Abstract
Immunotherapies are effective for cancer treatment but are limited in ‘cold’ tumor microenvironments due to a lack of infiltrating CD8+ T cells, key players in the anti-cancer immune response. The onset of the COVID-19 pandemic sparked the widespread use of mRNA-formulated vaccines and is well documented that vaccination induces a Th1-skewed immune response. Here, we evaluated the effects of an intratumoral injection of the mRNA COVID-19 vaccine in subcutaneous melanoma tumor mouse models. Tumor growth and survival studies following a single intratumoral injection of the COVID-19 vaccine showed significant tumor suppression and prolonged survival in established B16F10 subcutaneous tumor-bearing mice. mRNA vaccine treatment resulted in a significant increase in CD8+ T cell infiltration into the tumor microenvironment, as observed using intravital imaging and flow cytometry. Further tumor growth suppression was achieved using additional mRNA vaccine treatments. Combination administration of mRNA vaccine with immune checkpoint therapies demonstrated enhanced effects, further delaying tumor growth and improving the survival time of tumor-bearing mice. This study demonstrates that mRNA vaccines may be used as adjuvants for immunotherapies.
Similar content being viewed by others
Introduction
CD8+ T cells are the major effector immune cells driving anti-cancer immune responses1,2. Studies correlate more favorable survival in melanoma patients with increased infiltration of CD8+ T cells within the tumor microenvironment3,4,5. The presence of intratumoral T cells are also another major positive predictive factor for immunotherapy response6,7. Tumors with high CD8+ T cell infiltration have demonstrated improved responses to immune checkpoint therapy compared to tumors where T cell infiltration is low8,9,10,11,12,13. Although immune checkpoint therapy has demonstrated a durable response in a subset of melanoma patients, the majority of patients do not respond to immunotherapy14,15,16,17,18. This is thought to occur due to the high prevalence of “cold” tumor microenvironments with low immune T cell infiltration in this type of cancer19,20.Development of tumor microenvironment modulating strategies that increase CD8+ T cell recruitment to the tumor site are needed to convert cold tumor microenvironments to hot, immune cell-rich microenvironments increasing anti-cancer responses21,22.
The use of chemokines and cytokines to attract T cells to the tumor site have been explored with varying results23,24,25,26. Feasibility of systemic administration of cytokines is limited due to the risk of inducing a cytokine storm – a condition where aberrant inflammation can cause damage to host tissues and systems27,28. Alternative indirect routes of boosting cytokine and chemokine responses are an attractive approach due to safety. Vaccines, using low doses of antigens to introduce immunological memory responses without exposure to a whole pathogen, can cause mild and transient increases in cytokines. Thus, interest has risen in the use of intratumoral, rather than systemic, injection of vaccines to enhance T cell recruitment locally to the tumor microenvironment. Several studies utilizing off-the-shelf vaccines have demonstrated promising effects29,30,31. Intratumoral injection of FDA-approved flu vaccine in combination with tetanus/pertussis/diphtheria vaccines demonstrated tumor regression with increased infiltration of immune cells29,30. There are reports of clinical regression of tumors following administration of the pertussis acellular vaccine31 and the COVID-19 vaccine32. In the latter case, intramuscular vaccination with mRNA COVID-19 vaccine resulted in spontaneous tumor regression with localized inflammatory phenotypes in the tumor microenvironment32. This interesting interplay between systemic vaccination and localized effects led to our hypothesis that immune responses due to mRNA COVID-19 vaccination could drive CD8+ T cells into the tumor if administered via an intratumoral injection route. Here, we demonstrate that the intratumoral injection of a mRNA COVID-19 vaccine elicits anti-tumor responses and enhances immunotherapy efficacy. These data pave the path to the next generation of cancer treatments and the development of cancer-related mRNA adjuvant-based therapies.
Results
mRNA COVID-19 immunization results in an increase of CD8+ T cells
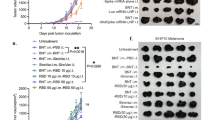
The onset of the COVID-19 pandemic sparked the widespread use of mRNA-formulated vaccines. COVID-19 SpikeVax (mRNA-1273) vaccine, has been extensively studied in both pre-clinical and clinical studies, and it is well documented that vaccination induces a Th1-skewed immune response33,34. To determine if mRNA-1273 induced a local immune response at the site of administration, we delivered a high-dose intramuscular injection of mRNA-1273 vaccine and observed a significant increase in CD8+ T cells in the muscle and lymph nodes 24 h compared to PBS control in healthy C57BL/6 mice (Fig. 1A & B). Given the induced localization of CD8+ T cells from mRNA-1273 vaccination, we hypothesized that mRNA-1273 would enhance CD8+ T cells into the tumor microenvironment. In vitro, we observed that B16F10 melanoma cells express the SARS-CoV-2 spike protein after incubation with mRNA-1273, with detectable protein levels at 24 h, 48 h and 72 h post incubation (Fig. 1C). Uptake of mRNA-1273 lipid nanoparticle by B16F10 cells corresponded to high production of CXCL10, a chemokine involved in T cell recruitment, in culture media (Fig. S1A)35,36,37,38.
mRNA-1273 vaccine immunization increases CD8 + T cell infiltration at the injection site and is uptaken by B16F10 melanoma cells. Flow cytometry of CD8+ T cells in the (A) muscle of injection site and (B) draining lymph node 24 h post immunization with 10 µg of mRNA-1273 or PBS control (n = 5 mice/group, n = 10 mice total). Data shown as mean (SD); unpaired student t test, **p = 0.0081, ***p < 0.001. (C) Cropped Western blot of spike protein in B16F10 cells at different timepoints after incubation with 200 µL (40 µg) mRNA-1273 (n = 3 independent experiments). Original full blots are presented in Supplementary Fig. 14.
A single intratumoral injection of mRNA COVID-19 vaccine significantly delays tumor growth
To evaluate the potential effects of mRNA-1273 vaccination on melanoma tumors, we utilized a subcutaneous B16F10 tumor model. When B16F10 tumors reached 5 mm in diameter, an intratumoral (IT) injection of 3 µg mRNA-1273 or empty lipid nanoparticle (LNP) without mRNA or PBS occurred. A statistically significant delay in tumor growth was observed in tumors treated with mRNA-1273 compared to those that received LNP or PBS (Fig. 2A – C); at day 8 post IT injection, LNP and PBS tumors were 2-fold greater in size compared to tumors treated with mRNA-1273. While a significant difference in tumor volume was observed between LNP and PBS at 4 days post treatment (Fig. 2B), this difference was lost by day 8 (Fig. 2C). With this delay in tumor growth, we also observed a significant increase in survival time for mice treated with mRNA-1273 compared to PBS (Fig. S1B). To confirm these findings in another melanoma model, we repeated these experiments using the YUMM1.G1 melanoma cell line39, and found that IT injection of 3 µg mRNA-1273 again significantly delayed tumor growth compared to PBS (Fig. S1C-E). These data suggest that an IT injection of mRNA-1273 results in tumor reduction.
mRNA-1273 vaccine intratumoral treatment reduces tumor growth and increases CD8+ T cells in the tumor. (A) B16F10 tumor growth curves and tumor volume measurements at (B) 4 days and (C) 8 days following a 20 µL intratumoral injection of 3 µg mRNA-1273, empty LNP or PBS (n = 5 mice/group, n = 15 mice total). Day 0 represents the time of IT injection when tumors reached 5 mm in diameter. (D) Representative intravital image of tumor (GFP, green) and CD8+ T cells (CD8a+, red) prior to (Pre) and 24 h post (24 h) 10 µL IT injection of 1.5 µg mRNA-1273 or PBS; scale bar represents 100 μm. (E) Corresponding quantification of the number of CD8+ T cells in the tumor (n = 5 mice treated with mRNA-1273, n = 4 mice treated with PBS, n = 9 mice total; n = 3 images per mouse per time point). Flow cytometry analysis of (F) CD8+ T cell subsets within the tumor 4 days post IT injection (n = 3 mice/group, n = 9 mice total). Data shown as mean (SD); one-way or two-way ANOVA followed by Tukey’s multiple comparison test, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
mRNA-1273 increases CD8+T cell in the tumor microenvironment
Using intravital imaging with skinfold window chamber murine models40,41, we evaluated changes in CD8+ T cell frequency within the tumor microenvironment following IT injection of mRNA-1273 or PBS (Fig. 2D). A significant increase in CD8+ T cells in B16F10 tumors 24 h post treatment with mRNA-1273 was observed (Fig. 2E). Similar to the subcutaneous tumor studies, a significant reduction in tumor size was observed as well in the intravital studies by quantifying the GPF area as a measure of B16F10 tumor size, where the fold change normalized to pre-treatment size was < 1 when treated with mRNA-1273, indicating a reduction in tumor size (Fig. S2A & B). Using flow cytometry, the immune cell infiltration profile was evaluated. Similar to the intravital imaging and subcutaneous tumor studies, a significant decrease in B16F10 tumor volume was observed 24 h post IT injection (Fig. S2C). No difference in CD3+ or CD11b+ cells were observed at 24 h following IT injection with mRNA-1273 or PBS (Fig. S2D). We further characterized the immune cell infiltrates in the tumor by flow cytometry at 4 days post IT injection when the largest difference in tumor volume between experimental groups was observed. At 4 days post IT injection, a decrease in CD3+ cells (p = 0.05) but not in CD11b+ cells were observed between mRNA-1273 and LNP (Fig. S2E). It should be noted that the IT injection itself appeared to enhance the infiltration of CD3+ or CD11b+ cells within the tumor as untreated B16F10 tumors had minimal immune infiltrates - less than 2% of CD45+ immune cells were CD3+ or CD11b+ at the experimental tumor endpoint (Fig. S2F). Interestingly, the majority of the immune cells within the tumor microenvironment at 24 h were CD3+CD8+ T cells where no differences in population size were observed in tumors treated with mRNA-1273 or PBS (Fig. S2G). Looking at the specific subsets of CD8+ T cells, a significant increase in CD8+CD44+PD-1+ T cells was observed in tumors treated with mRNA-1273 (Fig. S2G). However, PBS administration resulted in a significant increase in CD8+GranzymeB+ T cells compared to mRNA-1273 treatment (Fig. S2G). While the CD4+ T cell population made up less than 1% of the immune infiltrate, we observed a significant increase in CD4+ T cells in tumors treated with mRNA-1273 (Fig. S2H). A significant increase in CD8+ T cells was observed in mRNA-1273 treated B16F10 tumors compared to LNP and PBS controls 4 days post IT injection (Fig. 2F). Approximately two-thirds of the CD8+ T cells from mRNA-1273 treatment were Granzyme B+, which was a significant increase of 7-fold and 10-fold when compared to LNP or PBS controls (Fig. 2F). Approximately half of these CD8+GranzymeB+ T cells were CD62L+ and was also significantly increased in B16F10 tumors treated with mRNA-1273 compared to LNP or PBS controls, 112-fold and 59-fold increase respectively (Fig. 2F). All CD8+ T cells lacked IFN-γ expression. At 4 days post IT injection, an increase in CD4+ T cells was seen in which a significant increase in CD4+CD44+CD62L+ (93-fold and 49-fold increase) and CD4+CD62L+ T cells (209-fold and 67-fold increase) were observed in B16F10 tumors treated with mRNA-1273 compared to LNP or PBS controls (Fig. S2I). The specificity of the CD8+ T cells within B16F10 tumors were also evaluated using TRP-2 staining by flow cytometry. A significant increase of 4.5-fold in TRP-2+CD8+ T cells was observed at 24 h but was lost by 4 days post IT mRNA-1273 treatment (Fig. S2J & K). Approximately 20% of CD8+ T cells were TRP-2+ at 24 h post IT injection of mRNA-1273 (Fig. S2J). There were no differences observed in T cell subsets in the spleen at 24 h or 4 days post IT injection of mRNA-1273, LNP or PBS (Fig. S3). No significant differences were seen in T cell subsets in the draining lymph node at 24 h post IT injection of mRNA-1273, PBS or LNP (Fig. S4A & B). Interestingly, there was a significant increase in CD8+ T cells, CD4+ T cells and CD4+CD44+ T cells in the draining lymph node following treatment with PBS compared to mRNA-1273 or LNP at 4 days post injection (Fig. S4C & D). These data demonstrate enhanced CD8+ and CD4+ T cell subset infiltration into the tumor microenvironment following IT treatment with the COVID-19 vaccine.
Multiple mRNA 1273 IT injections lead to enhanced reduction of tumor growth
While a significant decrease in tumor volume following a single mRNA-1273 IT injection was observed (Fig. 2A), we did not observe long-term survival or complete tumor regression. We evaluated the anti-tumor responses following multiple mRNA-1273 IT injections. IT injections began when tumors reached 5 mm in diameter and were administered every 3 days for a total of 3 doses. A significant delay in tumor growth was observed following three 20 µL IT injections of 3 µg mRNA-1273 compared to LNP or PBS controls (Fig. 3A). At 8 days post injection, a statistically significant delay in tumor growth was observed in tumors treated with mRNA-1273 compared to LNP or PBS as well as between LNP and PBS (Fig. S5A). A significant increase in survival was observed when B16F10 tumors were treated with three IT doses of mRNA-1273 compared LNP or PBS (Fig. 3B). No differences in survival were observed between treatment with three IT doses of LNP or PBS (Fig. 3B). Compared to a single dose of mRNA-1273, three IT injections significantly decreased tumor volume at day 8 (Fig. 3C). While we observe an increase in the median survival from 27 to 34 days for a single mRNA-1273 dose compared to multiple mRNA-1273 injections, this increase was not found to be statistically significant using a Log-rank test (Fig. S5B). One mouse developed ulcerations exceeding 0.5 mm in length, likely due to multiple IT injections, necessitating euthanasia before the tumor reached the endpoint size of over 1.5 mm. This earlier endpoint may account for the lack of significant differences in survival when comparing single versus multiple administrations of mRNA-1273. Whether an IT injection of mRNA-1273 could induce abscopal effects, where treatment of one tumor induces positive effects on a secondary distal tumor, was also evaluated. Mice were implanted with tumors on the right and left flanks, where only the right flank was treated with multiple mRNA-1273 IT injections or PBS control. A significant reduction in both the IT mRNA-1273 treated right and untreated left tumor was observed compared to PBS treated control mice at day 8 post IT treatment (Fig. 3D & S5C). However, the tumor reduction in the abscopal studies was not similarly delayed as that observed when a single tumor was treated with multiple mRNA-1273 doses (Fig. S5D). These data show that multiple intratumoral treatments with the COVID-19 vaccine significantly reduced tumor volume and induced partial abscopal effects.
Enhanced tumor volume reduction results from using multiple mRNA-1273 IT injections. B16F10 (A) tumor growth curves and (B) survival curves following three 20 µL IT injections of 3 µg mRNA-1273, empty LNP or PBS (n = 5 mice treated with mRNA-1273 and LNP, n = 3 mice treated with PBS, n = 13 mice total); hazard ration (HR) of mRNA-1273/LNP is 0.043 with log-rank (Mantel-Cox) test, **p = 0.0035, HR of mRNA-1273/PBS is 0.024 with log-rank (Mantel-Cox) test, **p = 0.0082 and HR of LNP/PBS is 0.16 with log-rank (Mantel-Cox) test, p > 0.5. (C) B16F10 tumor volume measurements at 8 days post IT injection comparing three doses of mRNA-1273 versus a single mRNA-1273 IT dose (single dose mRNA-1273 IT data is same dataset as shown in Fig. 2C, n = 5 mice/group, n = 10 mice total). Abscopal effects of mRNA-1273 was evaluated using mice implanted with tumors on the right and left flanks. (D) B16F10 tumor volume measurements of treated right and untreated left flank tumors at day 8 post IT injection using three 20 µL IT injections of 3 µg mRNA-1273 or PBS (n = 5 mice treated with mRNA-1273 and n = 3 mice treated with PBS, n = 8 mice total). Data shown as mean (SD); unpaired student t test or two-way ANOVA followed by Tukey’s multiple comparison test, *p < 0.05, **p < 0.01, ***p < 0.001.
mRNA COVID-19 vaccine enhances immune checkpoint therapy response
Given the increase in CD8+ and CD4+ T cells from IT administration of COVID-19 vaccine, we evaluated the possibility of mRNA-1273 IT treatment synergizing with immune checkpoint therapy (ICT). Four days post IT injection with mRNA-1273 or LNP, ICT treatment with anti-PD-1 and anti-CTLA-4 followed for three doses 3 days apart. A significant reduction in B16F10 tumor volume was observed from combination mRNA-1273 and ICT (Fig. 4A & S6A). It should be noted that combination PBS and ICT control group reached tumor endpoint, tumor diameter ≥ 15 mm, before ICT treatments were completed (data not shown). A significant increase in survival was observed when B16F10 tumors were treated with this combination of mRNA-1273 and ICT compared to LNP with ICT (Fig. 4B). However, no difference in tumor growth curves or survival was observed between a single IT mRNA-1273 with or without ICT (Fig. S6B & C). We also evaluated if adjusting the ICT treatment schedule enhanced immunotherapy responses. A single IT injection of mRNA-1273 followed by ICT treatment 1 day later did not demonstrate a significant delay in B16F10 tumor growth compared to PBS and ICT treatment (Fig. S6D & E). However, a single IT injection of mRNA-1273 followed by ICT treatment 1 day later did significantly delay YUMM1.G1 tumor growth compared to PBS with ICT (Fig. S6F). We then combined multiple mRNA 1273 injections with ICT treatments starting 1 day post IT injection for a total of 3 treatments, and also observed a significant decrease in tumor volume from three doses of mRNA-1273 and ICT compared to combination LNP and ICT (Fig. 4C, S6G & H). A significant increase in survival time was observed from multiple treatments of combination mRNA-1273 and ICT (Fig. 4D). Multiple mRNA-1273 treatments further enhanced ICT efficacy as tumor growth was significantly reduced compared to multiple IT mRNA-1273 administration alone (Fig. S6I – K). In addition, the combination of multiple mRNA-1273 treatments with ICT increased survival time significantly compared to multiple mRNA-1273 treatments alone (Fig. S6L). We additionally immunized mice intramuscularly with mRNA-1273 prior to IT treatment with the COVID-19 vaccine to evaluate whether educating the immune system to the spike protein would further enhance melanoma reduction (Fig. S7). However, immunizing mice with mRNA-1273 vaccine did not enhance the anti-tumor effects of IT mRNA-1273 treatments when combined with ICT (Fig. S7E & F). How treatment using combination IT COVID-19 vaccine with ICT affected the tumor immune composition was evaluated. A significant increase in CD3+ cells but not CD11b+ cells was observed at tumor endpoint analyzed by flow cytometry in tumors treated with combination mRNA-1273 and ICT (Fig. S8A). A significant increase in CD8+ T cells was seen with approximately 25% of the CD8+ cells being IFNy+. Roughly 90% of the CD8+ T cells were also CD44+, where the majority of these cells were PD-1+. Interestingly, the endpoint tumors from combination of multiple LNP treatments with ICT had an increase in CD8+CD62L+, CD8+CD62L+PD-1+, CD8+CD44+CD62L+, and CD8+CD44+CD62L+PD-1+ T cells when compared to the mRNA-1273 with ICT treatment group (Fig. S8B). There were no differences seen in the CD4+ T cells subsets (Fig. S8C). These data demonstrate that IT administration of the COVID-19 vaccine enhanced immunotherapy efficacy.
Enhanced therapeutic effects of mRNA-1273 and immune checkpoint therapy (ICT). B16F10 (A) tumor growth curves and (B) survival curves following a single 20 µL IT injections of 3 µg mRNA-1273 or empty LNP combined with ICT 4 days later (n = 4 mice/group, n = 8 mice total); *p = 0.043, **p = 0.0016. PBS + ICT control group reached tumor endpoint before ICT treatments were completed. HR of mRNA-1273 + ICT/LNP + ICT is 0.050 with log-rank (Mantel-Cox) test, *p = 0.011. B16F10 (C) tumor growth curves (*** p < 0.001, ****p < 0.0001) and (D) survival curves following 3 doses of a 20 µL IT injections of 3 µg mRNA-1273 or empty LNP followed by ICT 1 day later (n = 5 mice/group, n = 10 mice total); HR of mRNA-1273 + ICT 3X/LNP + ICT 3X is 0.043 with log-rank (Mantel-Cox) test, *p = 0.0039. Tumor growth data shown as mean (SD), unpaired student t test comparing tumor volume post treatment.
IT injection of mRNA-1273 does not result in systemic cytokine responses or transcriptional changes in the tumor
Changes in cytokines in the tumor, lymph node, and sera at 48 h following an IT injection of mRNA-1273 was evaluated to determine if there were any systemic responses. Interestingly, no significant differences were observed between mRNA-1273 or PBS treated tumor-bearing mice in the tumor, lymph node or sera among proinflammatory cytokines at the 48 h timepoint (Fig. 5A). Similarly, even after multiple doses of IT mRNA-1273, we did not measure seroconversion of anti-SARS-CoV-2 RBD IgG. Conversely, as expected we measured high titers of anti-RBD two weeks after a single dose of intramuscular administered mRNA-1273 (Fig. S9). This data suggests that IT injection of mRNA-1273 does not induce a systemic cytokine response.
IT injection of mRNA-1273 does not alter cytokines or have transcriptional effects. (A) Cytokine levels of tumor, inguinal lymph node, and sera at 48 h after IT injection with 3 µg mRNA-1273 (mRNA) or PBS. Heat map shown depicts mean pg/ml of each cytokine log10 transformed (n = 5 mice/group, n = 10 mice total). tSNE plot of (B) immune cell types separated by treatment groups and (C) the cells defined by their single-cell transcriptome analysis from combined mRNA-1273 and PBS IT injections. (D) The proportions of cell projections in tumors treated with mRNA-1273 or PBS IT injections. (B–D) scRNAseq analysis was performed using pooled samples (n = 5 mice/group, n = 10 mice total), prior to library preparation.
The transcriptional effects of mRNA-1273 using single cell RNA sequencing (scRNAseq) was also characterized. We observed the largest differences in tumor size at 4 days post IT injection, thus, we hypothesized that cytotoxic CD8+ T cell activity would be increased at this timepoint. Subcutaneous tumors from tumor-bearing mice IT injected with mRNA-1273 or PBS were harvested, pooled, and live cell sorted prior to library preparation and sequencing, to remove low-quality reads. As expected, mRNA-1273 treated tumor size and total cell numbers were reduced compared to tumors treated with PBS (Fig. S10). Our analysis found 1178 cells detected from the samples (Fig. 5B). We identified that 729 cells were from PBS treated tumors and 449 were belonging to mRNA-1273 treated tumors. IT injection of mRNA-1273 did not result in significant changes in cellular population clustering between PBS and mRNA-1273 treated tumors at this timepoint (Fig. 5C & D). An analysis comparing the proportion of identified clusters did not reveal increases in CD8+ T cells (Fig. 5D). Furthermore, no differences in proinflammatory genes such as Cxcl10 were observed (data not shown). Together the cytokine and transcriptomic analyses suggest that IT injection of mRNA-1273 does not induce a sustained proinflammatory response systemically or in the tumor microenvironment.
Discussion
Historically, infectious disease vaccines have been repurposed to support cancer treatments due to the inflammatory milieu which results. In one example, the highly inflammatory Bacillus Calmette-Guerin vaccine has been used to treat bladder cancer in a tumor-nonspecific manner. In another, clinical reports have shown a correlation between recent influenza vaccination and a reduced likelihood of post-operative metastatic disease due to improved natural killer cell activity. Administering canonical infectious disease vaccines into tumors has also shown early success, directing the inflammatory responses into the tumor microenvironment to guide anti-cancer functions42,43,44. mRNA vaccines, utilized to control the COVID-19 pandemic are known to induce strong inflammatory responses including CD8+ T cell responses. The reactogenicity of these vaccines is likely inherent to the RNA package itself, the lipids which stimulate IL-6 production, and residual DNA template contaminant left in the formulation post-purification. Thus, we aimed to consider how mRNA COVID-19 vaccination could impact melanoma progression in a preclinical murine model. Our study proposes that intratumoral (IT) injection of the mRNA-1273 COVID-19 vaccine (Moderna) impedes melanoma growth by enhancing CD8+ T cells into the tumor microenvironment, significantly prolonging survival. A single dose treatment of mRNA-1273 IT significantly decreases tumor volume in two murine melanoma models, both of which are resistant to immune checkpoint therapy (ICT). Specifically, our study shows that IT treatment using mRNA-1273: (1) reduces tumor volumes on its own, (2) reduces tumors that are ICT resistant, and (3) improves outcomes from anti-PD-1 and anti-CLTA-4 ICT.
Our findings reveal that in vitro, B16F10 melanoma cells effectively internalize the mRNA-1273 vaccine LNPs and exhibit robust expression of the encoded SARS-CoV-2 Spike antigen (Fig. 1). In line with prior studies, expression of the spike antigen correlated with high expression of the C-X-C motif chemokine ligand 10 (CXCL10) in cell culture medium. CXCL10 downstream of IFN-γ has been implicated many times as an immunological signature following mRNA vaccination45,46. Studies have demonstrated that T cell recruitment into tumor sites are predominately driven by CXCL1032,33,34,35. High CXCL10 expression after mRNA-1273 vaccination in clinical trial cohorts has been implicated in the induction of immune memory responses including production of neutralizing antibodies which are an integral correlate of protection for COVID-19 vaccines34,46. In vitro, co-stimulation with recombinant CXCL10 protein and model antigens can elicit cytotoxic effector responses upon subsequent challenge47. Despite this relationship and our observation in vitro, IT treatment with mRNA-1273 compared to PBS resulted in minimal differences in production of CXCL10 and other cytokines both locally (in the tumor microenvironment) as well as systemically (in the draining lymph node and serum) in vivo (Fig. 5A). Interestingly, we still observed a reduction in tumor volume occurred as early as 24 h post mRNA-1273 IT injection with a significant increase in CD8+ T cells into the tumor microenvironment. A major proportion of these intratumoral CD8+ T cells were identified as activated CD8+ GranzymeB+ T cells, which were significantly increased by the mRNA-1273 treatment. While mRNA-1273 initially induced the generation of B16F10-specific CD8+TRP-2+ T cells at 24 h post IT injection, these melanoma-specific T cell subsets proved transient, and were undetectable 4 days later. Despite significant suppression of tumor growth, long-term tumor control was not achieved following mRNA-1273 treatments. Once mRNA-1273 IT injections stopped, tumor outgrowth occurred.
Given the lack of long-term tumor regression in our model, re-challenge studies to demonstrate the presence of memory T cells could not be conducted. The transient nature of melanoma-specific T cells following mRNA-1273 treatment combined with the minimal induced systemic immune changes may contribute to the observed lack of sustained, durable anti-cancer responses. A caveat to these studies was that only a single B16F10 antigen, TRP-2, was evaluated to quantify a melanoma-specific response. Surprisingly, seroconversion of antibodies against the mRNA-1273 antigen was not observed, as evidenced by anti-RBD ELISA performed using serum from IT vaccinated mice. Future studies could consider quantitating anti-TRP2 antibodies as well as antibodies and T cells specific to additional B16F10 neoantigens to measure a more diverse pool of anti-cancer immunity correlates.
The remarkable clinical successes achieved by the use of immunotherapy to improve the lives of cancer patients underscore the indispensable role of the immune system in cancer treatment. Yet, while immunotherapies have yielded notable successes, they have, thus far, provided benefits to only a fraction of patients. Tumors exhibiting substantial infiltration of CD8+ T cells into the microenvironment have shown enhanced responsiveness to ICT8,9,10,11,12,13. Our study evaluates the potential of mRNA-1273 as an adjuvant to enhance ICT outcomes by increasing the T cell response in an otherwise “cold” tumor model. Improved responses from combination treatment with mRNA-1273 and ICT appears to be time dependent (Fig. 4 & S6). Enhanced responses were only observed using multiple mRNA-1273 IT injections followed by ICT one day later, repeated in combination for a total of three doses. No further tumor reduction was observed when ICT was started 4 days post mRNA-1273 administration in B16F10 tumors. We hypothesize that this improved anti-tumor response from multiple mRNA-1273 treatments where ICT began one day later may be a result of the enhanced B16F10-specific CD8+ T cell infiltration that was observed at 24 h post mRNA-1273 IT injection.
Surprisingly, our data demonstrated a significant increase in CD3+ T cell subsets into the tumor and the draining lymph node following treatment with PBS compared to mRNA-1273 or LNP at 4 days post injection. It is possible that the injection of PBS into the tumor created a proinflammatory stress response to the injection itself. However, while we measured an increase in infiltrating CD3+ T cells due to the PBS injection compared to untreated tumors, we did not observe tumor suppression in PBS treated mice. In addition to the tumor suppression observed in mRNA-1273 treated mice, we measured a significant increase in TRP-2 specific cells which was absent in control animals’ tissues. Therefore, while the stress response due to PBS injection may have induced a measurable CD3+ T cell response, this did not translate to enhanced CD8+ T cells or tumor suppression as we observed with mRNA-1273 treatment.
mRNA vaccines are generally recognized as “self-adjuvanted” However, it is clear that lipids used to form the LNP, the mRNA itself, as well as dsDNA and dsRNA carried over from in vitro transcription, can all have effects on the immunogenicity of the vaccine. We observed that control LNP caused a decreased in tumor volume at early timepoints (Fig. 2B) that was later lost at later timepoints (Fig. 2C); however, only COVID-19 IT immunization resulted in improved survival (Fig. 3B) and enhanced ICT outcomes (Fig. 4). These data suggest there may be some effects of the mRNA cargo and the background contaminants that are known to be in commercial mRNA vaccines due to the production process. Several studies demonstrate that the ionizable lipid component contributes to the induction of chemokines and pro-inflammatory cytokines, including C-C motif chemokine ligand 2 (CCL2), CCL3, CCL4, granulocyte-macrophage colony-stimulating factor (GM-CSF), C-X-C motif chemokine ligand 2 (CXCL2), CXCL10, interleukin (IL)-1β, IL-6, tumor necrosis factor (TNF), and interferon (IFN)-γ. Thus, varying the LNP composition may further enhance immune activation and anti-tumor responses, potentially enhancing therapeutic efficacy48,49,50,51,52. mRNA controls expressing non-immunological proteins, such as fluorescent reporters could be used in future studies to investigate the contribution of the mRNA cargo on the immune response.
Many groups have shown that the mRNA-1273 vaccine induces strong Th1 responses33. While relevant for mediating anti-viral responses, Th1 immunity is also useful for other targets including cancer. Th1 T cells exist in the tumor microenvironment at very small numbers, highlighting the therapeutic importance of selectively enhancing this subtype and avoiding its suppression by Th2 dominance which arises from canonical antigen presentation pathways53. Disis et al. showed the importance of selecting Th1 epitopes during neoantigen vaccination to generate durable T cell responses53. While broadly CD8+ cytotoxic effector T cells were an important cell population to identify in these pilot experiments, the Th1 T cell subtype and additional effector cells of the Th1 inflammatory profile such as macrophages and CD4+ T cells would be important to profile in future experiments. A modest increase in IL-12, a cytokine implicated in coordinating Th1 responses, in mRNA-1273-treated tumors was observed (Fig. 5), but this finding requires further analysis. However, it must be noted that one caveat of our study is that we did not investigate Th1 mediated immunity and its potential contribution to the effects we observed.
scRNAseq data did not reveal significant differences in the cellular populations recruited to the tumors. However, given that tumors were significantly smaller 4 days post IT injection with mRNA-1273, immune cell driven changes to the tumor microenvironment may have occurred at earlier timepoints which could not have been measured during our transcriptomic analysis timepoint. For example, we observed in our intravital imaging studies that at 48 h post IT mRNA-1273 treatment, areas where the tumor had been eradicated (as evidenced by minimal GFP fluorescence), had minimal detectable CD8+ T cells, despite these areas having detectable cell levels 24 h earlier. Furthermore, the scRNAseq analysis removed cells with low quality read data, and before RNA sequencing, we removed any cells positive for 7-AAD viability stain. It is possible that these quality control steps, constrained potentially subtle changes in the tumor transcriptome. Future investigations into mRNA vaccines should examine early changes within the timeframe of 2 to 6 h, as research indicates that LNPs are internalized by cells and initiate antigen expression within this window54.
Our studies demonstrate a significant reduction in tumor volume which corresponds to enhanced infiltration of activated CD8+ T cells into the tumor microenvironment after IT administration of an mRNA COVID-19 vaccine. mRNA COVID-19 vaccines are administered by the intramuscular route, however, our immunization studies (Fig. S7) suggest that intramuscular injection of the COVID-19 vaccine would not elicit similar anti-tumor responses as IT treatment. Recently, a Phase II clinical trial demonstrated prolonged recurrence-free survival in melanoma patients using an intramuscular injection of an mRNA-based individualized neoantigen vaccine combined with the immune checkpoint, pembrolizumab55. Studies have evaluated therapeutic immunization using TRP-2, a tumor-associated antigen of B16F10 cells, to drive CD8+ T cell responses in mice56,57. Wang et al., developed an mRNA-encoded TRP-2 vaccine which could significantly decrease tumor volume in mice, but TRP-2 mRNA alone did not fully cure mice from the tumor58. Furthermore, Castle et al. showed that non-TRP-2 B16F10 specific neoantigen peptide vaccines decreased tumor burden in murine tumor models but still were unable to elicit long-term durability59. We recognize that our studies are limited by the absence of tumor-specific neoantigens. Studies are underway to evaluate B16F10-specific neoantigen mRNA vaccines combined with immune-potentiating mRNA vaccines to enhance both the infiltration and activation of melanoma-specific T cells within the tumor microenvironment. These studies are also designed to compare how IT versus the intramuscular administration route affects infiltration of tumor-specific T cells. We anticipate that highly technical bioinformatic selection of antigens will enhance therapeutic outcomes. Additionally, mRNA vaccines can be further formulated to stimulate enhanced responses that combat cancer. These findings have important implications for mRNA vaccines in priming patients to respond to existing immunotherapies.
Materials and methods
Cells
B16F10 and YUMM1.G1 melanoma cells were purchased from the American Type Culture Collection (ATCC, VA, USA). B16F10 click beetle green luciferase and green fluorescent protein (CBG-GFP) cells were previously described26,37,39,40. B16F10 cells were cultured in DMEM supplemented with 10% heat-inactivated fetal bovine serum (FBS). YUMM1.G1 were cultured in DMEM: F12 supplemented with 10% heat-inactivated FBS and 1% NEAA. Cell cultures were grown at 37ºC in a humidified 5% CO2 atmosphere. All cell lines tested negative for mycoplasma.
In vitro studies
2 × 105 B16F10 cells were seeded in a 6-well plate. B16F10 cells were cultured up to 96 h after inoculation with 40 µg of mRNA-1273 SARS-CoV-2 vaccine. Cell culture supernatants and cell lysates were collected every 24 h. Lysates were prepared with RIPA Buffer and HALT proteinase inhibitor (Thermo Fisher Scientific, MA, USA). SARS-CoV-2 Spike protein production was confirmed by western blot of lysates. Cell lysate concentrations were normalized by total protein concentration. Western blots were prepared using Bolt 4–12% Bis-Tris Plus Protein Gels (Invitrogen, CA, USA) and transferred to PVDF membrane using iBlot 2 transfer. Western blot membrane was blocked using 3% BSA in tris-phosphate buffer. Spike protein positive control was confirmed by sera from convalescent SARS-Cov-2 challenged mice, and Spike-S1 subunit (Sino Biological, PA, USA, part no. 40591-V08H). Western blot image in Fig. 1C was processed equally across entire image and to controls using Adobe Photoshop where brightness was increased by 91% and contrast was increased by 44%.
In vivo subcutaneous tumor model
All the animal studies were performed after approval from the Institutional Animal Care and Use Committee (IACUC) at West Virginia University (protocol #: 2109047227). All methods performed in this study were in accordance with the IACUC guidelines and regulation. These guidelines are equivalent to the ARRIVE guidelines and therefore all methods were performed in accordance with the ARRIVE guidelines. 5 × 104 B16F10 cells were injected subcutaneously on the right flank of female 8-week-old C57BL/6 mice (Charles River Laboratories, NY, USA). Before tumor cell injection, the fur was removed on the right flank. Tumors were measured by calipers every 3–4 days once palpable. Following institutional animal guidelines, animals were euthanized once tumors reached 1.5 cm in diameter or were ulcerated greater than 0.5 cm. When tumors reached 5 mm in diameter, mice were randomized and treated with an IT injection of 20 µL of 3 µg mRNA-1273, empty LNP or PBS control occurred. mRNA-1273 (also known as Spikevax®, Moderna) vaccine was obtained for research from WVU Medicine Pharmacy. Empty LNP without mRNA was prepared using the using the flash nanoprecipitation approach as described elsewhere60. The composition of empty LNP is similar to that of mRNA-1273 LNP vaccine (Fig. S12). Empty LNP were 120 nm in size when analyzed using dynamic light scattering. Animals were treated with ICT (1st dose was 200 µg Anti-CTLA-4 and 200 µg anti-PD-1 followed by 2 doses of 100 µg Anti-CTLA-4 and 100 µg anti-PD-1 administered intraperitoneally every 3 days). Anti-CTLA-4 (9D9), and anti-αPD-1 (RMP1-14) were obtained from Bio X cell (Bio X cell, NH, USA). For the prime/boost studies, mice were immunized intramuscularly with mRNA-1273 at six weeks old. Two weeks post prime, B16F10 tumor inoculation occurred. Four weeks post prime (two weeks post B16F10 cell injection), an intramuscular mRNA-1273 boost occurred. When tumors reached 5 mm in diameter, approximately, one week following boost (three weeks post B16F10 cell injection), IT administration of with 3 doses of mRNA-1273 without or with ICT, or PBS occurred.
Flow cytometry
At 24 h following a 50 µL injection of 50 µg mRNA-1273 vaccine or PBS control using an intramuscular injection, 8-week old C57BL/6 mice were euthanized by pentobarbital injection. The muscle where the site of injection occurred and the inguinal lymph node were analyzed by flow cytometry following intramuscular immunization. At various timepoints, tumor-bearing mice were euthanized by carbon dioxide asphyxiation. Single cell suspension of cells were harvested from the spleen, lymph nodes, and tumor. Tumors and muscle were dissociated using a mouse tumor dissociation kit, or skeletal muscle dissociation kit, respectfully (Miltenyi Biotec, CA, USA). Red blood cells were lysed using 1X red blood cell lysis buffer (Thermo Fisher Scientific, CA, USA). Remaining cells were resuspended in cell staining buffer (BioLegend, CA, USA) at a concentration of 2 × 105 – 1 × 106 cells per 100 µl. Fc receptors are blocked using 10 µg/ml ChromPure of mouse IgG (Jackson ImmunoResearch Inc., PA, USA) per 106 cells in a 100 µl volume for 10 min on ice. Cells were washed with cell staining buffer and incubated with antibody mix (Table S1) for 15–20 min on ice in the dark. Cells were fixed using 200 µl Fixation buffer (Thermo Fisher Scientific, CA, USA) for 20 min at 4ºC in the dark. Cells are washed 2 times with 1X PermWash (BD Bioscience, CA, USA) and permeabilized using Permeabilization solution (BD Bioscience, CA, USA). Intracellular antibodies (Table S1) were incubated for 20 min at room temperature in the dark. Stained cells were analyzed using the Cytek® Aurora (Cytek, MD, USA) within 2 weeks of staining. Data was analyzed using FCS express (De Novo Software, CA, USA) \where the gating strategy is provided in Figure S13.
Intravital imaging
Skin window chamber implantation, imaging and analysis were previously described using 8-week old male and female C57BL/6 mice40,41. Briefly, following skin window chamber implantation and B16F10 cell inoculation, microscopy occurred on day 2 post implantation. Skin window chamber microscopy was done using a Nikon confocal AX microscopy (Nikon, NY, USA). 50 µL of 0.15 µg CD8a-APC (Miltenyi, CA, USA) antibody in PBS was administered through retro-orbital injection. Microscopic imaging occurred 30 min post antibody injection using 3 fields of view per mouse at 20X objective. B16F10 tumors were visualized within the skin window chamber using GFP. Following background imaging, an IT injection of 10 µL of 1.5 µg mRNA-1273 or PBS control occurred. 24 h post injection, intravital microscopy occurred again. Images were analyzed using the Nikon Elements software (Nikon, NY, USA) using the automated measurement analysis macro. An individual threshold was applied to each imaging channel and positive CD8a-APC positive cells were quantified using the following automated measurement parameters; size: 5 μm to 10 μm, circularity: 0 to 1, smooth: 5X, fill holes: on and exclude objects touching frame borders. The average of three fields of view from each imaging time point for each animal was calculated.
Cytokine analysis
Blood draws by submandibular bleed were performed on subcutaneous tumor-bearing mice at various timepoints during treatment and stored at − 80 °C until analysis. Serum was prepared using serum separator microtiter tubes (BD Biosciences, NJ, USA). Cytokines were analyzed using a V-Plex Mouse Cytokine 19-Plex kit (Mesoscale Diagnostics, MD, USA), following the manufacturer’s protocol.
Single cell RNA sequencing
At 4 days post IT injection of mRNA-1273 or PBS, mice were euthanized by carbon dioxide asphyxiation. Tumors were smashed and washed with PBS through a 70 µm cell strainer to achieve a single cell suspension. Samples were pooled per group and combined at equal proportion per individual sample. Single cell suspensions were stained with 7-AAD (STEMCELL Technologies, BC, Canada), and viable cells sorted using BD FACSAria™ III Cell Sorter (BD Biosciences, NJ, USA). RNA was isolated using Chromium Next GEM Single Cell 5’ Kit (10X Genomics) following manufacturers protocol. Read coverage was calculated at 2 billion paired reads per 5’ library samples. scRNAseq analysis was conducted using Loupe Browser (7.0.1). Low quality reads were filtered using the following thresholds: Unique molecular identifiers (UMI) 500 − 50,000; genes per barcode 300–7000; and less than 20% mitochondrial UMIs. The cells lacking Ptprc gene expression assigned as tumor cells. To assign major immune cell types to each cluster, we annotated each cluster based on its top 10 upregulated, differentially expressed genes compared to other clusters. Top 10 upregulated genes in each cell cluster were shown in Supplementary Figure S11.
Quantification of anti-SARS-CoV-2 RBD IgG antibodies
Serum was collected after the final timepoint (~ 20 days from initial dose) from the multiple (3X) mRNA-1273 IT (3 µg) treated mice. Non-tumor bearing six-week-old, C57BL/6 mice received one IM dose of 5 µg mRNA-1273. After 14 days serum was collected from submandibular bleeds. Anti-SARS-CoV-2 RBD IgG levels were quantified using ELISA61 from sera of the two cohorts. High-binding microtiter plates were coated with 2 µg/ml of WA-1 S RBD (Sino Biological, PA, USA), and incubated for 24 h at 4 °C. The next day plates were washed 3 times with wash buffer (0.1% Tween-20 + PBS). Plates were blocked using 3% nonfat milk in wash buffer and incubated at room temperature for 1 h. After 3X washes, sera were serial diluted in 1% nonfat milk and wash buffer, using 2-fold dilutions starting at 1:50 dilution. Samples were incubated for 1 h at room temperature while shaking. Plates were then washed 4X, and secondary antibody added (goat anti-mouse IgG HRP, Novus Biological, CO, USA, NB7539; 1:2000) and incubated for 1 h while shaking at room temperature. Following 5X wash, plates were developed using TMB substrate (Biolegend, CA, USA) for 15 min. Reaction was stopped using 2 N sulfuric acid and absorbance read at 450 nm on a BioTek Synergy H1 Multimode reader (Agilent, CA, USA). Serum antibody levels were quantified using area under curve analysis in GraphPad prism (GraphPad Software, Inc, CA, USA).
Statistical analyses
Graphs were made and statistical analyses were performed using GraphPad Prism V10.1.0 (GraphPad Software, Inc, CA, USA). Data were expressed as mean ± SD. For analysis of three or more groups or variables, analysis of variance (ANOVA) tests were performed followed by Tukey’s or Šídák’s multiple comparisons test as indicated in figure legends. Standard comparison of survival curves used the log rank (Mantel-Cox) analysis. Analysis of differences between two normally distributed paired test groups were performed using a Student’s t-test. p values were considered statistically significant if p < 0.05.
Data availability
The datasets generated during the current study are available in the NCBI SRA repository, accession: PRJNA1160311. All other data relevant to the study are included in the article, uploaded as supplementary information.
References
Raskov, H., Orhan, A., Christensen, J. P. & Gögenur, I. Cytotoxic CD8 + T cells in cancer and cancer immunotherapy. Br. J. Cancer. 124, 359–367. https://doi.org/10.1038/s41416-020-01048-4 (2021).
Bos, R. & Sherman, L. A. CD4 + T-Cell help in the Tumor Milieu is required for recruitment and cytolytic function of CD8+ T lymphocytes. Cancer Res. 70, 8368–8377. https://doi.org/10.1158/0008-5472.can-10-1322 (2010).
Thomas, N. E. et al. Tumor-infiltrating lymphocyte Grade in primary melanomas is independently associated with melanoma-specific survival in the population-based genes, environment and melanoma study. J. Clin. Oncol. 31, 4252–4259. https://doi.org/10.1200/jco.2013.51.3002 (2013).
DONIZY, P. et al. Paucity of Tumor-infiltrating lymphocytes is an unfavorable prognosticator and predicts Lymph Node metastases in cutaneous melanoma patients. Anticancer Res. 35, 351–358 (2015).
Zhao, Y. et al. A leukocyte infiltration score defined by a gene signature predicts melanoma patient prognosis. Mol. Cancer Res. 17, 109–119. https://doi.org/10.1158/1541-7786.mcr-18-0173 (2019).
Yuan, Y. et al. Development and validation of a CD8 + T cell infiltration-related signature for Melanoma patients. Front. Immunol. 12 https://doi.org/10.3389/fimmu.2021.659444 (2021).
Edwards, J. et al. CD103 + tumor-resident CD8 + T cells are Associated with Improved Survival in Immunotherapy-Naïve Melanoma patients and Expand significantly during Anti-PD-1 treatment. Clin. Cancer Res. 24, 3036–3045. https://doi.org/10.1158/1078-0432.Ccr-17-2257 (2018).
Hodi, F. S. et al. Immunologic and clinical effects of antibody blockade of cytotoxic T lymphocyte-associated antigen 4 in previously vaccinated cancer patients. Proc. Natl. Acad. Sci. U S A. 105, 3005–3010. https://doi.org/10.1073/pnas.0712237105 (2008).
Tumeh, P. C. et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature 515, 568–571. https://doi.org/10.1038/nature13954 (2014).
Sade-Feldman, M. et al. Defining T Cell states associated with response to checkpoint immunotherapy in melanoma. Cell 175, 998–1013e1020. https://doi.org/10.1016/j.cell.2018.10.038 (2018).
Curran, M. A., Montalvo, W., Yagita, H. & Allison, J. P. PD-1 and CTLA-4 combination blockade expands infiltrating T cells and reduces regulatory T and myeloid cells within B16 melanoma tumors. Proc. Natl. Acad. Sci. U S A. 107, 4275–4280. https://doi.org/10.1073/pnas.0915174107 (2010).
Curran, M. A., Kim, M., Montalvo, W., Al-Shamkhani, A. & Allison, J. P. Combination CTLA-4 blockade and 4-1BB activation enhances tumor rejection by increasing T-cell infiltration, proliferation, and cytokine production. PLoS ONE. 6, e19499. https://doi.org/10.1371/journal.pone.0019499 (2011).
Daud, A. I. et al. Tumor immune profiling predicts response to anti-PD-1 therapy in human melanoma. J. Clin. Invest. 126, 3447–3452. https://doi.org/10.1172/JCI87324 (2016).
Wolchok, J. et al. (ed, D.) CheckMate 067: 6.5-year outcomes in patients (pts) with advanced melanoma. J. Clin. Oncol. 39 9506–9506 https://doi.org/10.1200/JCO.2021.39.15_suppl.9506 (2021).
Larkin, J. et al. Five-year survival with combined Nivolumab and Ipilimumab in advanced melanoma. N. Engl. J. Med. 381, 1535–1546. https://doi.org/10.1056/NEJMoa1910836 (2019).
Wolchok, J. D. et al. Overall survival with combined Nivolumab and Ipilimumab in Advanced Melanoma. N Engl. J. Med. 377, 1345–1356. https://doi.org/10.1056/NEJMoa1709684 (2017).
Wolchok, J. D. et al. Nivolumab plus Ipilimumab in advanced melanoma. N. Engl. J. Med. 369, 122–133. https://doi.org/10.1056/NEJMoa1302369 (2013).
Wolchok, J. D., Rollin, L. & Larkin, J. Nivolumab and Ipilimumab in Advanced Melanoma. N. Engl. J. Med. 377, 2503–2504. https://doi.org/10.1056/NEJMc1714339 (2017).
Liu, Y. T. & Sun, Z. J. Turning cold tumors into hot tumors by improving T-cell infiltration. Theranostics 11, 5365–5386. https://doi.org/10.7150/thno.58390 (2021).
Ouyang, P. et al. Overcoming cold tumors: A combination strategy of immune checkpoint inhibitors. Front. Immunol. 15, 1344272. https://doi.org/10.3389/fimmu.2024.1344272 (2024).
Wang, L. et al. Hot and cold tumors: Immunological features and the therapeutic strategies. MedComm (2020). 4, e343. https://doi.org/10.1002/mco2.343 (2023).
Sharma, P. & Allison, J. P. The future of immune checkpoint therapy. Science 348, 56–61. https://doi.org/10.1126/science.aaa8172 (2015).
Mahvi, D. M. et al. Intratumoral injection of IL-12 plasmid DNA – results of a phase I/IB clinical trial. Cancer Gene Ther. 14, 717–723. https://doi.org/10.1038/sj.cgt.7701064 (2007).
Intratumoral Injection of DNA Encoding Human Interleukin 12 into Patients with Metastatic Melanoma. Clinical efficacy. Hum. Gene Ther. 16, 35–48. https://doi.org/10.1089/hum.2005.16.35 (2005).
Mauldin, I. S. et al. Intratumoral interferon-gamma increases chemokine production but fails to increase T cell infiltration of human melanoma metastases. Cancer Immunol. Immunother. 65, 1189–1199. https://doi.org/10.1007/s00262-016-1881-y (2016).
Cebon, J. S. et al. Results of a randomized, double-blind phase II clinical trial of NY-ESO-1 vaccine with ISCOMATRIX adjuvant versus ISCOMATRIX alone in participants with high-risk resected melanoma. J. Immunother. Cancer. 8, e000410. https://doi.org/10.1136/jitc-2019-000410 (2020).
Fajgenbaum, D. C. & June, C. H. Cytokine storm. N. Engl. J. Med. 383, 2255–2273. https://doi.org/10.1056/nejmra2026131 (2020).
Suntharalingam, G. et al. Cytokine storm in a phase 1 trial of the Anti-CD28 monoclonal antibody TGN1412. N. Engl. J. Med. 355, 1018–1028. https://doi.org/10.1056/nejmoa063842 (2006).
Newman, J. H. et al. Intratumoral injection of the seasonal flu shot converts immunologically cold tumors to hot and serves as an immunotherapy for cancer. Proc. Natl. Acad. Sci. U S A. 117, 1119–1128. https://doi.org/10.1073/pnas.1904022116 (2020).
Tähtinen, S. et al. Exploiting preexisting immunity to enhance oncolytic cancer immunotherapy. Cancer Res. 80, 2575–2585. https://doi.org/10.1158/0008-5472.CAN-19-2062 (2020).
Tran, T., Burt, D., Eapen, L. & Keller, O. R. Spontaneous regression of metastatic melanoma after inoculation with tetanus-diphtheria-pertussis vaccine. Curr. Oncol. 20, e270–273. https://doi.org/10.3747/co.20.1212 (2013).
Sousa, L. G. D. et al. Spontaneous tumor regression following COVID-19 vaccination. J. Immunother. Cancer. 10, e004371. https://doi.org/10.1136/jitc-2021-004371 (2022).
DiPiazza, A. T. et al. COVID-19 vaccine mRNA-1273 elicits a protective immune profile in mice that is not associated with vaccine-enhanced disease upon SARS-CoV-2 challenge. Immunity 54, 1869–1882e1866. https://doi.org/10.1016/j.immuni.2021.06.018 (2021).
Bergamaschi, C. et al. Systemic IL-15, IFN-γ, and IP-10/CXCL10 signature associated with effective immune response to SARS-CoV-2 in BNT162b2 mRNA vaccine recipients. Cell. Rep. 36, 109504. https://doi.org/10.1016/j.celrep.2021.109504 (2021).
Reschke, R. et al. Immune cell and tumor cell-derived CXCL10 is indicative of immunotherapy response in metastatic melanoma. J. Immunother. Cancer. 9, e003521. https://doi.org/10.1136/jitc-2021-003521 (2021).
Antonicelli, F. et al. CXCL10 reduces melanoma proliferation and invasiveness in vitro and in vivo. Br. J. Dermatol. 164, 720–728. https://doi.org/10.1111/j.1365-2133.2010.10176.x (2011).
Mikucki, M. E. et al. Non-redundant requirement for CXCR3 signalling during tumoricidal T-cell trafficking across tumour vascular checkpoints. Nat. Commun. 6, 7458. https://doi.org/10.1038/ncomms8458 (2015).
Harlin, H. et al. Chemokine expression in melanoma metastases associated with CD8 + T-cell recruitment. Cancer Res. 69, 3077–3085. https://doi.org/10.1158/0008-5472.CAN-08-2281 (2009).
Meeth, K., Wang, J. X., Micevic, G., Damsky, W. & Bosenberg, M. W. The YUMM lines: A series of congenic mouse melanoma cell lines with defined genetic alterations. Pigment Cell. Melanoma Res. 29, 590–597. https://doi.org/10.1111/pcmr.12498 (2016).
Liu, T. W., Gammon, S. T., Fuentes, D. & Piwnica-Worms, D. Multi-modal multi-spectral intravital macroscopic imaging of signaling dynamics in real time during tumor-immune interactions. Cells 10 https://doi.org/10.3390/cells10030489 (2021).
Liu, T. W., Gammon, S. T. & Piwnica-Worms, D. Multi-modal multi-spectral intravital microscopic imaging of signaling dynamics in real-time during tumor-immuneInteractions. Cells 10 https://doi.org/10.3390/cells10030499 (2021).
Mukherjee, N., Wheeler, K. M. & Svatek, R. S. Bacillus Calmette-Guérin treatment of bladder cancer: A systematic review and commentary on recent publications. Curr. Opin. Urol. 29, 181–188. https://doi.org/10.1097/mou.0000000000000595 (2019).
Tai, L. H. et al. Perioperative Influenza Vaccination reduces postoperative metastatic disease by reversing surgery-induced dysfunction in natural killer cells. Clin. Cancer Res. 19, 5104–5115. https://doi.org/10.1158/1078-0432.Ccr-13-0246 (2013).
Vandeborne, L., Pantziarka, P., Van Nuffel, A. M. T. & Bouche, G. Repurposing infectious diseases vaccines against Cancer. Front. Oncol. 11, 688755. https://doi.org/10.3389/fonc.2021.688755 (2021).
Rosati, M. et al. Rapid transient and longer-lasting innate cytokine changes associated with adaptive immunity after repeated SARS-CoV-2 BNT162b2 mRNA vaccinations. Front. Immunol. 14 https://doi.org/10.3389/fimmu.2023.1292568 (2023).
Hellgren, F. et al. Modulation of innate immune response to mRNA vaccination after SARS-CoV-2 infection or sequential vaccination in humans. JCI Insight. 9 https://doi.org/10.1172/jci.insight.175401 (2024).
Krathwohl, M. D. & Anderson, J. L. Chemokine CXCL10 (IP-10) is sufficient to trigger an immune response to injected antigens in a mouse model. Vaccine 24, 2987–2993. https://doi.org/10.1016/j.vaccine.2005.11.032 (2006).
Alameh, M. G. et al. Lipid nanoparticles enhance the efficacy of mRNA and protein subunit vaccines by inducing robust T follicular helper cell and humoral responses. Immunity 54, 2877–2892e2877. https://doi.org/10.1016/j.immuni.2021.11.001 (2021).
Ndeupen, S. et al. The mRNA-LNP platform’s lipid nanoparticle component used in preclinical vaccine studies is highly inflammatory. iScience 24, 103479. https://doi.org/10.1016/j.isci.2021.103479 (2021).
Tahtinen, S. et al. IL-1 and IL-1ra are key regulators of the inflammatory response to RNA vaccines. Nat. Immunol. 23, 532–542. https://doi.org/10.1038/s41590-022-01160-y (2022).
Moradian, H., Roch, T., Anthofer, L., Lendlein, A. & Gossen, M. Chemical modification of uridine modulates mRNA-mediated proinflammatory and antiviral response in primary human macrophages. Mol. Ther. Nucleic Acids. 27, 854–869. https://doi.org/10.1016/j.omtn.2022.01.004 (2022).
Waickman, A. T. et al. mRNA-1273 vaccination protects against SARS-CoV-2–elicited lung inflammation in nonhuman primates. JCI Insight. 7 https://doi.org/10.1172/jci.insight.160039 (2022).
Disis, M. L., Watt, W. C. & Cecil, D. L. Th1 epitope selection for clinically effective cancer vaccines. Oncoimmunology 3, e954971. https://doi.org/10.4161/21624011.2014.954971 (2014).
Maugeri, M. et al. Linkage between endosomal escape of LNP-mRNA and loading into EVs for transport to other cells. Nat. Commun. 10, 4333. https://doi.org/10.1038/s41467-019-12275-6 (2019).
Weber, J. S. et al. Individualised neoantigen therapy mRNA-4157 (V940) plus pembrolizumab versus pembrolizumab monotherapy in resected melanoma (KEYNOTE-942): A randomised, phase 2b study. Lancet 403, 632–644. https://doi.org/10.1016/S0140-6736(23)02268-7 (2024).
Wang, Y., Zhang, L., Xu, Z., Miao, L. & Huang, L. mRNA vaccine with antigen-specific checkpoint blockade induces an enhanced immune response against established melanoma. Mol. Ther. 26, 420–434. https://doi.org/10.1016/j.ymthe.2017.11.009 (2018).
Oberli, M. A. et al. Lipid nanoparticle assisted mRNA delivery for potent cancer immunotherapy. Nano Lett. 17, 1326–1335. https://doi.org/10.1021/acs.nanolett.6b03329 (2017).
Wang, R. F., Appella, E., Kawakami, Y., Kang, X. & Rosenberg, S. A. J. T. J. O. E. M. identification of TRP-2 as a human tumor antigen recognized by cytotoxic T lymphocytes. J. Exp. Med. 184, 2207–2216 (1996).
Castle, J. C. et al. Exploiting the mutanome for tumor vaccination. Cancer Res. 72, 1081–1091. https://doi.org/10.1158/0008-5472.Can-11-3722 (2012).
Misra, B. et al. Flash nanoprecipitation assisted self-assembly of ionizable lipid nanoparticles for nucleic acid delivery. Nanoscale 16, 6939–6948. https://doi.org/10.1039/D4NR00278D (2024).
Wong, T. Y. et al. Intranasal administration of BReC-CoV-2 COVID-19 vaccine protects K18-hACE2 mice against lethal SARS-CoV-2 challenge. NPJ Vaccines. 7, 36. https://doi.org/10.1038/s41541-022-00451-7 (2022).
Acknowledgements
Imaging experiments and image analysis were performed in the West Virginia University (WVU) Animal Models & Imaging Facility which has been supported by the WVU Cancer Institute, the WVU HSC Office of Research and Graduate Education, and National Institute of Health Grants P20GM121322 and U54GM104942. Flow cytometry and isolation experiments and analysis were performed in the WVU Flow Cytometry & Single Cell Core Facility, RRID: SCR_017738 which has been supported by National Institute of Health National Institute of General Medical Sciences CoBRE P20GM121322, S10 equipment grant OD028605 and OD016165, WVCTSI Grant GM104942.
Funding
This work was supported in part by a NIH/NIGMS CoBRE award (5P20GM121322).
Author information
Authors and Affiliations
Author notes
These authors contributed equally: Dylan T. Boehm and Kaitlyn M. Landreth.
Contributions
DB and KML performed all in vitro and in vivo experiments, quantified and analyzed the data, provided critical intellectual input, and wrote and edited the manuscript. ESK performed the bioinformatics analysis of the scRNAseq data, prepared Fig. 5 and edited the manuscript. KSL assisted with immunization studies and edited the manuscript. SB and BM provided empty LNP for control studies. SB edited the manuscript. FHD supervised the study, quantified and analyzed the data, provided critical intellectual input, and edited the manuscript. TWL supervised the study, quantified and analyzed the data, provided critical intellectual input, wrote and edited the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Boehm, D.T., Landreth, K.M., Kilic, E.S. et al. Intratumoral administration of mRNA COVID-19 vaccine delays melanoma growth in mice. Sci Rep 15, 5337 (2025). https://doi.org/10.1038/s41598-025-89930-0
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-89930-0