Abstract
This study utilized two-sample MR to investigate causality between genetically predicted inflammatory markers and the risk of IDB. This research leveraged publicly available GWAS summary statistics to collect data on inflammatory cytokines and IDB. The IVW method was primarily employed for causal inference, supplemented by weighted median, mode-based estimation, and MR-Egger regression. Stringent sensitivity methods included Cochran’s Q test, MR-Egger regression, MR-PRESSO, and leave-one-out analyses to assess the robustness of the findings. This study selected 452 instrument variables (IVs) related to inflammatory factors. The IVW analysis revealed that GROa and RANTES/CCL5 exhibited causal relationships with IDB. Additionally, after removing outliers, significant causal associations were observed for IL-1ra and IL-9. Notably, the causal associations of RANTES/CCL5 and IL-9 with IBD remained significant after FDR correction. Upon integrating the findings from all sensitivity analyses, it is unlikely that heterogeneity and pleiotropy substantially influenced the observed relationships, underscoring the robustness of our findings. Our MR analysis identified the causal roles of specific inflammatory cytokines such as GROa, RANTES/CCL5, IL-1ra and IL-9 in the development of IDB. These findings deepen our understanding of the complex regulatory mechanisms involving inflammation in breast diseases and suggest directions for future research on biological pathways linking inflammation with IDB.
Similar content being viewed by others
Introduction
Inflammatory disorders of the breast (IDB) encompass a range of conditions characterized by inflammation, which can present with symptoms such as pain, heat, and redness. These disorders can be debilitating, leading to prolonged morbidity and varying in severity from benign to aggressive malignancies1,2. The spectrum of IDB can be categorized into infectious mastitis, non-infectious mastitis, and mastitis associated with underlying malignancy2,3. Additionally, they may manifest with nonspecific symptoms that can complicate early diagnosis and necessitate appropriate treatment4,5. Recognizing the risk factors for inflammatory breast disorders is essential for timely diagnosis and intervention, which are critical for enhancing patient outcomes.
Inflammatory cytokines, which include chemokines, growth factors, interleukins, and other related molecules, are integral regulators of the immune response6,7. They have been identified as key players in the pathogenesis of various diseases, including inflammatory breast disorders6,7,8. A growing body of observational evidence suggests a significant association between specific inflammatory cytokines and the development of IDB. For example, Ibrahim et al. reported that cytokine array profiling of cancer-associated adipose tissue ex-vivo cultures from obese inflammatory breast cancer (IBC) patients revealed a significantly higher secretion of a panel of 28 cytokines compared to non-IBC patients9. Li and co-workers employed cytokine microarray detection to discern a pronounced upregulation in the expression levels of cytokine factors, notably interleukin-1β (IL-1β), monokine-induced by γ-interferon (MIG), macrophage inflammatory protein (MIP)-1α, MIP-1β, and tumor necrosis factor receptor 2 (TNF RII), in patients diagnosed with idiopathic granulomatous mastitis (IGM) relative to control subjects10. Furthermore, Iwase and colleagues elucidated that within the top 15 canonical pathways activated in IBC, the IL-7 signaling pathway, in conjunction with pathways such as ERK/MAPK and PDGF, is intricately linked to the estrogen receptor signaling pathway, thereby distinguishing it from non-IBC cases11. Despite these suggestive associations, establishing a definitive causal link between inflammatory cytokines and inflammatory breast disorders is challenging due to the limitations inherent in observational studies, such as confounding factors and the possibility of reverse causality.
Mendelian randomization (MR) is an innovative epidemiological method that utilizes naturally occurring genetic variations as instrumental variables to infer causality. This approach takes advantage of the random distribution of genetic variants during meiosis, which reduces many of the biases found in traditional observational research12,13. MR provides a robust framework for investigating the potential causal effects of inflammatory factors on inflammatory breast disorders, free from the influence of confounding or reverse causality. This study employs a two-sample MR design to explore the causal relationship between inflammatory cytokines and IDB, focusing on the impact of these cytokines on the development of the disorders. By integrating genome-wide association study (GWAS) data on 41 inflammatory cytokines and outcomes related to inflammatory breast disorders, our investigation aims to provide a more reliable basis for causal inference than purely observational studies.
Methods
Study design
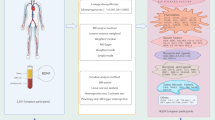
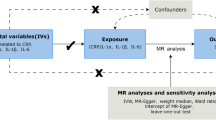
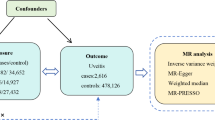
Our MR study, as depicted in Fig. 1, adheres to the MR-STROBE guidelines14 and is designed to explore the potential causal associations between inflammatory cytokines and IDB. The two-sample MR approach is predicated on three key assumptions. Assumption 1: the instrumental variables (IVs) directly affect the exposure; Assumption 2: IVs are not associated with confounders; Assumption 3: IVs influence risk of the outcome directly through the exposure, not through other pathways15.
Data sources
Outcome data for IDB were procured from the FinnGen Consortium, encompassing 1,880 cases and 211,699 controls. Data for the 41 inflammatory cytokines were sourced from a prior investigation16, encompassing chemokines, growth factors, interleukins, and additional cytokines. Table S1 provides a comprehensive overview. All data originated from peer-reviewed studies or publicly accessible GWAS summary data, with ethical approval and informed consent already obtained. This study did not necessitate separate ethical clearance.
Instrumental variable selection
Genetic instrumental variables were initially sought for each cytokine and IDB trait with a stringent significance threshold of P < 5 × 10− 8. However, due to insufficient single nucleotide polymorphisms (SNPs) meeting this threshold, the criterion was adjusted to a more lenient significance level of P < 5 × 10− 6. These selected SNPs were independently associated with their respective exposures within populations of European ancestry17. SNPs with substantial linkage disequilibrium (LD; r² < 0.001 within a 10,000 kb window) were excluded, retaining only the SNP with the most pronounced P-value18. In cases where an IV was absent in the outcome summary data, proxy SNPs with high LD with the original IV (R² > 0.8) were identified. The predictive power of each SNP as an IV was quantified using the F-statistic, calculated as follows: F = R2×(N − 2)/(1 − R2), where R2 is the proportion of variance in the exposure explained by the SNP in the IV, ensuring adequate predictive strength (F > 10)19 Finally, we used an online web tool (https://sb452.shinyapps.io/power/) to calculate the statistical power of each cytokine. GWAS Catalog (https://www.ebi.ac.uk/gwas/) was utilized to further assess whether the IVs might be associated with confounding factors or risk factors for IDB20.
Mendelian randomization analysis
The random-effects inverse variance weighted (IVW) method was utilized as the primary analytical technique to estimate the causal impact of inflammatory factors on IDB, with Odds Ratios (OR) and corresponding 95% Confidence Intervals (CI) being calculated21. To ensure the robustness of the findings, alternative MR methods were employed, including MR-Egger regression, weighted median, and weighted mode estimators. The MR-Egger method, which accounts for an intercept term, provides unbiased causal effect estimates even in the presence of potential pleiotropic bias22. The weighted median method presupposes that half of the IVs are valid, thus estimating the causal link between exposure and outcome23.
Sensitivity analysis
Sensitivity analyses were conducted to assess potential violations of the MR assumptions due to horizontal pleiotropy. Cochran’s Q statistic was calculated to measure heterogeneity in effect sizes, with a P-value > 0.05 indicating low heterogeneity and suggesting that the variation among IV estimates is random and minimally impactful on the IVW results24. The MR-Egger regression was used to evaluate the influence of horizontal pleiotropy on the estimated association, with a nonsignificant intercept term indicating the absence of pleiotropy that could bias the results25. Additionally, the MR pleiotropy residual sum and outlier (MR-PRESSO) method was employed to identify and exclude outlier SNPs (P < 0.05)26. Steiger tests were incorporated to examine causal directions. Leave-one-out analyses were performed to ensure the robustness and consistency of the findings, demonstrating that the conclusions remain stable when individual genetic variants are sequentially omitted27.
In this study, we applied both unadjusted and adjusted thresholds for statistical significance. An unadjusted P-value threshold of P < 0.05 was used for initial significance testing. To account for multiple comparisons, P-values were further adjusted using the Benjamini-Hochberg method, maintaining a significance threshold of P < 0.05 for FDR control. To bolster the interpretability of our findings, we employed visualization techniques such as scatter plots and diagrams illustrating the results of sensitivity analyses. The computational framework for all analyses was established using the “TwoSampleMR” package within the R statistical environment, specifically version 4.0.5, ensuring a robust and standardized approach to our data evaluation.
Results
Instrumental variable selection
In our MR analysis, after rigorous quality control measures, 452 SNPs were identified as IVs for inflammatory cytokines as exposures (Table S2). For IDB as an outcome, 17 SNPs (rs145902143, rs80341932, rs118158560, rs9450351, rs10892381, rs13143163, rs116615337, rs6900267, rs9793308, rs74966328, rs111913416, rs11700536, rs56134659, rs10903540, rs115360066, rs73479333, rs112783231)not matched in the summary and were excluded. The mean F-statistic for the IVs was 39.14, with a minimum of 11.16 and a maximum of 788.95, indicating no weak instrument bias in our analysis and the post hoc statistical power analysis. The power analysis showed moderate power (≥ 50%) for RANTES/CCL5, GROa, and SDF1a in detecting significant associations (Table S3). IVs related to confounding factors were excluded from the analysis (Table S4). Notably, after excluding IVs associated with confounding factors, there were insufficient SNPs for further analysis between P10, MCP3, IL-12p70, IL-8, and IDB.
Causal effects of inflammatory cytokines on IDB
The IVW analysis indicated a negative causal relationship between GROa and the risk of IDB (OR 0.86, 95% CI 0.74–0.99, P = 0.04, FDR = 0.099), although MR Egger, weighted mode, and weighted median methods did not establish a causal association (all P > 0.05) (Table 1; Table S5; Figrue 2 A; Figrue 3 A). A similar negative causal relationship was observed for RANTES/CCL5 (OR 0.83, 95% CI 0.71–0.98, P = 0.026, FDR = 0.048), supported by the weighted median (OR 0.78, 95% CI 0.623–0.987, P = 0.0329, FDR = 0.048) (Table 1; Table S5; Figs. 2B and 3B). Cochran’s Q test did not detect significant heterogeneity, and MR-Egger analysis showed no evidence of directional pleiotropy affecting risk estimates for the associations between GROa, RANTES/CCL5, and IDB (Table 2). The MR-presso test did not identify any outlier SNPs or horizontal pleiotropy (Table 3).
Scatter plots of Mendelian randomization models: exploring the potential associations between IDB and GROa (A), RANTES/CCL5 (B), IL-1ra (C), and IL-9 (D). SNP, single-nucleotide polymorphism; IDB, inflammatory disorders of the breast; MIG, Monokine induced by gamma interferon; PDGFbb, Platelet-derived growth factor BB.
Forest plot of MR effect size for potential relationship between IDB and GROa (A), RANTES/CCL5 (B), IL-1ra (C), and IL-9 (D). SNP, single-nucleotide polymorphism; IDB, inflammatory disorders of the breast; MR, Mendelian randomization; MIG, Monokine induced by gamma interferon; PDGFbb, Platelet-derived growth factor BB.
Additionally, primary analysis did not reveal significant causal associations between IL-1ra (OR 0.92, 95% CI 0.65–1.30, P = 0.633, FDR = 0.633), IL-9 (OR 0.87, 95% CI 0.59–1.26, P = 0.453, FDR = 0.566) and IDB, with their associations showing significant heterogeneity (Tables 1 and 2). After the removal of outliers (rs11869294 for IL-1ra, rs61867538 for IL-9) identified through MR-PRESSO (Table 3), the causal associations between IL-1ra (OR 0.78, 95% CI 0.62–0.99, P = 0.038, FDR = 0.095) (Table 1; Figs. 2C and 3C), IL-9 (OR 0.68, 95% CI 0.52–0.89, P = 0.005, FDR = 0.013) and IDB turned significant (Table 1; Figs. 2D and 3D). In addition, the associations between IL-1ra and IL-9 with IDB exhibited no evidence of significant heterogeneity after outliers were removed (Table 2). However, the association between IL-13 and IDB remained insignificant even after the removal of outliers (rs12623722 and rs27949) according to the leave-one-out analysis, even though no pleiotropy existed after their removal (Tables 1 and 2). In addition, as IL-10 exhibited significant heterogeneity (Table 2), its association with IDB were examined via random effect IVW method. The results indicated that IL-10 was not causally associated with IDB via fixed-effect IVW (OR 1.17, 95% CI 0.79–1.75, P = 0.432, FDR = 0.719) or random-effect IVW method (OR 1.17, 95% CI 0.79–1.75, P = 0.432, FDR = 0.719) (Table 1).
No other inflammatory factors investigated showed a statistically significant relationship with IDB (Table 1). Sensitivity analyses revealed no evidence of heterogeneity, pleiotropy, or outlier SNPs. (Tables 2 and 3).
Through MR Steiger tests, we confirmed the consistency in causal directions of all inflammatory factors on IDB (Table 4). The symmetrical distribution of funnel plots in Fig. 4A-D, suggesting that the estimates of the relationships between GROa, RANTES/CCL5, IL-Ira and IL-9 and IDB were not influenced by any single outlier SNP. The consistency of their associations was further confirmed by leave-one-out sensitivity analyses, as depicted in Fig. 5A-D, with no significant alteration in the observed relationships upon the exclusion of any single SNP, underscoring the reliability of our study’s conclusions.
Funnel plot of IVW model and MR-Egger model for potential relationship between IDB and GROa (A), RANTES/CCL5 (B), IL-1ra (C), and IL-9 (D). SNP, single-nucleotide polymorphism; IDB, inflammatory disorders of the breast; MR, Mendelian randomization; MIG, Monokine induced by gamma interferon; PDGFbb, Platelet-derived growth factor BB.
Discussion
The findings from this MR study shed new light on the causal relationship between inflammatory cytokines and the development of IDB. By bypassing the biases often encountered in traditional observational studies, our research presents compelling evidence for potential causal relationships between specific cytokines and the risk of IDB. Notably, the negative causal associations observed for GROa, RANTES/CCL5, IL-1ra and IL-9 suggest that these cytokines may exert a protective influence against the onset of IDB, a finding that merits further exploration.
GROa, also known as CXCL1, is a member of the CXC chemokine family. It mainly acts as a chemoattractant, especially for neutrophils, and is involved in inflammation, angiogenesis, and tumorigenesis. While direct studies linking GROa specifically to IDB are limited, its involvement in inflammatory breast cancer implies potential relevance. For instance, research shows that GROa exerts pro-survival and anti-apoptotic effects on breast cancer cells, which are crucial for chemoresistance and radioresistance28. It also induces the migration and epithelial-to-mesenchymal transition (EMT) of breast cancer cells by activating the extracellular signal-regulated kinase (ERK) MAPK pathway, leading to increased expression of matrix metalloproteinase 2 (MMP2) and MMP929. Additionally, GROa plays a role in angiogenesis by acting directly on endothelial cells and indirectly by increasing VEGF expression in breast cancer cells30. In contrast, our study revealed that GROa served as a protective factor for IDB, though the OR was relatively low, suggesting its limited role in IDB. The discrepancy may stem from the fact that IDB may differ significantly from inflammatory breast cancer. The protective effect observed in our study might indicate that GROa contributes to a different immune response or cellular environment in IDB, potentially mitigating inflammation or promoting tissue repair rather than exacerbating tumor growth. This discrepancy highlights the complexity of chemokine functions and suggests that the dual roles of GROa in both promoting and protecting against disease processes warrant further investigation to fully understand its implications in various pathological contexts.
RANTES/CCL5 serves a dual function as a T cell chemoattractant and an immune-regulatory molecule. It exerts its effects by signaling through specific G Protein-Coupled Receptors (GPCRs), namely CCR1, CCR3, and CCR5. Research by Maillard and colleagues has indicated that the biological impacts of RANTES/CCL5 are contingent upon the syndecan-4/PKCα signaling pathway31. Here, our findings indicated that RANTES/CCL5 was negatively associated with IDB risks, albeit with a relatively low OR, suggesting its limited role in this condition. It is conceivable that in IDB, RANTES/CCL5 may exert a protective influence by modulating inflammatory-associated cellular signaling pathways, which could involve the inhibition of pro-inflammatory cytokines such as TNF-α and IL-6s32. In addition, RANTES/CCL5 is involved in the recruitment of regulatory T cells (Tregs) to sites of inflammation. Tregs play a vital role in suppressing excessive immune responses and maintaining immune tolerance33. By attracting these cells, RANTES can help mitigate the inflammatory processes characteristic of IBD. However, further research is required to elucidate the detailed mechanisms by which RANTES/CCL5 inhibits IDB.
IL-1ra is a crucial anti-inflammatory cytokine that plays a significant role in regulating inflammatory responses by competitively binding to the IL-1 receptor (IL-1R). Our findings reveal that IL-1ra was negatively associated with the risk of IDB after outliers removal. Elevated levels of IL-1 are often observed in inflammation, contributing to inflammatory disorders. However, there has been limited report on the direct connection of IL-1ra and IDB. It can be postulated that dysregulation of the IL-1/IL-1ra axis can lead to exacerbated inflammatory responses in various diseases, including those affecting breast tissue. For instance, insufficient IL-1ra may fail to counteract the effects of elevated IL-1 levels, potentially leading to persistent inflammation and contributing to the pathogenesis of IDB34,35. On the other hand, IL-9 exerts a protective effect against IDB after outliers were removed. IL-9 is a pleiotropic cytokine primarily produced by CD4 + T helper cells and stimulates the growth of various immune cells. Although specific studies directly connecting IL-9 to IBD are limited, its roles in inflammation and immune modulation suggest potential implications. Given that IL-9 is involved in promoting inflammation through its effects on T cells and mast cells, it may influence the inflammatory microenvironment characteristic of IBD36. Despite these insights, further longitudinal studies are essential to elucidate the mechanisms involving GROa, RANTES/CCL5, IL-1ra and IL-9 in IDB and to assess their potential as therapeutic targets. Our findings contribute to the understanding of the complex interplay of inflammatory cytokines with breast diseases, emphasizing the need for ongoing research to clarify their individual roles and mechanisms of action.
Our research also failed to identify a causal relationship between IDB and other inflammatory mediators, such as IL-1β, MIG, IL-4, IL-10, MIP1, etc. However, the plausibility of a relationship between these factors and IDB remains a subject of interest. For instance, Li et al. utilized cytokine microarray detection to measure and analyze differentially expressed cytokine factors between patients with IGM and control subjects. Their findings revealed a significant increase in the expression of cytokines in IGM patients compared to controls, including IL-1β, MIG, MIP1α, MIP1β, and TNF RII10. Additionally, Du et al. employed univariate and multivariate analysis to demonstrate that IL-4, IL-10, and INF-α were independent diagnostic factors for abscess formation in granulomatous lobular mastitis (GLM). They further developed a predictive model for GLM abscess formation based on inflammatory markers, offering a novel strategy for the early diagnosis and treatment of GLM during the purulent phase37. Similarly, Mohamed et al. found that cytokine profiling of CD14 + cells isolated from IBC patients showed a marked increase in the secretion of TNF-α, MCP1/CC-chemokine ligand 2, IL-8, and IL-10, compared to those from non-IBC patients6. Rubbo and his team investigated immune markers in subclinical mastitis (SCM) breast milk samples, finding higher levels of inflammatory markers (TNF-α, IL-6, IL-8, IL-17, RANTES, etc.) and Th1-related cytokines (IL-2R, IL-12p40/70, IFN-α, IFN-γ, CXCL-9, and IP-10) associated with SCM, which was observed in 23% of women38.
Collectively, these studies underscore the necessity for continued research into the interplay between inflammatory mediators and IDB. While our study suggests that inflammatory cytokines such as GROa, RANTES/CCL5, IL-1ra and IL-9 are associated with the development of IDB, it is essential to consider the clinical translation of these findings. The potential for these cytokines to serve as biomarkers for disease progression and as therapeutic targets in clinical settings should be considered in the following research. Such efforts could explore the use of specific anti-cytokine therapies, which have already shown promise in other inflammatory conditions.
Our study benefits from the comprehensive evaluation of a wide array of inflammatory cytokines, providing a nuanced perspective on cytokine effects on breast diseases. However, the limitations of this study have to be addressed. Firstly, caution is advised when extending the conclusion of this study to other populations as this study was solely based on European ancestry. Future research should aim to include diverse ethnic groups to offer a more comprehensive understanding of the causal relationships being investigated. Secondly, the number of IVs employed in the analysis varied from 3 to 20, potentially impacting the MR findings due to the restricted amount of IVs. However, this is unlikely to mislead the study as the F-statistics of each IV exceeds 10. Thirdly, the absence of individual information hinders further categorization of patients into finer subgroups based on disease stages. Lastly, the statistical power for other exposure factors, aside from RANTES/CCL5 and GROa, was relatively low, thereby leading to an increased probability of type II errors.
Data availability
All data generated or analyzed during this study are included in this article and supplementary information files.
References
Scott, D. M. Inflammatory diseases of the breast. Best Pract. Res. Clin. Obstet. Gynaecol. 83, 72–87. https://doi.org/10.1016/j.bpobgyn.2021.11.013 (2022).
Lepori, D. Inflammatory breast disease: the radiologist’s role. Diagn. Interv Imaging. 96, 1045–1064. https://doi.org/10.1016/j.diii.2015.07.006 (2015).
Kamal, R. M., Hamed, S. T. & Salem, D. S. Classification of inflammatory breast disorders and step by step diagnosis. Breast J. 15, 367–380. https://doi.org/10.1111/j.1524-4741.2009.00740.x (2009).
Kasales, C. J. et al. Nonpuerperal mastitis and subareolar abscess of the breast. AJR Am. J. Roentgenol. 202, W133–139. https://doi.org/10.2214/AJR.13.10551 (2014).
Leong, P. W., Chotai, N. C. & Kulkarni, S. Imaging features of inflammatory breast disorders: A pictorial essay. Korean J. Radiol. 19, 5–14. https://doi.org/10.3348/kjr.2018.19.1.5 (2018).
Mohamed, M. M. et al. Cytokines secreted by macrophages isolated from tumor microenvironment of inflammatory breast cancer patients possess chemotactic properties. Int. J. Biochem. Cell. Biol. 46, 138–147. https://doi.org/10.1016/j.biocel.2013.11.015 (2014).
Liu, C. et al. Cytokines: from clinical significance to quantification. Adv. Sci. (Weinh). 8, e2004433. https://doi.org/10.1002/advs.202004433 (2021).
Winkler, M. Role of cytokines and other inflammatory mediators. BJOG 110 (Suppl 20), 118–123. https://doi.org/10.1016/s1470-0328(03)00062-4 (2003).
Ibrahim, A. S., El-Shinawi, M., Sabet, S., Ibrahim, S. A. & Mohamed, M. M. Role of adipose tissue-derived cytokines in the progression of inflammatory breast cancer in patients with obesity. Lipids Health Dis. 21, 67. https://doi.org/10.1186/s12944-022-01678-y (2022).
Li, F. et al. Evaluation of significantly changed chemokine factors of idiopathic granulomatous mastitis in non-puerperal patients. FASEB J. 38, e23745. https://doi.org/10.1096/fj.202400114RRR (2024).
Iwase, T. et al. Quantitative hormone receptor (HR) expression and gene expression analysis in HR + inflammatory breast cancer (IBC) vs non-IBC. BMC Cancer. 20, 430. https://doi.org/10.1186/s12885-020-06940-z (2020).
Ebrahim, S. & Davey Smith, G. Mendelian randomization: can genetic epidemiology help redress the failures of observational epidemiology? Hum. Genet. 123, 15–33 (2008).
Emdin, C. A., Khera, A. V. & Kathiresan, S. Mendelian Randomization JAMA 318, 1925–1926, doi:https://doi.org/10.1001/jama.2017.17219 (2017).
Skrivankova, V. W. et al. Strengthening the reporting of observational studies in epidemiology using Mendelian randomization: the STROBE-MR statement. JAMA 326, 1614–1621. https://doi.org/10.1001/jama.2021.18236 (2021).
Davies, N. M., Holmes, M. V. & Davey Smith, G. Reading Mendelian randomisation studies: a guide, glossary, and checklist for clinicians. BMJ 362, k601. https://doi.org/10.1136/bmj.k601 (2018).
Ahola-Olli, A. V. et al. Genome-wide association study identifies 27 loci influencing concentrations of Circulating cytokines and growth factors. Am. J. Hum. Genet. 100, 40–50. https://doi.org/10.1016/j.ajhg.2016.11.007 (2017).
Wang, S. et al. Assessment of the relationship between generalized convulsive epilepsy and systemic inflammatory regulators: a bidirectional Mendelian randomization study. Front. Neurol. 14, 1206290. https://doi.org/10.3389/fneur.2023.1206290 (2023).
Lawlor, D. A., Harbord, R. M., Sterne, J. A. C. & Timpson, N. Davey Smith, G. Mendelian randomization: using genes as instruments for making causal inferences in epidemiology. Stat. Med. 27, 1133–1163 (2008).
Bowden, J. et al. Assessing the suitability of summary data for two-sample Mendelian randomization analyses using MR-Egger regression: the role of the I2 statistic. Int. J. Epidemiol. 45, 1961–1974. https://doi.org/10.1093/ije/dyw220 (2016).
Zeng, W. et al. Causal associations between human gut microbiota and osteomyelitis: a Mendelian randomization study. Front. Cell. Infect. Microbiol. 14 https://doi.org/10.3389/fcimb.2024.1338989 (2024).
Burgess, S., Bowden, J., Fall, T., Ingelsson, E. & Thompson, S. G. Sensitivity analyses for robust causal inference from Mendelian randomization analyses with multiple genetic variants. Epidemiology 28, 30–42 (2017).
Bowden, J., Davey Smith, G. & Burgess, S. Mendelian randomization with invalid instruments: effect Estimation and bias detection through Egger regression. Int. J. Epidemiol. 44, 512–525. https://doi.org/10.1093/ije/dyv080 (2015).
Bowden, J., Davey Smith, G., Haycock, P. C. & Burgess, S. Consistent Estimation in Mendelian randomization with some invalid instruments using a weighted median estimator. Genet. Epidemiol. 40, 304–314. https://doi.org/10.1002/gepi.21965 (2016).
Bowden, J. et al. A framework for the investigation of Pleiotropy in two-sample summary data Mendelian randomization. Stat. Med. 36, 1783–1802. https://doi.org/10.1002/sim.7221 (2017).
Burgess, S. & Thompson, S. G. Interpreting findings from Mendelian randomization using the MR-Egger method. Eur. J. Epidemiol. 32, 377–389. https://doi.org/10.1007/s10654-017-0255-x (2017).
Verbanck, M., Chen, C. Y., Neale, B. & Do, R. Publisher correction: detection of widespread horizontal Pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat. Genet. 50, 1196. https://doi.org/10.1038/s41588-018-0164-2 (2018).
Wang, Q. et al. Shorter leukocyte telomere length is associated with adverse COVID-19 outcomes: A cohort study in UK biobank. EBioMedicine 70, 103485. https://doi.org/10.1016/j.ebiom.2021.103485 (2021).
Yang, B. et al. Tumor-associated macrophages/C-X-C motif chemokine ligand 1 promotes breast cancer autophagy-mediated chemoresistance via IGF1R/STAT3/HMGB1 signaling. Cell Death Dis. 15, 743. https://doi.org/10.1038/s41419-024-07123-5 (2024).
Ciummo, S. L. et al. The C-X-C motif chemokine ligand 1 sustains breast Cancer stem cell Self-Renewal and promotes tumor progression and immune escape programs. Front. Cell. Dev. Biology. 9, 689286. https://doi.org/10.3389/fcell.2021.689286 (2021).
Acharyya, S. et al. A CXCL1 paracrine network links cancer chemoresistance and metastasis. Cell 150, 165–178. https://doi.org/10.1016/j.cell.2012.04.042 (2012).
Proudfoot, A. E. et al. The BBXB motif of RANTES is the principal site for heparin binding and controls receptor selectivity. J. Biol. Chem. 276, 10620–10626. https://doi.org/10.1074/jbc.M010867200 (2001).
Maillard, L. et al. RANTES/CCL5 mediated-biological effects depend on the syndecan-4/PKCalpha signaling pathway. Biol. Open. 3, 995–1004. https://doi.org/10.1242/bio.20148227 (2014).
Zeng, Z., Lan, T., Wei, Y. & Wei, X. CCL5/CCR5 axis in human diseases and related treatments. Genes Dis. 9, 12–27. https://doi.org/10.1016/j.gendis.2021.08.004 (2022).
Arend, W. P., Malyak, M., Guthridge, C. J. & Gabay, C. Interleukin-1 receptor antagonist: role in biology. Annu. Rev. Immunol. 16, 27–55. https://doi.org/10.1146/annurev.immunol.16.1.27 (1998).
Fang, Z., Jiang, J. & Zheng, X. Interleukin-1 receptor antagonist: an alternative therapy for cancer treatment. Life Sci. 335, 122276. https://doi.org/10.1016/j.lfs.2023.122276 (2023).
Goswami, R. & Kaplan, M. H. A brief history of IL-9. J. Immunol. (Baltimore Md. : 1950). 186, 3283–3288. https://doi.org/10.4049/jimmunol.1003049 (2011).
Du, N. N., Feng, J. M., Shao, S. J., Wan, H. & Wu, X. Q. Construction of a Multi-Indicator model for abscess prediction in granulomatous lobular mastitis using inflammatory indicators. J. Inflamm. Res. 17, 553–564. https://doi.org/10.2147/JIR.S443765 (2024).
Tuaillon, E. et al. Subclinical mastitis occurs frequently in association with dramatic changes in inflammatory/anti-inflammatory breast milk components. Pediatr. Res. 81, 556–564. https://doi.org/10.1038/pr.2016.220 (2017).
Author information
Authors and Affiliations
Contributions
(I) Conception and design: Binyi Li. (II) Administrative support: Hongbo Ge. (III) Provision of study materials or patients: Hongxia We. (IV) Collection and assembly of data: Ying Qian. (V) Data analysis and interpretation: Hongxia Wei. (VI) Manuscript writing: All authors. (VII) Final approval of manuscript: All authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
This article is a mendelian randomization study. The data for this study were obtained from publicly available databases and published literature data and does not require ethical approval and written informed consent.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Wei, H., Ge, H., Qian, Y. et al. Genetic determinants of inflammatory cytokines and their causal relationship with inflammatory disorders of breast: a two-sample Mendelian randomization study. Sci Rep 15, 7300 (2025). https://doi.org/10.1038/s41598-025-91723-4
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-91723-4