, progression, metastasis and response to therapy of tumors31. CD8 + T cells, as the cytotoxic T lymphocytes that can recognize tumor-associated antigens and then destroy tumor cells。Besides,
IFN-γ secreted by CD8 cells has been found to be associated with inhibition of tumor angiogenesis by reprogramming tumor-associated macrophages from M2-to M1-like types32. Macrophages can be divided into classical M1 macrophages and M2 macrophages, depending on signals from the surrounding microenvironment. Dendritic cells stimulate angiogenesis by secreting angiogenic factors and cytochemokines to promote the proangiogenic activity of T lymphocytes and trans-differentiation into endothelial cells33. Mast cell recruitment in TIME leads to impaired proinflammatory pathways that contribute to tumor progression, contribute to angiogenesis, promote metastasis, and recruit tumor-associated macrophages to promote tumor responses34. In our study, individuals in the high-risk group had reduced infiltration levels of CD8 T cells, γδT cells, M1 macrophages, M2 macrophages, resting dendritic cells, and resting mast cells, which may be related to increased angiogenesis. High-risk HCC patients had reduced immune cell infiltration and lower survival rates, likely due to increased angiogenesis. The angiogenesis model score can reflect the tumor immune microenvironment of HCC and show a more suppressive immunophenotype. However, immune cell composition is calculated based on various algorithms and remains inaccurate compared to IHC and flow cytometry. Even so, tumor immune microenvironment analysis is still helpful to understand more about tumor immunity. Overall, the angiogenesis model score was significantly correlated with tumor infiltrating cells, suggesting that it may contribute to immune regulation involved in HCC progression.
When constructing the risk score model, 13 genes significantly associated with HCC prognosis were identified by LASSO Cox regression analysis. These 13 genes each have different coefficients in the model, which reflect the degree to which the gene expression level contributes to the risk score. Among them, four genes, ATP2A3, AEBP1, PNMA1 and PLAT, had relatively large absolute values of coefficients in the model, indicating that they had a relatively important impact on the calculation of risk scores and patient risk stratification, and were therefore prioritized as subjects for follow-up studies. By analyzing angiogenesis genes at the single-cell level, it was found that AEBP1 was mainly expressed in innate lymphoid cells and mast cells. ATP2A3 and PNMA1 were widely expressed in different cell subsets. PLAT is only expressed in mast cells. These four genes showed significant differences in single cell analysis.
AEBP1 is a transcriptional repressor involved in the regulation of key biological processes, including lipogenesis, inflammation, macrophage cholesterol homeostasis, and atherogenesis35. AEBP1 mRNA and protein expression has been reported to be upregulated in various cancer samples, supporting a role for AEBP1 in cancer progression. AEBP1 is highly expressed in gastric cancer36, colorectal cancerand37 their cell lines, and the up-regulation of AEBP1 expression is significantly correlated with poor prognosis and low OS rate of patients. Zhang38 found that the expression level of AEBP1 was increased in liver fibrosis, which was associated with poor prognosis of patients. Recently, Yorozu39 found that AEBP1 expression is elevated in tumor endothelial cells within colorectal cancer tissues, and that upregulation of AEBP1 might contribute to tumor angiogenesis. ATP2A3 alterations have been reported in various tumor types and are associated with human susceptibility to a variety of cancers due to regulation of gene transcription and cell proliferation40. ATP2A3 expression is reduced or absent in various cancer tissues, and high ATP2A3 expression can be used as a good prognostic marker for patients with colorectal and gastric cancer41. PLAT is a tumor-related angiogenic factor, which plays a role in tumor progression by activating angiogenesis, cell invasion and mediating cell proliferation42. In addition, Lin43 used whole-human genome oligonucleotide microarrays to detect differentially expressed genes in primary cultured hepatocellular carcinoma cells with an aggressive phenotype (HA 22T/VGH cell line) and found that PLAT had the effect of promoting proliferation, migration and angiogenesis, thereby promoting tumor invasion. PLAT expression is influenced by epigenetic mechanisms44. Recently, a study showed45 that PNMA1 is an oncogene in HCC that contributes to cell proliferation, migration, and invasion.
In the risk scoring model we developed, the genes ATP2A3, AEBP1, PNMA1, and PLAT exhibited large absolute values of coefficients in the model, which had important effects on the calculation of risk scores and risk stratification of patients. Additionally, these four genes demonstrated significant differences in single-cell analysis. Literature review revealed that these genes exert potent biological effects associated with angiogenesis or hepatocellular carcinoma (HCC), with well-defined functional implications. Consequently, their expression levels were assessed via qRT-PCR in three pairs of HCC tissues and corresponding peritumoral tissues. Our findings indicated that ATP2A3, AEBP1, PNMA1, and PLAT were markedly upregulated in HCC tissues relative to adjacent tissues, corroborating the results from bioinformatics analysis. Furthermore, AEBP1 displayed significant differential expression at both the transcriptomic and proteomic levels, and functional analysis suggested that AEBP1 might facilitate angiogenesis and tumor progression by modulating the tumor microenvironment. Therefore, we used wet experiments to verify the role of AEBP1 in HCC. In HCCLM3 cells, the proliferation rate is slowed down after knocking down AEBP1. Tanswell experiment shows that the migration and invasion of HCCLM3 cells are inhibited after knocking down AEBP1. Therefore, we speculate that AEBP1, by promoting angiogenesis, leads to the progression of HCC and thus to the shortened survival of patients. High expression of AEBP1 gene in HCC is significantly associated with poor prognosis of patients. AEBP1 can be a potential therapeutic target for HCC. However, further experiments are needed to prove this.
This study has limitations, including the need for further validation of the 13-gene model in larger clinical datasets. This model then represents almost the best prognostic model associated with angiogenesis by integrating a large number of angiogenesis-related genes but excludes some important genes, such as VEGFA. At the same time, the inherent weakness of assuming only one phenotype to establish a prognostic signature is unavoidable, as many other fundamental prognostic genes in HCC may also be excluded. In addition, the sample size used for wet validation was relatively small, which may limit the statistical power and generalizability of the findings. At the same time, due to limited experimental resources, we only verified the role of AEBP1 in HCC, and the specific mechanism needs to be further studied.
Conclusion
We constructed a risk score model based on 13 angiogenesis-related genes, which serves as a novel prognostic biomarker for hepatocellular carcinoma (HCC) and has the potential to guide the development of more personalized treatment strategies for HCC patients. In addition, knockdown of AEBP1 in HCCLM3 cells inhibited the proliferation, migration and invasion of HCCLM3 cells, indicating that AEBP1 is a potential therapeutic target for HCC.
Data availability
The dataset analysed during the current study are available in the GEO Database (http://www.ncbi.nih/geo/), TCGA Database (http://www.portal.gdc.cancer.gov), GnneCards database (http://www.genecards.org/) and Molecular Signatures Database (MSiDB, http://www.broad.mit.edu/gsea/msigdb). All data analysed during this study are included in the supplementary file.
References
Bray, F. et al. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 74, 229–263. https://doi.org/10.3322/caac.21834 (2024).
Llovet, J. M. et al. Hepatocellular carcinoma. Nat. Rev. Dis. Primers 7, 6. https://doi.org/10.1038/s41572-020-00240-3 (2021).
Chidambaranathan-Reghupaty, S., Fisher, P. B. & Sarkar, D. Hepatocellular carcinoma (HCC): Epidemiology, etiology and molecular classification. Adv. Cancer Res. 149, 1–61. https://doi.org/10.1016/bs.acr.2020.10.001 (2021).
Toh, M. R. et al. Global epidemiology and genetics of hepatocellular carcinoma. Gastroenterology 164, 766–782. https://doi.org/10.1053/j.gastro.2023.01.033 (2023).
Couri, T. & Pillai, A. Goals and targets for personalized therapy for HCC. Hepatol. Int. 13, 125–137. https://doi.org/10.1007/s12072-018-9919-1 (2019).
Oura, K., Morishita, A., Hamaya, S., Fujita, K. & Masaki, T. The roles of epigenetic regulation and the tumor microenvironment in the mechanism of resistance to systemic therapy in hepatocellular carcinoma. Int. J. Mol. Sci. https://doi.org/10.3390/ijms24032805 (2023).
Mossenta, M. et al. New insight into therapies targeting angiogenesis in hepatocellular carcinoma. Cancers https://doi.org/10.3390/cancers11081086 (2019).
Yao, C. et al. Angiogenesis in hepatocellular carcinoma: Mechanisms and anti-angiogenic therapies. Cancer Biol. Med. 20, 25–43. https://doi.org/10.20892/j.issn.2095-3941.2022.0449 (2023).
Llovet, J. M. et al. Sorafenib in advanced hepatocellular carcinoma. N. Engl. J. Med. 359, 378–390. https://doi.org/10.1056/NEJMoa0708857 (2008).
Yan, T. et al. The advanced development of molecular targeted therapy for hepatocellular carcinoma. Cancer Biol. Med. 19, 802–817. https://doi.org/10.20892/j.issn.2095-3941.2021.0661 (2022).
Ribatti, D., Annese, T., Ruggieri, S., Tamma, R. & Crivellato, E. Limitations of anti-angiogenic treatment of tumors. Transl. Oncol. 12, 981–986. https://doi.org/10.1016/j.tranon.2019.04.022 (2019).
Dong, Z. R. et al. TMPRSS4 drives angiogenesis in hepatocellular carcinoma by promoting HB-EGF expression and proteolytic cleavage. Hepatology 72, 923–939. https://doi.org/10.1002/hep.31076 (2020).
Wen, Y. et al. Bclaf1 promotes angiogenesis by regulating HIF-1α transcription in hepatocellular carcinoma. Oncogene 38, 1845–1859. https://doi.org/10.1038/s41388-018-0552-1 (2019).
Zhen, Z., Shen, Z., Hu, Y. & Sun, P. Screening and identification of angiogenesis-related genes as potential novel prognostic biomarkers of hepatocellular carcinoma through bioinformatics analysis. Aging 13, 17707–17733. https://doi.org/10.18632/aging.203260 (2021).
Zhao, S. et al. Expression of hub genes of endothelial cells in glioblastoma-A prognostic model for GBM patients integrating single-cell RNA sequencing and bulk RNA sequencing. BMC Cancer 22, 1274. https://doi.org/10.1186/s12885-022-10305-z (2022).
Zhu, G. et al. Prognostic value of ferroptosis-related genes in patients with lung adenocarcinoma. Thorac. Cancer 12, 1890–1899. https://doi.org/10.1111/1759-7714.13998 (2021).
Yan, C., Liu, Q. & Jia, R. Construction and validation of a prognostic risk model for triple-negative breast cancer based on autophagy-related genes. Front. Oncol. 12, 829045. https://doi.org/10.3389/fonc.2022.829045 (2022).
Cai, J. et al. Development and validation of a novel endoplasmic reticulum stress-related lncRNA prognostic signature and candidate drugs in breast cancer. Front. Genet. 13, 949314. https://doi.org/10.3389/fgene.2022.949314 (2022).
Xie, S. et al. Construction of a hypoxia-immune-related prognostic model and targeted therapeutic strategies for cervical cancer. Int. Immunol. 34, 379–394. https://doi.org/10.1093/intimm/dxac017 (2022).
Zhang, Y., Li, T., Liu, H. & Wang, L. Function and prognostic value of basement membrane-related genes in lung adenocarcinoma. Front. Pharmacol. 14, 1185380. https://doi.org/10.3389/fphar.2023.1185380 (2023).
Ma, Y. et al. Comprehensive molecular analyses of a TNF family-based gene signature as a potentially novel prognostic biomarker for cervical cancer. Front. Oncol. 12, 854615. https://doi.org/10.3389/fonc.2022.854615 (2022).
Zhang, J. et al. T cell-related prognostic risk model and tumor immune environment modulation in lung adenocarcinoma based on single-cell and bulk RNA sequencing. Comput. Biol. Med. 152, 106460. https://doi.org/10.1016/j.compbiomed.2022.106460 (2023).
Kasprzak, A. & Adamek, A. Role of endoglin (CD105) in the progression of hepatocellular carcinoma and anti-angiogenic therapy. Int. J. Mol. Sci. https://doi.org/10.3390/ijms19123887 (2018).
Huang, A., Yang, X. R., Chung, W. Y., Dennison, A. R. & Zhou, J. Targeted therapy for hepatocellular carcinoma. Signal Transd. Target. Ther. 5, 146. https://doi.org/10.1038/s41392-020-00264-x (2020).
Ribatti, D., Vacca, A., Nico, B., Sansonno, D. & Dammacco, F. Angiogenesis and anti-angiogenesis in hepatocellular carcinoma. Cancer Treatm. Rev. 32, 437–444. https://doi.org/10.1016/j.ctrv.2006.06.002 (2006).
Teuwen, L. A., Geldhof, V. & Carmeliet, P. How glucose, glutamine and fatty acid metabolism shape blood and lymph vessel development. Dev. Biol. 447, 90–102. https://doi.org/10.1016/j.ydbio.2017.12.001 (2019).
Mullen, N. J. & Singh, P. K. Nucleotide metabolism: A pan-cancer metabolic dependency. Nat. Rev. Cancer 23, 275–294. https://doi.org/10.1038/s41568-023-00557-7 (2023).
Yang, S. et al. A novel purine and uric metabolism signature predicting the prognosis of hepatocellular carcinoma. Front. Genet. 13, 942267. https://doi.org/10.3389/fgene.2022.942267 (2022).
Song, L. et al. Nucleoside/nucleotide reverse transcriptase inhibitors attenuate angiogenesis and lymphangiogenesis by impairing receptor tyrosine kinases signalling in endothelial cells. Br. J. Pharmacol. 175, 1241–1259. https://doi.org/10.1111/bph.14036 (2018).
Sangro, B., Sarobe, P., Hervás-Stubbs, S. & Melero, I. Advances in immunotherapy for hepatocellular carcinoma. Nat. Rev. Gastroenterol. Hepatol. 18, 525–543. https://doi.org/10.1038/s41575-021-00438-0 (2021).
Flecken, T. et al. Immunodominance and functional alterations of tumor-associated antigen-specific CD8+ T-cell responses in hepatocellular carcinoma. Hepatology 59, 1415–1426. https://doi.org/10.1002/hep.26731 (2014).
Kim, H. J., Ji, Y. R. & Lee, Y. M. Crosstalk between angiogenesis and immune regulation in the tumor microenvironment. Arch. Pharm. Res. 45, 401–416. https://doi.org/10.1007/s12272-022-01389-z (2022).
David Dong, Z. M., Aplin, A. C. & Nicosia, R. F. Regulation of angiogenesis by macrophages, dendritic cells, and circulating myelomonocytic cells. Curr. Pharm. Des. 15, 365–379. https://doi.org/10.2174/138161209787315783 (2009).
Lichterman, J. N. & Reddy, S. M. Mast cells: A new frontier for cancer immunotherapy. Cells https://doi.org/10.3390/cells10061270 (2021).
Majdalawieh, A. F., Massri, M. & Ro, H. S. AEBP1 is a novel oncogene: Mechanisms of action and signaling pathways. J. Oncol. 2020, 8097872. https://doi.org/10.1155/2020/8097872 (2020).
Liu, J. Y. et al. AEBP1 promotes epithelial-mesenchymal transition of gastric cancer cells by activating the NF-κB pathway and predicts poor outcome of the patients. Sci. Rep. 8, 11955. https://doi.org/10.1038/s41598-018-29878-6 (2018).
Xing, Y. et al. AEBP1, a prognostic indicator, promotes colon adenocarcinoma cell growth and metastasis through the NF-κB pathway. Mol. Carcinogen. 58, 1795–1808. https://doi.org/10.1002/mc.23066 (2019).
Zhang, W. et al. Fibroblast-specific adipocyte enhancer binding protein 1 is a potential pathological trigger and prognostic marker for liver fibrosis independent of etiology. J. Digest. Dis. 24, 550–561. https://doi.org/10.1111/1751-2980.13230 (2023).
Yorozu, A. et al. Upregulation of adipocyte enhancer-binding protein 1 in endothelial cells promotes tumor angiogenesis in colorectal cancer. Cancer Sci. 111, 1631–1644. https://doi.org/10.1111/cas.14360 (2020).
Themistocleous, S. C. et al. Clinical significance of P-class pumps in cancer. Oncol. Lett. 22, 658. https://doi.org/10.3892/ol.2021.12919 (2021).
Hernández-Oliveras, A. et al. Histone deacetylase inhibitors promote ATP2A3 gene expression in hepatocellular carcinoma cells: p300 as a transcriptional regulator. Int. J. Biochem. Cell Biol. 113, 8–16. https://doi.org/10.1016/j.biocel.2019.05.014 (2019).
Ortiz-Zapater, E. et al. Tissue plasminogen activator induces pancreatic cancer cell proliferation by a non-catalytic mechanism that requires extracellular signal-regulated kinase 1/2 activation through epidermal growth factor receptor and annexin A2. Am. J. Pathol. 170, 1573–1584. https://doi.org/10.2353/ajpath.2007.060850 (2007).
Lin, Z. Y. & Chuang, W. L. Genes responsible for the characteristics of primary cultured invasive phenotype hepatocellular carcinoma cells. Biomed. Pharmacother. 6, 454–458. https://doi.org/10.1016/j.biopha.2012.04.001 (2012).
Lin, C. Y. et al. ADAM9 promotes lung cancer progression through vascular remodeling by VEGFA, ANGPT2, and PLAT. Sci. Rep. 7, 15108. https://doi.org/10.1038/s41598-017-15159-1 (2017).
Liu, P., Chen, B., Gu, Y. & Liu, Q. PNMA1, regulated by miR-33a-5p, promotes proliferation and EMT in hepatocellular carcinoma by activating the Wnt/β-catenin pathway. Biomed. Pharmacother. 108, 492–499. https://doi.org/10.1016/j.biopha.2018.09.059 (2018).
Acknowledgements
This work was financially supported by the National Natural Science Foundation of China [82260363]; Guizhou Provincial Department of Science and Technology, Guizhou Science and Technology Plan Project "14th Batch of Outstanding Young Scientific and Technological Talents Program", QKH Platform Talent-YQK[2023]032; Guizhou Province Health Committee of Science and Technology Fund [gzwkj2023-069, gzwki2021-382]; Guizhou Medical University Affiliated Hospital, "2023 Excellent Reserve Talents of Discipline"[gyfyxkrc-2023-06]; Doctoral Starting Fund of Guizhou Medical University[gyfybsky-2021-7];Key Laboratory for Cancer Prevention and treatment of Guizhou Province:QKHPT[2025]031.
Author information
Authors and Affiliations
Contributions
GDG and LY equally performed the majority of the work. GDG and LY performed bioinformatic analyses. LY conducted wet-lab experiments. JTP provided bioinformatics technical support. LY and JTP prepared the figures. DQH proofread the manuscript. HZ and DQH designed the research and summarized the data. GDG wrote the paper. All authors contributed to the article and approved the submitted version.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval
The study involving human participants were approved by the Ethics Review Committee of Guizhou Medical University (NO.2022375). The all patients provided their written informed consent in accordance with the Declaration of Helsinki.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Gao, D., Lu, Y., Jiang, T. et al. To construct and validate a risk score model of angiogenesis-related genes to predict the prognosis of hepatocellular carcinoma. Sci Rep 15, 4660 (2025). https://doi.org/10.1038/s41598-025-87459-w
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-87459-w