Abstract
Loss of synaptic activity correlates best with cognitive dysfunction in Alzheimer’s disease (AD). We have previously shown that mild inhibition of mitochondrial complex I with the small molecule tricyclic pyrone compound CP2 restores long-term potentiation and cognitive function assessed by electrophysiology and behavior tests in multiple mouse models of AD. Using serial block-face scanning electron microscopy and three-dimensional electron microscopy reconstruction, we examined the effect of CP2 treatment on synapses, and the distribution and morphology of synaptic mitochondria in the hippocampus of APP/PS1 mice. Structural data confirmed the loss of synapses in APP/PS1 compared to non-transgenic (NTG) littermates. Mitochondrial distribution assessed in pre- and postsynaptic compartments was significantly altered in AD model demonstrating increased presence of mitochondria around dendritic spines compared to NTG mice, indicating the loss of mitochondrial ability to support synaptic function. CP2 treatment restored distribution of synaptic mitochondria and the number of synapses to the NTG control levels. Improved synaptic function in CP2-treated APP/PS1 mice was supported by RNA-seq analysis indicating upregulation of genes involved in axonal guidance, dendritic maturation and synaptic function, and Western blot analysis of brain tissue. Taken together, functional, imaging, biochemistry and structural findings further support the potential of targeting mitochondria as a therapeutic approach for AD.
Similar content being viewed by others
Introduction
Alzheimer’s disease (AD) is a neurodegenerative disorder characterized by progressive cognitive deficits, memory loss, and secondary complications leading to death. Among numerous pathological findings of AD, increased synaptic damage and decreased number of synapses are the best correlates of cognitive dysfunction1. Synapses, the specialized junctions between neurons, play a critical role in cognitive processes. Understanding the neuronal structure, distribution, and function of synaptic compartments and organelles associated with synapses is therefore essential for elucidating the pathophysiology and potential treatments of AD. Synapses involve several components that mediate signal transmission within the brain. At the axonal presynaptic terminal, vesicles are positioned for depolarization-induced release of neurotransmitters into the synaptic cleft, between the presynaptic and postsynaptic neurons. Upon release, neurotransmitters diffuse and bind to the receptors on the postsynaptic membrane located on the dendritic spines of the neighboring neuron2. Dendritic spines (DS) are structurally characterized by a head that contains proteins necessary for signal reception, and a neck that connects the spine to the dendrite2. DS exhibit numerous shapes and sizes, which vary depending on the location and maturation of the spine. Initially, immature DS are small, highly plastic, and filopodia-shaped. Immature DS have weak signaling capabilities and are pruned or stabilized through an activity-dependent mechanism2. As immature DS become stabilized through repeated synaptic potentiation, they mature into larger spines with a mushroom-like head. These mature DS have a greater transmission capacity and can remain stable for months to years before degrading2. Through this process of dynamic spinogenesis and maturation, DS are able to evolve and strengthen neuronal communication based on environmental input. DS maturation thus plays a key role in synaptic plasticity, the very process theorized to facilitate learning and the formation and consolidation of memory. Interestingly, patients with AD have been found to have a greater number of immature DS, potentially contributing to their overall decrease in functional and stable synapses3.
Historically, excessive accumulation of amyloid beta (Aβ) plaques and hyperphosphorylated Tau (pTau) protein has been used to explain synaptic and DS dysfunction in the brains of AD patients4. As the accumulation of these abnormal proteins increases inside and around the neuron, a corresponding decrease in DS quantity and size, synaptic vesicle mobilization, mitochondrial localization at synapses, synapse stability, and cognitive function are observed5,6,7. Furthermore, APP/PS1 transgenic mice that carry familial AD mutations in the amyloid-beta precursor protein (APP) and presenilin 1 (PS1) genes and display significant Aβ intraneuronal and extracellular accumulation, have less mature DS and more dendritic abnormalities including shaft atrophy, neurite breakage, and reduced spine density8,9,10. These findings provided strong reason to pursue treatments that mitigate Aβ plaques and pTau formation in patients with AD. However, since immunotherapies targeting these pathogenic proteins have yielded inconclusive results, often with only marginal improvement in quality of life and substantial side effects11,12, attention has shifted to other aspects of AD etiology, such as the role of mitochondrial function in synaptic support and neuronal health.
Mitochondria are crucial for synaptic function. After forming at the neuronal cell body via a process known as biogenesis, mitochondria are distributed throughout a neuron13. Mitochondria are transported along microtubules and populate the presynaptic and postsynaptic compartments where energy demands are increased14. At the synapse, mitochondria provide ATP for neurotransmitter packaging, exocytosis of synaptic vesicles, neurotransmitter reuptake, calcium (Ca2+) buffering via the Ca2+-ATPase, and modulation of the activity of ion channels, pumps and receptors15. Mitochondria are also capable of sequestering Ca2+ via the mitochondrial Ca2+ uniporter, which is necessary to maintain Ca2+ homeostasis in the synapse and prevent excitotoxicity15. Furthermore, mitochondrial influence in the neuron extends far beyond their local role at the synapse. Through the release of reactive oxygen species (ROS), Ca2+ ions, and metabolites, mitochondria can communicate with the nucleus, endoplasmic reticulum (ER), and other organelles to maintain energy homeostasis, especially during conditions of stress15. In AD, dysfunctional mitochondria may lead to reduced ATP synthesis, overproduction of ROS, disrupted Ca2+ homeostasis, and disturbed signaling, all potentially impacting overall neuronal health16. In fact, reduced brain glucose metabolism, an indication of mitochondrial dysfunction, is one of the earliest findings of AD pathogenesis17. While some studies attribute early mitochondrial dysfunction in AD to Aβ and pTau accumulation, others have suggested that mitochondrial dysfunction not only precedes but modulates levels of APOE, Aβ and pTau18,19,20,21. Regardless, both mechanisms warrant deeper research into mitochondria and their vast molecular and intracellular relationships in the neuron.
To adapt to everchanging energy demands, mitochondria undergo dynamic cycles of fusion and fission. This process, deemed mitochondrial dynamics, is essential for organelles to maintain homogeneous function, even under stress conditions22. For example, if a mitochondrion becomes damaged, it may fuse with another mitochondrion to compensate and repair itself or undergo fission to segregate dysfunctional regions of the organelle for degradation process known as mitophagy. Additionally, mitochondrial dynamics allow organelles to better fit variably sized regions of the neuron, as smaller mitochondria are often required to properly position at presynaptic terminals22 while mitochondria in dendrites are usually longer. Notably, mitochondria found in neurons of AD brains have significant changes in morphology ranging from short and wide with overall increases in size, to extensively fragmented with decreased size19,22. These observations, first noted using transmission electron microscopy (TEM), lacked spatial resolution and dimensional complexity, making comprehensive analysis challenging. Through recent advances in serial block-face scanning electron microscopy (SBFSEM), however, three-dimensional (3D) reconstructions of the brain tissue have revealed new AD-associated mitochondrial morphologies, including mitochondria-on-a-string (MOAS). MOAS consist of multiple teardrop-shaped mitochondrial bodies connected by a thin double membrane23, which are easily missed when tissue is interrogated using conventional 2D TEM. MOAS have been observed under normal conditions in multiple organisms and in postmortem brain tissue of AD patients and mouse models of AD24,25. The formation of MOAS is believed to be due to the arrest of the late stages of fission, however the exact mechanism is not clear23,24,26. In one study that induced MOAS morphology in mice through transient global cerebral ischemia, the MOAS phenotype was reversed following reperfusion, suggesting energetic stress-associated mitochondrial morphological changes may be salvageable26. These findings emphasize the need to apply advanced imaging techniques such as SBFSEM to reveal details of mitochondrial morphology, distribution and function in neurons and the importance of mitochondria as a potential therapeutic target for AD.
In search of disease-modifying therapies for AD, we have identified a small molecule tricyclic pyrone compound (code name CP2) that penetrates the blood-brain barrier and accumulates in mitochondria where it inhibits complex I (MCI) of the electron transport chain27,28,29,30. CP2 inhibits roughly 15% of MCI activity at treatment-relevant concentrations, meaning significant ROS production or glycolytic switching is avoided31. Chronic CP2 treatment of transgenic AD mice initiated at various stages of disease, including in utero, presymptomatic, and symptomatic, showed significant reduction of Aβ and pTau, inflammation, oxidative stress, neurodegeneration, and cognitive dysfunction27,28,29. Notably, CP2 improved neuronal glucose metabolism, restoring energy homeostasis and overall synaptic function27,28. The mechanism of CP2 action includes activation of the adaptive stress response which is largely mediated through activation of AMP-activated protein kinase (AMPK), a crucial regulator of energy metabolism, autophagy, mitochondrial biogenesis, and neuroinflammatory suppression in the brain27,28,2932,33,−34. While CP2 has been shown to safely improve synaptic and mitochondrial function when administered to APP/PS1 mice after the development of cognitive dysfunction and neurodegeneration28, structural changes of synaptic domains have yet to be thoroughly analyzed. Here, together with Western blot analysis and next generation RNA sequencing (RNA-seq) in hippocampal tissue, we utilized SBFSEM with 3D EM reconstruction to determine the effect of CP2 treatment on synaptic compartments, mitochondrial morphology and distribution in respect to synaptic structure in APP/PS1 mice.
Results
Chronic CP2 treatment restores the number of synapses in hippocampus of APP/PS1 mice
We have previously shown that CP2 treatment improved synaptic function in mouse models of AD with Aβ and pTau pathologies27,28,29. Treatment of 3xTgAD or APP/PS1 mice with CP2 resulted in augmented long-term potentiation (LTP), increased levels of synaptic proteins synaptophysin and PSD95, integral to synaptic function, and improved cognitive outcomes in behavior tests27,28,29. We also observed an enhancement of mitochondrial dynamics and function, particularly the restoration of axonal trafficking essential for the delivery of mitochondria to the distant sites of synapses28,29. Collectively, these mechanisms contributed to blocking the ongoing neurodegeneration in APP/PS1 mice28. To visualize the effect of CP2 treatment on synaptic densities and mitochondria distribution in axons and dendrites in regard to different type of synapses in the hippocampus from CP2- or vehicle-treated APP/PS1 mice compared to age- and sex-matched non-transgenic (NTG) littermates, we utilized our well-established protocol that allows for the preservation of brain tissue and mitochondria for the SBFSEM examination35. This protocol was designed to ensure the consistency of tissue dissection in order to compare similar brain regions across samples, to avoid the introduction of artifacts during fixation and processing for accurate analysis of morphological characteristics35.
APP/PS1 and NTG female mice were treated with vehicle or CP2 (25 mg/kg/day) via drinking water ad lib starting at 9 months of age till 24 months of age (Fig. 1a)28. Mice were sacrificed and brain tissue from the CA1 hippocampal region was removed and immediately processed for SBFSEM as we described previously35. For SBFSEM and 3D EM reconstructions, due to the labor intense process, we randomly selected one mouse from vehicle-treated NTG group, vehicle-treated APP/PS1 group, and CP2-treated APP/PS1 group. Each tissue block face was serially sectioned (280–320 sections) and images were acquired for 3D EM reconstruction of pre- and postsynaptic compartments and synaptic mitochondria (Figs. 1b and 2a and b).
We first examined the effect of CP2 treatment on the number of synapses in NTG and APP/PS1 mice. Quantification of synapses in each tissue was performed by counting synaptic membrane densities within three independently selected regions of interest (ROI) defined as 5 μm x 5 μm x 3.75 μm (Fig. 1c). Since the cellular organization of brain tissue is very complex and could be unique to a particular region, the selection of three ROIs from the same block ensured the consistency of the analysis. All ROIs were of the same size and included regions that excluded cell bodies containing neuropils only. ROIs were randomly selected throughout the depth of the image volume (Fig. 1d).
Treatment paradigm and a workflow for SBFSEM and 3D EM reconstruction of synapses and mitochondria in dendrites and axons in hippocampal mouse brain tissue. (a) A timeline of chronic vehicle or CP2 administration to NTG and APP/PS1 female mice. Brain tissue was dissected after 14 months of treatment and processed for the analyses. (b) Serial block-face scanning electron microscopy (SBFSEM) with subsequent 3D EM reconstruction was done in the CA1 hippocampal region using Reconstruct and Amira software. Mitochondria (yellow), dendrites (red), axon (blue), and synaptic densities (green) were traced through consecutive slices generated using SBFSEM (left) to create 3D reconstructions (right). (c) Synapses were counted using Espina software in three regions of interest (ROI, red) with the dimension of 1000 pixels x 1000 pixels x 50 consecutive serial sections. ROIs excluded cell bodies and included only areas with neuropils. Voxel size was 5 nm x 5 nm x 75 nm (x, y, z). Each ROI has dimensions of 5 μm x 5 μm x 3.75 μm and total volume of 93.75 μm3. Scale bar, 2 μm. (d) Synaptic densities (green) were identified and counted using the ESPINA software in selected ROIs from vehicle or CP2 treated NTG and APP/PS1 mice. Micrographs depict representative synapses in 2D and 3D format in a NTG mouse. Scale bar, 400 nm.
Each synapse was first characterized as symmetric or asymmetric36. The symmetric synapses are defined by a synaptic density that is similar in size between pre- and postsynaptic compartments (Fig. 2a), while the asymmetric synapses have a prominent postsynaptic density (PSD) that is larger than the presynaptic density (Fig. 2b)37. The asymmetric and symmetric synapses relate to excitatory or inhibitory signaling, respectively37. An example of the 3D EM reconstruction of the asymmetric synapse within five consecutive sections is shown in Fig. 2c.
Analysis of synaptic structure using consecutive SBFSEM sections. Representative images of symmetric (a) and asymmetric (b) synapses. Symmetric synapses have similar width of pre- and postsynaptic densities associated with the synaptic junction. Asymmetric synapses have a thicker postsynaptic density compared to the presynaptic density. Black arrows indicate pre- and postsynaptic densities. Red arrows indicate synapses. SV, synaptic vesicles located in axons next to presynaptic densities. M, axonal mitochondrion. Scale bar, 160 nm (a) and 140 nm (b). (c) Representative serial sections of an asymmetric synapse with the visibly thick postsynaptic density highlighted in green. Red asterisk, axonal mitochondrion; yellow hashtag, dendritic mitochondrion. Scale bar, 400 nm.
We found that most synapses were asymmetric in all treatment groups (96% in NTG, 95% in APP/PS1, 98% in APP/PS1 + CP2) (Fig. 3b). In the NTG mouse, from 564 synapses counted across three ROI, 543 (96%) were asymmetric and 21 (4%) symmetric. In the vehicle-treated APP/PS1 mouse, from 190 synapses counted across three ROI, 181 (95%) were asymmetric and 9 (5%) were symmetric. In the APP/PS1 mouse treated with CP2, out of 505 synapses across three ROI, 496 (98%) were asymmetric and 9 (2%) were symmetric (Fig. 3b, Supplementary Table 1). Therefore, only asymmetric synapses were included in the analysis. We found that compared to the NTG littermate, the number of synapses was significantly reduced in the APP/PS1 mouse brain (Fig. 3a, c). CP2 treatment in the APP/PS1 mouse increased the number of synapses to the level observed in the NTG littermate (Fig. 3a, c). The 3D EM visualization of synapses in the brain tissue of the CP2-treated APP/PS1 mouse confirms previous observations demonstrating an improvement of synaptic function and LTP after CP2 treatment in APP/PS1 and 3xTgAD mice27,28,29.
Chronic CP2 treatment restores the number of synapses in the hippocampus of the APP/PS1 mouse to the observed in a NTG mouse. (a) Representative 3D EM reconstructions of synaptic densities in NTG, APP/PS1, APP/PS1 + CP2 mice. The analysis was done using asymmetric synapses only. Synaptic densities are colored in green. Reconstruction was performed using SBFSEM images from three ROIs of the CA1 region using Espina software. Scale bar, 800 nm. (b) The number of asymmetric (blue bars) vs. symmetric (red bars) synapses in each ROI. Since most synapses were asymmetric, the symmetric synapses were excluded from the further analyses. (c) Quantitative analyses of synapses from NTG, APP/PS1 (A/P) and APP/PS1 + CP2 mice. Each data point represents one ROI. Each ROI has dimensions of 5 μm x 5 μm x 3.75 μm and the total volume of 93.75 μm3. Data were generated from one mouse per group using three ROIs per each mouse.
CP2 treatment in APP/PS1 mice restores mitochondrial distribution at synapses
Mitochondria are an essential component of synapses providing energy for synaptic activity38. Mitochondrial morphology is different in dendritic (postsynaptic) and axonal (presynaptic) compartments39. In axons, mitochondria are small and sparse while in dendrites they are larger and form filamentous networks39,40,41. Mitochondrial localization to synapses is dependent on multiple factors, including axonal trafficking, calcium signaling, and the fidelity of fission/fusion machinery40. We have shown that in APP/PS1 mice, axonal trafficking is inhibited42 and CP2 treatment restored that deficiency29. Similarly, CP2 treatment improved mitochondrial dynamics enhancing biogenesis and turnover, which relies on robust fission/fusion cycles28. Therefore, we next examined the localization of mitochondria to dendritic and axonal synaptic compartments in NTG and vehicle- or CP2-treated APP/PS1 mice. We conducted the analyses to establish the distribution of mitochondria in synapses based on the maturation of DS randomly selecting ten thin, mushroom or stubby spines43 from one of the ROIs in NTG, APP/PS1 and APP/PS1 + CP2 mice (Fig. 4a, Supplementary Table 2). Mitochondria were taken into the analyses and reconstructions if they were located within 1 μm of the base of the DS and in the presynaptic axonal terminal (Fig. 4b). Mitochondria were classified using criteria established previously for the analyses of mitochondria-ER communication44: Type I with the length less than 0.5 μm; Type II with the length between 0.5 and 5 μm; Type III with the length over 5 μm; and Type IV MOAS, multiple teardrop-shaped mitochondria (~ 0.5 μm in diameter) connected by a thin double membrane up to 5 μm long (Fig. 4c).
We found that in the dendrites of the NTG mouse, all DS were primarily supported by Type III mitochondria (Fig. 4d). However, stubby DS were also associated with Type II mitochondria. MOAS were present at all DS in a relatively small number (Fig. 4d). The distribution of mitochondria in dendrites of the APP/PS1 mouse was strikingly different compared to the NTG (Fig. 4d). In all types of DS in the APP/PS1 mouse, we found a presence of Type I and Type II mitochondria with reduced abundance of Type III. MOAS remained present only in thin dendrites of the APP/PS1 mouse (Fig. 4d). We also found that the number of mitochondria at mushroom DS of the APP/PS1 mouse, the most mature type of DS, doubled compared to the NTG. In the APP/PS1 mouse treated with CP2, all DS were predominantly associated with MOAS and mitochondria Type III (Fig. 4d). After CP2 treatment, the number of mitochondria per each type of DS was the same as in the NTG mouse. These data indicate that synapses in the APP/PS1 mouse were increasingly associated with greater number of shorter mitochondria in all DS compared to a NTG mouse, and CP2 treatment rescued this phenotype. However, CP2 treatment increased the number of MOAS in all DS compared to the NTG mouse (Fig. 4d).
Mitochondrial morphology and distribution in axonal and dendritic synaptic compartments in the CA1 region. (a) Three types of DS were analyzed based on their level of maturation: thin with the spine head width < 1 μm, mushroom with the spine head width > 1 μm, and stubby with length to width ratio (LWR) < 1. (b) Analysis of mitochondrial distribution around synapses was done using organelles located within 1 μm of the base of the DS and in the presynaptic axonal terminal. D, dendrite; A, axon. (c) Mitochondria were classified using the following criteria: Type I with the length less than 0.5 μm; Type II with the length between 0.5 and 5 μm; Type III with the length over 5 μm; and Type IV MOAS, multiple teardrop-shaped mitochondria (~ 0.5 μm in diameter) connected by a thin double membrane up to 5 μm long. (d, e) Mitochondria distribution in dendrites (d) and axons (e) associated with a particular type of DS. Note that only mitochondria Type I and II were present in axonal presynaptic terminals. Ten spines of each type were taken into the analyses from three ROI per group. Color scheme for mitochondria in (d, e): grey, Type I; white, Type II; blue, Type III; red, Type IV MOAS.
The analysis of synaptic mitochondria in axonal shafts in respect to different types of DS revealed that in the NTG mouse, all DS were supported with mitochondria Type I and II (Fig. 4e). In the APP/PS1 mouse, the number of mitochondria was lower in stubby DS with complete loss of mitochondria Type I. Mushroom DS were associated with axonal mitochondria Type II only. CP2 treatment resulted in increased number of mitochondria at stubby DS and the shift to mitochondria Type I for all DS (Fig. 4e).
We next calculated the ratio of dendritic vs. axonal mitochondria per specific type of DS (Fig. 5a, b). Data suggest that in the NTG mouse, there were more mitochondria in dendrites vs. axons per synapse with the ratio of dendritic/axonal mitochondria of 1.7–2 regardless of the DS type. In the APP/PS1 mouse, the ratio increased significantly reaching 6 for the stubby DS. This indicates that more mitochondria have to be present in dendrites to support a synapse. CP2 treatment in an APP/PS1 mouse normalized this ratio for all DS to the levels observed in the NTG mouse (Fig. 5a, b). These data suggest that in NTG mice, synapses are supported with a permanent presence of longer mitochondria in dendrites but an occasional presence of shorter ones in axons with the average ratio between dendritic/axonal organelles of 2 per all types of DS. In the APP/PS1 mouse, the number of dendritic mitochondria were increased, especially in mushroom DS, and decreased in stubby DS, while the number of axonal mitochondria was decreased, especially in stubby DS, compared to the NTG mouse (Figs. 4d, e and 5a, b). The 3D EM reconstructions of mitochondrial localization around different types of DS is shown in Fig. 5c, d. Note the lack of mitochondria in axonal terminals and the mitochondrial network consisting of Type I, II and III in dendrites of the APP/PS1 mouse. CP2 treatment rescued mitochondrial distribution around DS to resemble the NTG mouse (Fig. 5a, b). Interestingly, after CP2 treatment, accumulation of dendritic mitochondria observed in an APP/PS1 mouse was replaced with MOAS (Fig. 5c–f).
Distribution of synaptic mitochondria in axons and dendrites in relationship to different type of DS. (a) Mitochondria distribution in axons and dendrites calculated per 10 synapses associated with different types of DS. The ratio (*) represents the number of dendritic mitochondria per axonal mitochondria per one DS of different type. (b) The ratio of dendritic/axonal mitochondria per the type of DS from (a) plotted for NTG (white), APP/PS1 (red) and APP/PS1 + CP2 (blue) mice. (c–f) Representative 3D EM reconstructions of dendrites (pink), axons (blue), and mitochondria (yellow) in the CA1 brain region of APP/PS1 (c, d) and APP/PS1 + CP2 (e, f) mice. #, thin DS; *, mushroom DS. Note mitochondria Type I and II in DS in (c,d) and MOAS in (e) and (f).
CP2 treatment in APP/PS1 mice improves expression of genes involved in synaptic function
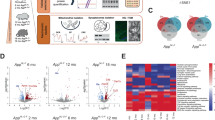
Labor-intensive 3D EM reconstruction of synapses and mitochondria limited the number of samples that we took into the analyses. Therefore, we confirmed whether CP2 improved mechanisms related to synaptic function in APP/PS1 mice using RNA-seq and Western blot analyses conducted in the brain tissue from multiple NTG and APP/PS1 mice treated with vehicle or CP2 within the same treatment cohorts as the mice used for 3D EM analyses28 (Fig. 1a). Principal component analysis (PCA) based on the RNA-seq data revealed clear separation among all groups of mice where CP2-treated APP/PS1 mice were separated from vehicle-treated counterparts (Fig. 6a). The comparison between vehicle-treated NTG and APP/PS1 mice identified 3320 differentially expressed genes (DEGs) while 1262 DEGs were identified between CP2- and vehicle-treated APP/PS1 mice28. The top biological processes associated with these DEGs included regulation of synaptic function and dendritic and axonal morphogenesis28. Out of 3,320 genes differentially affected in vehicle-treated APP/PS1 vs. NTG mice, the expression of 567 genes was reverted by CP2 treatment to the levels observed in NTG mice. Gene function enrichment analysis showed that pathways upregulated by CP2 included dendrite morphogenesis and spine maturation (Fig. 6b), axonal extension and guidance (Fig. 6c), and regulation of synaptic assembly and transmission (Fig. 6d, e). These data are consistent with the improved cognition and LTP shown in CP2-treated APP/PS1 mice using a battery of behavior tests and electrophysiology28.
Validation of RNA-seq data and structural findings were further conducted using Western blot analysis (Fig. 6f, g, Supplementary Fig. 1). In APP/PS1 mice, levels of post-synaptic protein PSD95, brain derived neurotrophic factor (BDNF) essential for neuronal function and survival45, and mitochondria specific deacetylase Sirtuin 3 (Sirt3) that is required for synaptic and neuronal functions46 were significantly reduced compared to NTG mice (Fig. 6f, g). CP2 treatment in APP/PS1 mice increased levels of synaptophysin, PSD95, Sirt3 and BDNF to the levels observed in NTG mice consistent with the observed restoration of the number of synapses and mitochondrial distribution reported here. Taken together, structural and functional data strongly support the notion that application of mitochondrial complex I inhibitor CP2 improves synaptic structure mitigating cognitive dysfunction in AD mouse models.
CP2 treatment improves gene expression related to synaptic function in brain tissue of APP/PS1 mice. (a) PCA based on the RNA-seq data generated on cortico-hippocampal brain tissue shows separated clusters of samples among three groups: NTG, green; APP/PS1 (AD), blue; APP/PS1 + CP2 (AD + CP2), orange. (b–e) Heat maps show changes in genes in pathways related to dendrite morphogenesis (b), regulation of axonal extension involved in axon guidance (c), synapse assembly (d) and synaptic transmission (e) after CP2 treatment in APP/PS1 mice. (f) Western blot analysis in the hippocampal tissue demonstrates increased levels of synaptophysin (Syn), post synaptic density 95 (PSD95), brain derived neurotrophic factor (BDNF), and sirtuin 3 (Sirt3) proteins in APP/PS1 mice after CP2 treatment. (g) Quantification of the Western blot from (f) All mice were 20 months of age treated with CP2 or vehicle for 12 months, n = 4–5 mice per group for RNA-seq and n = 6–8 per group for Western blot. Data are presented as mean ± S.E.M. A one-way ANOVA with Fisher`s LSD post-hoc test was used for statistical analysis. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. The original analyses of the RNAseq data and Western blots were reported in our previous publication28 and are repurposed here without major alterations.
Discussion
In this study, we extended our preclinical work demonstrating that mild mitochondrial complex I inhibition with the small molecule CP2 results in the restoration of synapses and mitochondrial distribution in the hippocampus of APP/PS1 mice. Previously, we have shown chronic CP2 treatment to improve synaptic activity, glucose metabolism, and cognition, while simultaneously reducing oxidative stress and inflammation in the brain of AD mice27,28. Here, we confirm and expand upon these findings using advanced SBFSEM and 3D EM reconstructions of synapses, providing a novel, structural and quantitative assessment of the impact of CP2 on synaptic and mitochondrial morphology and distribution. While structural analyses were conducted in one mouse per group, the collective evidence generated within the same cohorts of mice provide strong support for the utility of SBFSEM as a complementary tool for in-depth visualization of the brain. It is important to emphasize that, given the current technological constraints, robust tools for reconstructing EM data from SBFSEM remain limited. Despite this limitation, the data generated in our study provide significant insights into the method’s potential and offer additional validation for the improvement of synaptic function by visualizing synapses in 3D space.
Synaptic density in the hippocampus of the APP/PS1 mouse was observed to be markedly decreased compared to the NTG control. Despite a popular understanding that APP/PS1 mice experience cognitive decline mediated through changes at the synapse, synaptic density is rarely imaged and analyzed at the power of SBFSEM. By demonstrating decreased synaptic density in the APP/PS1 hippocampus, this model is further validated as a useful tool for drug discovery for AD since synaptic destruction is the best correlate of cognitive decline1. Importantly, this significant decrease in synaptic density was fully restored to NTG levels in the APP/PS1 mouse treated with CP2. Furthermore, the ratio of symmetric to asymmetric synapses was unaffected, with asymmetric (excitatory) synapses making up the vast majority of synaptic types observed between treatment and control groups. These findings reinforce safety of CP2 treatment in respect to the synapse, as synaptic density was restored without observed alterations in synaptic structure.
Transcriptomics, protein analysis, electrophysiology assays, and behavioral testing conducted in the cohorts of the same mice as were taken into the SBFSEM study28 correlate with our structural observations at the synaptic level, and further elucidate the restorative effects of CP2 at the synapse. RNA-seq data generated in APP/PS1 mice treated with CP2 reveal significant upregulation of synaptic genes known to be suppressed in AD. Identified upregulated genes involved in synapse assembly included Neurexin 1 (NRXN1), which facilitates proper alignment of the trans-synaptic complex, and Ephrin type-B receptor 2 (Ephb2), which promotes the development of excitatory synapses. Upregulated genes responsible for dendrite morphogenesis, axonal extension, and synaptic transmission included Down syndrome cell-adhesion molecule (DSCAM), involved in regulating neurite arborization and dendrite self-avoidance, BTB domain-containing protein 3 (BTBD3), which supports dendritic guidance toward the axon terminal, and Glutamate receptor 3 (GRIA4), a mediator of synaptic excitatory neurotransmission, metabotropic glutamate receptors 7 and 21 (GRMN7 and GRM2), which facilitate the formation of LTP, and Double C2 protein (Doc2a), which contributes to spontaneous excitatory and inhibitory release. Western blot analysis in the brain tissue of APP/PS1 mice treated with CP2 demonstrated increased levels of proteins indicative of synapse restoration including synaptophysin, a vesicular protein associated with presynaptic density, and postsynaptic density protein 95 (PSD95), a scaffolding protein associated with postsynaptic density. Furthermore, field excitatory postsynaptic potentials (fEPSPs) recorded in hippocampal slices showed that CP2-treated APP/PS1 mice exhibited significantly improved basal synaptic transmission and LTP compared to vehicle-treated APP/PS1 mice28. Finally, behavioral testing of CP2-treated APP/PS1 mice revealed improved motor coordination (rotarod), spatial memory (Morris Water Maze), and attention (Novel Object Recognition), among other cognitive enhancements. Together, these results support findings generated using SBFSEM, suggesting CP2 not only restored synaptic density, but also synaptic function.
3D EM reconstructions of mitochondria distribution at synapses in NTG and APP/PS1 mice revealed profound changes that support the loss of mitochondrial function in AD that could affect synaptic activity. The presence of mitochondria at the vehicle-treated APP/PS1 mouse dendrites was significantly increased, potentially indicating loss of individual mitochondrial capacity to support the synapse. CP2 treatment restored this errant mitochondrial distribution, reducing mitochondrial quantity at the dendrites to the NTG levels. Interestingly, the MOAS phenotype was significantly increased at the dendrites of the APP/PS1 mouse treated with CP2 compared to both vehicle-treated NTG and APP/PS1 mice. While the exact impact of MOAS structure on mitochondrial function is unknown, evidence suggests MOAS may represent an adaptive morphological configuration, mediated through fission arrest, that allows for enhanced metabolic exchange between neighboring mitochondria to maintain function under stress28,29,30,31. MOAS is often described in association with AD, and therefore decreased synaptic function and cognitive abilities29,30. However, despite the observed increase in MOAS, the same CP2-treated APP/PS1 mice still demonstrated significantly improved neuronal metabolism, synaptic function, and cognitive performance28. One possible explanation is that MOAS may represent an intermediate, reversible phenotype, which, although typically associated with pathology, might under certain conditions contribute to the stabilization of mitochondrial networks and support cellular adaptation in response to metabolic stress26,47. The observed increase in MOAS following CP2 treatment might therefore reflect a transitional state towards restored mitochondrial health rather than a fixed pathological outcome. It will be important to elucidate whether MOAS formation is unique to different neuronal compartments since our previous studies focused on mitochondria-ER communication demonstrated reduced levels of MOAS in hippocampus of APP/PS1 mice after CP2 treatment44. Further studies are warranted to elucidate the role of MOAS in the context of AD to determine the role in the therapeutic restoration of synaptic and cognitive functions.
Transcriptomics, proteomic analyses, and metabolic profiling underscore the improvement of mitochondrial function mediated by CP2, correlating with the observed restoration of mitochondrial quantity within dendrites. RNA-seq analysis revealed significant upregulation of numerous mitochondrial genes in CP2-treated APP/PS1 mice, including peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PGC-1α), Sirtuin 3 (SIRT3), and mitochondrial transcription factor A (TFAM), which play critical roles in mitochondrial biogenesis, stability, and function28. Western blot analysis additionally revealed increased expression of glucose transporters Glut 3 and 4, as well as a decreased ratio of phosphorylated vs. total pyruvate dehydrogenase (PDH), which together indicate improved glucose uptake and utilization28. Lastly, complementary metabolomic profiling and31P NMR spectroscopy demonstrated well-regulated ATP production with increased levels of N-acetyl aspartate (NAA) and citrate, reflecting improved tricarboxylic acid cycle activity and overall neuronal health28. Collectively, these findings suggest that CP2 treatment significantly restored mitochondrial function, thereby reducing mitochondrial quantity at the synapse to that of the NTG controls.
Furthermore, the analyses of the same data set conducted previously using SBFSEM and 3D EM reconstruction combined with Western blot analysis, RNA-seq, and metabolomics assessment demonstrated that CP2 treatment also restores mitochondrial morphology and mitochondria-endoplasmic reticulum (ER) communication, reducing ER and unfolded protein response (UPR) stress in the APP/PS1 mouse brain44. CP2 treatment in APP/PS1 mice reduced mitochondria-ER contact sites (MERCS) known to facilitate abnormal lipid and calcium homeostasis, accumulation of Aβ and pTau, abnormal mitochondrial dynamics, and apoptosis44. The multifaceted neuroprotective cascade associated with CP2 treatment further emphasizes the important role mitochondria play in multiple cellular mechanisms, justifying these organelles as promising therapeutic target for complex neurodegenerative diseases.
In conclusion, examining neuronal architecture at electron microscope resolution allows for a deeper understanding of how CP2 treatment affects mitochondrial shape, distribution, and synaptic structure in APP/PS1 mice. CP2 treatment restored synaptic density and mitochondrial distribution, supported by transcriptomic, proteomic, and functional analyses. Our results provide multimodal validation of the CP2 efficacy in improving synaptic and mitochondrial health in this prominent AD model. Due to the resource-intensive nature of the SBFSEM protocol, our 3D EM analyses were conducted on a single mouse per treatment group. Despite the limited sample size, these findings demonstrate the detailed insights SBFSEM can offer in AD drug discovery, providing strong structural confirmation of neuronal changes.
Materials and methods
All experiments with mice were approved by the Mayo Clinic Institutional Animal Care and Use Committee, IACUC Protocol A00001186-16-R18. We confirm that our research complies with all pertinent ethical regulations, including the ARRIVE (Animal Research: Reporting of In Vivo Experiments) guidelines and the National Institutes of Health’s Guide for the Care and Use of Laboratory Animals. This study was reported in accordance with the ARRIVE guidelines. All Methods were carried out in accordance with relevant guidelines and regulations.
CP2 synthesis
CP2 was synthesized by the Nanosyn, Inc biotech company (http://www.nanosyn.com) as described48 and purified using HPLC. NMR spectra were used for authentication and to ensure the lack of batch-to-batch variation. Standard operating procedures (SOP) were developed and followed for CP2 preparation, storage, and administration to mice.
CP2 treatment in APP/PS1 and NTG mice
All experiments described in this study were conducted using tissue obtained from the previous study described here28. The following female mice were used: double transgenic APP/PS149 and their non-transgenic (NTG) littermates. Genotypes were determined by PCR as described in49. All animals were kept on a 12 h–12 h light-dark cycle, with a regular feeding and cage-cleaning schedule. Mice were randomly selected to study groups based on their age and genotype. NTG and APP/PS1 female mice (n = 16–21 per group) received either CP2 (25 mg/kg/day in 0.1% polyethylene glycol [PEG] dissolved in drinking water ad lib) or vehicle-containing water (0.1% PEG) from 9 to 23 months of age. Mice were housed 5 per cage, water consumption and weight were monitored weekly.
Tissue collection
At the end of the study, mice were sacrificed via cervical dislocation to avoid any effect of anesthetic on molecular or structural changes in the brain tissue35. Rapid removal of the entire brain took less than 2 min. The hemispheres from each mouse were dissected using SOP to ensure the same brain regions were subjected to the Western blot and RNAseq analyses (cortico-hippocampal regions), and electron microscopy examination (CA1 hippocampal region).
Three-dimensional (3D) EM using serial block face scanning electron microscopy (SBFSEM)
Images for 3D EM reconstructions were obtained using a VolumeScope 2 SEM™ (Thermo Fisher Scientific) electron microscope. Hippocampal CA1 region was dissected from vehicle and CP2-treated NTG and APP/PS1 mice using a protocol designed to ensure the consistency of tissue dissection for accurate analyses of the same brain region35. Tissue was cut into 2 mm3 pieces, and immersion-fixed in neutral 2.5% glutaraldehyde + 2.5% paraformaldehyde in 0.1 M cacodylate buffer + 2 mM CaCl2. After a 24 h fixation, tissues were prepared for SBFSEM using the protocol developed by the National Center for Microscopy and Imaging Research (La Jolla, CA)50: (1) samples were rinsed 4 × 3 min in 0.1 M cacodylate buffer + 2 mM CaCl2, (2) incubated in 2% osmium tetroxide in 0.15 M cacodylate buffer for 1.5 h rotating at RT, (3) incubated in 2% osmium tetroxide + 2% potassium ferrocyanide in 0.1 M cacodylate for 1.5 h rotating at RT, (4) rinsed in H2O 4 × 3 min, (5) incubated in 1% thiocarbohydrazide (TCH) 45 min at 50 °C, (6) rinsed in H2O 4 × 3 min, (7) incubated in fresh 2% osmium tetroxide in H2O 1.5 h rotating at RT, (8) rinsed in H2O 4 × 3 min, (9) incubated in 1% aqueous uranyl acetate overnight at 4 °C, (10) further incubation in uranyl acetate 1 h at 50 °C, (11) rinsed in H2O 4 × 3 min, (12) incubated in lead aspartate 1 h at 50 °C, (13) rinsed in H2O 4 × 3 min, (14) dehydrated through ethanol series (60, 70, 80, 95, 100, 100%) 10 min each, (15) two rinses in 100% acetone 10 min each, (16) resin 1:2, 1:1, 3:1 in acetone 0.5 h, 1 h, 2 h, overnight in 100% resin. Samples were embedded into the Durcapan hard resin (EMS, Hatfield, PA), and allowed to polymerize at a minimum of 24 h prior to trimming and mounting. Tissue was trimmed of all surrounding resin and adhered to 8 mm aluminum pins (Ted Pella Inc., Redding, CA) using EpoTek silver epoxy (EMS, Hatfield, PA). A square tower (0.5 mm) was trimmed for the tissue using a Diatome ultratrim knife (EMS, Hatfield, PA, USA), and the entire pin was coated with gold palladium. A VolumeScope 2 SEM™ (Thermo Fisher, Inc., Waltham, MA, USA) with a starting energy of 1.5 kV was used to obtain serial block-face images for 3D EM reconstructions. 75 nm thick serial sections were removed from the block face while acquirring images through the z-axis. Reconstruct51, ESPINA52 and Amira 2020.2 (Thermo Fisher, Inc., Waltham, MA, USA) software were used to perform 3D reconstructions of dendrites, axons, and mitochondria, measure the number of synapses, and analyze mitochondrial associations with dendritic spines and axons, respectively.
Quantitative analysis and image segmentation using 3D EM
Tissue from one mouse per group was randomly selected and analyzed. For each mouse, the image stack acquired for 3D EM reconstructions consisted of 280–320 consecutive serial sections. Each image was processed in ImageJ53 to enhance the contrast, align or sharpen edges to make neuropils and organelle boundaries distinguishable. Images were then exported into Reconstruct in TIF format for 3D reconstruction. Manual segmentation of axons, dendrites, synapses, and mitochondria was performed by tracing consecutive serial images to generate 3D voxel segmentations with dimensions of 5 nm x 5 nm x 75 nm. Images were exported from ImageJ with just a Gaussian blur filter in TIF format into Espina52 (https://cajalbbp.es/espina/) for synaptic counting analysis and in JPEG format into Amira 2020.2 (Thermo Fisher, Inc., Waltham, MA, USA) for dendritic spine and mitochondria analysis with the same voxel dimensions. Espina software was used primarily due to its automated segmentation capabilities as the analysis, manual segmentation, and quantification of synapses are arduous and labor intensive.
The assessment of synaptic density
To evaluate the number of synapses, three regions of interest (ROI) were selected from each block in areas free of cell bodies. Each ROI consisted of 1000 × 1000 pixels spanning over 50 consecutive serial sections. The resulting three ROIs had the same dimensions of 5 μm x 5 μm x 3.75 μm. Synapses were counted using Espina Freehand Segmentation and the Grey Level Segmentation tools. Synapses that were only partially included in the ROI were excluded from the analysis. Synapses were identified by the presence of pre- and postsynaptic membrane densities and presynaptic vesicles as described in previous studies (Fig. 2a)37,54. Synapses were then categorized as either asymmetrical (excitatory) if they had a prominent postsynaptic density (PSD) that was larger than the presynaptic density or symmetrical (inhibitory) if the PSD was thin and similar in size to the presynaptic density37,55,56. For each synapse, the evaluation of consecutive images was done to define the origin of the synapse (symmetric vs. asymmetric). For each mouse, the number of synapses for each ROI was used as a single data point.
The assessment of mitochondrial morphology
The assessment of mitochondrial morphology in relationship to synapses in dendrites and axons was done in one of the three ROIs used to assess the number of synapses. The selection of the ROI taken into the analysis was random. For each ROI, the analysis was done using 10 synapses formed on dendritic spines of 3 different types: mushroom, thin, and stubby (30 in total). If 10 synapses for each spine type could not be found within the ROI, the search extended into the entire image stack produced by SEM. The randomly selected spines protruding from the dendrites were segmented, measured, and categorized using the following criteria57: mushroom (head > 0.6 μm); thin (head < 0.6 μm); stubby (length < 1 μm; length : width ratio < 1).
Mitochondrial morphology was established for organelles located within 1 μm of the base of the dendritic spine protrusion and within the presynaptic terminal. For categorizing mitochondrial morphology, the classification of mitochondrial types was adapted from58: Type I: round-shaped, less than 0.5 μm long; Type II: medium size, between 0.5 and 5 μm; Type III: elongated, longer than 5 μm; Type IV mitochondria-on-a-string (MOAS), multiple teardrop-shaped mitochondria (~ 0.5 μm in diameter) connected by a thin double membrane up to 5 μm long23. The length of each mitochondrion located within the designated area was recorded together with the number of mitochondria per dendritic spine located in axon and dendrite. For NTG and APP/PS1 + CP2 mice, data analyses included 30 spines. For APP/PS1 + Veh mouse, the total number of spines was 28 since only 8 stubby spines could be found in the entire image stack.
Next-generation RNA sequencing
The original analyses of the RNAseq data with corresponding Tables were reported in our previous publication28. Brain tissue, encompassing the hippocampal and cortical regions, from APP/PS1 and NTG mice treated with vehicle of CP2 for 14 months (n = 5 per group) were lysed in QIAzol (Qiagen cat. # 79306) followed by RNA isolation using miRNeasy (Qiagen cat. # 217004) according to the manufacturer’s instructions. The quantity and quality of RNA were measured using a NanoDrop spectrophotometer and Agilent 2100 Bioanlyzer, respectively. All RINs (RNA integrity numbers) had a value greater than eight. Total RNA (200 ng) was used to generate libraries using TruSeq RNA Library Prep Kit v2 (Illumina). All samples were sequenced at the Mayo Clinic Medical Genome Facility (MGF) Sequencing Core by Illumina HiSeq 4000 with paired end 101-bp read length. Approximately 50 million single fragment reads were acquired per sample. MAP-RSeq v 2.1.1, a comprehensive computational pipeline developed by the Mayo Clinic’s Division of Biomedical Statistics and Informatics, was used to analyze RNA-Sequencing data. MAP-RSeq uses a variety of publicly available bioinformatics tools tailored by methods developed in-house. The main outputs of the MAP-RSeq workflow are the gene counts, expressed single nucleotide variants (eSNVs), gene fusion candidates, and quality control plots. The aligning and mapping of reads was performed using TopHat2 against the mm10 reference genome. The gene and exon counts were generated by FeatureCounts using the gene definitions files from Ensembl. ‘-O’ option within FeatureCounts was used to account for expression derived from regions shared by multiple genomic features. FeatureCounts was also executed for quantifying expression on a per exon basis by utilizing the ‘-f’ option. RseqQC59 was used to create a variety of quality control plots to ensure the results from each sample were reliable and could be collectively used for differential expression analysis. Upon the completion of all the deliverables from MAP-RSeq, a html document was created tying everything together in one interactive document. The R bioinformatics package DeSeq2 was used for differential gene expression analysis. The criteria for the selection of significant differentially expressed genes was P value < 0.05. Gene function enrichment was determined using the Database for Annotation, Visualization and Integrated Discovery (DAVID v. 6.8)60. Mouse transcriptional factors were downloaded from the Riken genome database and mapped to our RNA-seq differential gene list. The clustering heatmaps were generated based on unsupervised hierarchical clustering using Pearson correlation distance. Most graphs were generated using customized R programs.
Western blot analysis
The original Western blot analyses were reported in our previous publication28. Levels of proteins were determined from the cortico-hippocampal region of the brain of vehicle and CP2-treated NTG and APP/PS1 mice (n = 3–4 mice per group) by Western blot analysis. Tissue was homogenized and lysed using 1× RIPA Buffer plus inhibitors. Total protein lysates (25 µg) were separated in equal volume on 4–20% Mini-PROTEAN TGX™ Precast Protein Gels (Bio-Rad, cat. # 4561096) and transferred to an Immun-Blot polyvinylidene difluoride membrane (PVDF cat. # 1620177). The following primary antibodies were used: Synaptophysin (1:200, Santa Cruz Biotechnology, Santa Cruz, CA, cat. # 17750), BDNF (1:200, Santa Cruz Biotechnology, Santa Cruz, CA, cat. # 546), PSD95 (1:1000, Cell Signaling Technology, cat. # 2507), Sirt3 (1:1000, Cell Signaling Technology, cat. # 5490), Tubulin (1:5000, Biovision, cat. # 3708). The following secondary antibodies were used: donkey anti-rabbit IgG conjugated with Horseradish Peroxidase (1:10000 dilution, GE Healthcare UK Limited, UK) and sheep anti-mouse IgG conjugated with Horseradish Peroxidase (1:10000 dilution, GE Healthcare UK Limited, UK). Band quantification was done using Image Lab™ v. 6.0.
Statistics
Since only one mouse per treatment group was used for 3D EM analyses, statistics were not applied to these data. Other data were analyzed using two-way ANOVA, the two-tailed unpaired and paired Student t test, where appropriate. Data are presented as: mean ± S.E.M. for each group.
Data availability
RNA-seq data generated in this study using mice that support the findings are depositedto public repository and are freely available at: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE149248 (GEO accession ID is GSE149248). Source data underlying heatmaps and Western blots presented in Fig. 6 are available in Supplementary Data to the previous publication28.
References
Barthet, G. & Mulle, C. Presynaptic failure in Alzheimer’s disease. Prog Neurobiol. 194, 101801. https://doi.org/10.1016/j.pneurobio.2020.101801 (2020).
Pchitskaya, E. & Bezprozvanny, I. Dendritic spines shape analysis-classification or clusterization? Perspective. Front. Synaptic Neurosci. 12, 31. https://doi.org/10.3389/fnsyn.2020.00031 (2020).
Reza-Zaldivar, E. E. et al. Dendritic spine and synaptic plasticity in Alzheimer’s Disease: a Focus on MicroRNA. Front. Cell. Dev. Biol. 8, 255. https://doi.org/10.3389/fcell.2020.00255 (2020).
Trejo-Lopez, J. A., Yachnis, A. T. & Prokop, S. Neuropathology of Alzheimer’s Disease. Neurotherapeutics 19, 173–185. https://doi.org/10.1007/s13311-021-01146-y (2022).
John, A. & Reddy, P. H. Synaptic basis of Alzheimer’s disease: focus on synaptic amyloid beta, P-tau and mitochondria. Ageing Res. Rev. 65, 101208. https://doi.org/10.1016/j.arr.2020.101208 (2021).
Ortiz-Sanz, C. et al. Early effects of Abeta Oligomers on Dendritic Spine Dynamics and Arborization in hippocampal neurons. Front. Synaptic Neurosci. https://doi.org/10.3389/fnsyn.2020.00002 (2020).
Zhou, L. et al. Tau association with synaptic vesicles causes presynaptic dysfunction. Nat. Commun. 8, 15295. https://doi.org/10.1038/ncomms15295 (2017).
Knafo, S. et al. Widespread changes in dendritic spines in a model of Alzheimer’s disease. Cereb. Cortex. 19, 586–592. https://doi.org/10.1093/cercor/bhn111 (2009).
Penzes, P., Cahill, M. E., Jones, K. A., VanLeeuwen, J. E. & Woolfrey, K. M. Dendritic spine pathology in neuropsychiatric disorders. Nat. Neurosci. 14, 285–293. https://doi.org/10.1038/nn.2741 (2011).
Tsai, J., Grutzendler, J., Duff, K. & Gan, W. B. Fibrillar amyloid deposition leads to local synaptic abnormalities and breakage of neuronal branches. Nat. Neurosci. 7, 1181–1183. https://doi.org/10.1038/nn1335 (2004).
Mullard, A. Alzheimer prevention hopes continue to dim. Nat. Rev. Drug Discov. https://doi.org/10.1038/d41573-020-00044-w (2020).
Panza, F., Lozupone, M., Logroscino, G. & Imbimbo, B. P. A critical appraisal of amyloid-beta-targeting therapies for Alzheimer disease. Nat. Rev. Neurol. 15, 73–88. https://doi.org/10.1038/s41582-018-0116-6 (2019).
Simmons, E. C., Scholpa, N. E. & Schnellmann, R. G. Mitochondrial biogenesis as a therapeutic target for traumatic and neurodegenerative CNS diseases. Exp. Neurol. 329, 113309. https://doi.org/10.1016/j.expneurol.2020.113309 (2020).
Sheng, Z. H. & Cai, Q. Mitochondrial transport in neurons: impact on synaptic homeostasis and neurodegeneration. Nat. Rev. Neurosci. 13, 77–93. https://doi.org/10.1038/nrn3156 (2012).
Devine, M. J. & Kittler, J. T. Mitochondria at the neuronal presynapse in health and disease. Nat. Rev. Neurosci. 19, 63–80. https://doi.org/10.1038/nrn.2017.170 (2018).
Bhatia, S. et al. Mitochondrial dysfunction in Alzheimer’s Disease: opportunities for Drug Development. Curr. Neuropharmacol. 20, 675–692. https://doi.org/10.2174/1570159X19666210517114016 (2022).
Kumar, V., Kim, S. H. & Bishayee, K. Dysfunctional Glucose Metabolism in Alzheimer’s Disease Onset and Potential Pharmacological Interventions. Int. J. Mol. Sci. https://doi.org/10.3390/ijms23179540 (2022).
Gabrielli, A. P. et al. Mitochondria profoundly Influence Apolipoprotein E Biology. J. Alzheimers Dis. 92, 591–604. https://doi.org/10.3233/JAD-221177 (2023).
Wang, W., Zhao, F., Ma, X., Perry, G. & Zhu, X. Mitochondria dysfunction in the pathogenesis of Alzheimer’s disease: recent advances. Mol. Neurodegener. 15, 30. https://doi.org/10.1186/s13024-020-00376-6 (2020).
Weidling, I. W. et al. Mitochondrial DNA manipulations affect Tau Oligomerization. J. Alzheimers Dis. 77, 149–163. https://doi.org/10.3233/JAD-200286 (2020).
Weidling, I. W. & Swerdlow, R. H. Mitochondria in Alzheimer’s disease and their potential role in Alzheimer’s proteostasis. Exp. Neurol. 330, 113321. https://doi.org/10.1016/j.expneurol.2020.113321 (2020).
Zhu, X., Perry, G., Smith, M. A. & Wang, X. Abnormal mitochondrial dynamics in the pathogenesis of Alzheimer’s disease. J. Alzheimers Dis JAD. 33 (Suppl 1), 253–262. https://doi.org/10.3233/JAD-2012-129005 (2013).
Zhang, L. et al. Altered brain energetics induces mitochondrial fission arrest in Alzheimer’s Disease. Sci. Rep. 6, 18725. https://doi.org/10.1038/srep18725 (2016).
Morozov, Y. M., Datta, D., Paspalas, C. D. & Arnsten, A. F. Ultrastructural evidence for impaired mitochondrial fission in the aged rhesus monkey dorsolateral prefrontal cortex. Neurobiol. Aging. 51, 9–18. https://doi.org/10.1016/j.neurobiolaging.2016.12.001 (2017).
Tyumentsev, M. A., Stefanova, N. A., Kiseleva, E. V. & Kolosova, N. G. Mitochondria with morphology characteristic for Alzheimer’s Disease patients are found in the brain of OXYS rats. Biochem. (Mosc). 83, 1083–1088. https://doi.org/10.1134/S0006297918090109 (2018).
Kirov, S. A., Fomitcheva, I. V. & Sword, J. Rapid neuronal ultrastructure disruption and recovery during spreading Depolarization-Induced cytotoxic edema. Cereb. Cortex. 30, 5517–5531. https://doi.org/10.1093/cercor/bhaa134 (2020).
Stojakovic, A. et al. Partial Inhibition of Mitochondrial Complex I Reduces Tau Pathology and Improves Energy Homeostasis and synaptic function in 3xTg-AD mice. J. Alzheimers Dis. 79, 335–353. https://doi.org/10.3233/JAD-201015 (2021).
Stojakovic, A. et al. Partial inhibition of mitochondrial complex I ameliorates Alzheimer’s disease pathology and cognition in APP/PS1 female mice. Commun. Biol. 4, 61. https://doi.org/10.1038/s42003-020-01584-y (2021).
Zhang, L. et al. Modulation of mitochondrial complex I activity averts cognitive decline in multiple animal models of familial Alzheimer’s Disease. EBioMedicine 2, 294–305. https://doi.org/10.1016/j.ebiom.2015.03.009 (2015).
Trushina, E., Trushin, S. & Hasan, M. F. Mitochondrial complex I as a therapeutic target for Alzheimer’s disease. Acta Pharm. Sin. B. 12, 483–495. https://doi.org/10.1016/j.apsb.2021.11.003 (2022).
Trushina, E., Nguyen, T. K. O. & Trushin, S. Modulation of mitochondrial function as a therapeutic strategy for neurodegenerative diseases. J. Prev. Alzheimers Dis. 10, 675–685. https://doi.org/10.14283/jpad.2023.108 (2023).
Eisner, V., Picard, M. & Hajnoczky, G. Mitochondrial dynamics in adaptive and maladaptive cellular stress responses. Nat. Cell. Biol. 20, 755–765. https://doi.org/10.1038/s41556-018-0133-0 (2018).
Logsdon, B. A. et al. Meta-analysis of the human brain transcriptome identifies heterogeneity across human AD coexpression modules robust to sample collection and methodological approach. bioRxiv https://doi.org/10.1101/510420 (2019).
Yang, Y. & Zhang, L. The effects of caloric restriction and its mimetics in Alzheimer’s disease through autophagy pathways. Food Funct. 11, 1211–1224. https://doi.org/10.1039/c9fo02611h (2020).
Hinton, A. Jr. et al. A Comprehensive Approach to Sample Preparation for Electron Microscopy and the Assessment of mitochondrial morphology in tissue and cultured cells. Adv. Biol. (Weinh). https://doi.org/10.1002/adbi.202200202 (2023).
Romaus-Sanjurjo, D. et al. Symmetric and asymmetric synapses driving neurodegenrative disorders. Symmetry 13, 2333. https://doi.org/10.3390/sym13122333 (2021).
Colonnier, M. Synaptic patterns on different cell types in the different laminae of the cat visual cortex. An electron microscope study. Brain Res. 9, 268–287. https://doi.org/10.1016/0006-8993(68)90234-5 (1968).
Duarte, F. V., Ciampi, D. & Duarte, C. B. Mitochondria as central hubs in synaptic modulation. Cell. Mol. Life Sci. 80, 173. https://doi.org/10.1007/s00018-023-04814-8 (2023).
Popov, V., Medvedev, N. I., Davies, H. A. & Stewart, M. G. Mitochondria form a filamentous reticular network in hippocampal dendrites but are present as discrete bodies in axons: a three-dimensional ultrastructural study. J. Comp. Neurol. 492, 50–65. https://doi.org/10.1002/cne.20682 (2005).
Virga, D. M. et al. Activity-dependent compartmentalization of dendritic mitochondria morphology through local regulation of fusion-fission balance in neurons in vivo. Nat. Commun. 15, 2142. https://doi.org/10.1038/s41467-024-46463-w (2024).
Seager, R., Lee, L., Henley, J. M. & Wilkinson, K. A. Mechanisms and roles of mitochondrial localisation and dynamics in neuronal function. Neuronal Signal. 4, NS20200008. https://doi.org/10.1042/NS20200008 (2020).
Trushina, E. et al. Defects in Mitochondrial Dynamics and Metabolomic Signatures of Evolving Energetic Stress in Mouse Models of Familial Alzheimer’s Disease. PLoS One 7, e32737. https://doi.org/10.1371/journal.pone.0032737 (2012).
Rochefort, N. L. & Konnerth, A. Dendritic spines: from structure to in vivo function. EMBO Rep. 13, 699–708. https://doi.org/10.1038/embor.2012.102 (2012).
Panes, J. et al. Partial inhibition of Complex I restores mitochondrial morphology and Mitochondria-ER communication in Hippocampus of APP/PS1 mice. Cells https://doi.org/10.3390/cells12081111 (2023).
Matrone, C., Ciotti, M. T., Mercanti, D., Marolda, R. & Calissano, P. NGF and BDNF signaling control amyloidogenic route and abeta production in hippocampal neurons. Proc. Natl. Acad. Sci. U. S. A. 105, 13139–13144. https://doi.org/10.1073/pnas.0806133105 (2008).
Mishra, Y. & Kaundal, R. K. Role of SIRT3 in mitochondrial biology and its therapeutic implications in neurodegenerative disorders. Drug Discov. Today. 28, 103583. https://doi.org/10.1016/j.drudis.2023.103583 (2023).
Vincent, A. E., Turnbull, D. M., Eisner, V., Hajnoczky, G. & Picard, M. Mitochondrial nanotunnels. Trends Cell. Biol. 27, 787–799. https://doi.org/10.1016/j.tcb.2017.08.009 (2017).
Hua, D. H. et al. Syntheses and bioactivities of tricyclic pyrones. Tetrahedron https://doi.org/10.1016/S0040-4020(03)00687-2 (2003).
Holcomb, L. et al. Accelerated Alzheimer-type phenotype in transgenic mice carrying both mutant amyloid precursor protein and presenilin 1 transgenes. Nat. Med. 4, 97–100 (1998).
Deerinck;, T. J., Bushong;, E. A., Ellisman;, M. & Thor, A. Preparation of Biological Tissues for Serial Block Face Scanning Electron Microscopy (SBEM) v2. Protocols https://doi.org/10.17504/protocols.io.36wgq7je5vk5/v2 (2022).
Fiala, J. C. Reconstruct: a free editor for serial section microscopy. J. Microsc. 218, 52–61. https://doi.org/10.1111/j.1365-2818.2005.01466.x (2005).
Morales, J. et al. Espina: a tool for the automated segmentation and counting of synapses in large stacks of electron microscopy images. Front. Neuroanat. 5, 18. https://doi.org/10.3389/fnana.2011.00018 (2011).
Schneider, C. A., Rasband, W. S. & Eliceiri, K. W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 9, 671–675. https://doi.org/10.1038/nmeth.2089 (2012).
Merchan-Perez, A., Rodriguez, J. R., Alonso-Nanclares, L., Schertel, A. & Defelipe, J. Counting synapses using FIB/SEM Microscopy: a true revolution for ultrastructural volume Reconstruction. Front. Neuroanat. 3, 18. https://doi.org/10.3389/neuro.05.018.2009 (2009).
Kwon, T. et al. Ultrastructural, molecular and functional mapping of GABAergic synapses on dendritic spines and shafts of neocortical pyramidal neurons. Cereb. Cortex. 29, 2771–2781. https://doi.org/10.1093/cercor/bhy143 (2019).
DeFelipe, J., Marco, P., Busturia, I. & Merchan-Perez, A. Estimation of the number of synapses in the cerebral cortex: methodological considerations. Cereb. Cortex. 9, 722–732. https://doi.org/10.1093/cercor/9.7.722 (1999).
Risher, W. C., Ustunkaya, T., Singh Alvarado, J. & Eroglu, C. Rapid golgi analysis method for efficient and unbiased classification of dendritic spines. PLoS One. 9, e107591. https://doi.org/10.1371/journal.pone.0107591 (2014).
Garza-Lopez, E. et al. Protocols for Generating surfaces and measuring 3D organelle morphology using Amira. Cells https://doi.org/10.3390/cells11010065 (2021).
Wang, L., Wang, S. & Li, W. RSeQC: quality control of RNA-seq experiments. Bioinformatics 28, 2184–2185. https://doi.org/10.1093/bioinformatics/bts356 (2012).
Huang da, W., Sherman, B. T. & Lempicki, R. A. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37, 1–13. https://doi.org/10.1093/nar/gkn923 (2009).
Acknowledgements
We thank Drs. A. Stojakovic, X. Li, M. Dehankar and S. Trushin for help with the RNAseq data analyses. This research was supported by grants from the National Institutes of Health NIA RF1AG55549, NINDS R01NS107265, RO1AG062135 (all to ET). Its contents are solely the responsibility of the authors and do not necessarily represent the official view of the NIH. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
E.T. conceived the study, received funding for the project. N.K and T.C. performed the research, analyzed and interpreted data, J.L.S. supervised SBFSEM experiments, E.W., N.K, J.L.S. and E.T. wrote the paper. All authors edited the manuscript and approved its publication.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Keller, N., Christensen, T.A., Wanberg, E.J. et al. Neuroprotective mitochondria targeted small molecule restores synapses and the distribution of synaptic mitochondria in the hippocampus of APP/PS1 mice. Sci Rep 15, 6528 (2025). https://doi.org/10.1038/s41598-025-90925-0
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-90925-0