Abstract
Periodontal inflammation is a chronic condition affecting the tissues surrounding teeth. Initiated by dental plaque, it triggers an immune response leading to tissue destruction. The AIM-2 inflammasome regulates this response, and understanding its peptide sequences could aid in developing targeted therapeutics. This study explores using transformers and graph attention networks (GAT) to treat periodontal inflammation. UniProt was used to download AIM-2 inflammasome proteins and FASTA sequences with 100%, 90%, and 50% similarity. DeepBio, a web service for developing deep-learning architectures, analyzed these sequences. Peptide sequence prediction methods were evaluated using a transformer, RNN-CNN, and GAT models. The transformer model achieved 84% accuracy, the GAT model 86%, and the RNN-CNN 64%. Both transformer and GAT models predicted peptide sequences more effectively than the RNN-CNN model, with the Transformer showing the highest class accuracy at 85%, followed by the GAT model at 80%. Models exhibited varying sensitivity and specificity, with the Transformer demonstrating superior performance in overall and class-specific peptide sequence prediction. AI-based peptide sequence prediction using transformers, GAT, and RNN-CNN shows promise for accurately predicting AIM-2 peptide sequences, with transformers and GAT outperforming RNN-CNN in accuracy and class accuracy.
Similar content being viewed by others
Introduction
Periodontal inflammation is a chronic inflammatory condition that affects the periodontium, including the gingiva, periodontal ligament, cementum, and alveolar bone, which support the teeth1,2. It begins with the accumulation of dental plaque, a biofilm composed of bacteria and their byproducts, which triggers an immune response. This response activates immune cells and releases proinflammatory cytokines such as IL-1 and TNF-α, destroying tissue. The inflammation extends into deeper periodontal tissues, causing tooth mobility, and if left untreated, can result in tooth loss. Periodontitis is also associated with systemic conditions such as cardiovascular disease, diabetes, and adverse pregnancy outcomes. Effective management of periodontal inflammation requires a multidisciplinary approach, including oral hygiene measures and lifestyle modifications3,4.
Inflammasomes play a critical role in the development and progression of periodontal inflammation, a condition characterized by the destruction of tissues surrounding the teeth. These multiprotein complexes produce proinflammatory cytokines, particularly interleukin-1β (IL-1β)5. The activation of inflammasomes begins with the recognition of danger signals or pathogen-associated molecular patterns (PAMPs) and damage-associated molecular patterns (DAMPs) by pattern recognition receptors (PRRs) expressed by immune cells. This recognition leads to activating proinflammatory cytokines, such as interleukin-6 (IL-6) and tumor necrosis factor-alpha (TNF-α), which recruit and activate immune cells at the site of inflammation.
The nucleotide-binding oligomerization domain-like receptor pyrin domain-containing 3 (NLRP3) inflammasome is particularly significant in periodontal inflammation. It comprises the sensor molecule NLRP3, an adaptor protein called apoptosis-associated speck-like protein containing a CARD (ASC), and the effector molecule caspase-1. The NLRP3 inflammasome can be activated by various stimuli, including microbial products and endogenous DAMPs1,6,7. Once activated, the NLRP3 inflammasome triggers the release of IL-1β and IL-18, exacerbating the inflammatory response in periodontal tissues. IL-1β promotes the production of other proinflammatory mediators such as IL-6, TNF-α, and matrix metalloproteinases (MMPs), leading to tissue destruction. IL-18 enhances the production of interferon-gamma (IFN-γ), further contributing to the inflammatory response in periodontal tissues2.
Dysregulation of inflammasome activation has been implicated in the development of periodontal disease. Studies have shown increased expression of NLRP3 and enhanced caspase-1 activity in patients with periodontitis compared to healthy individuals. Genetic variations in inflammasome-related genes have also been associated with an increased risk of severe periodontitis8. Targeting the inflammasome pathway may be a potential therapeutic strategy for managing periodontal inflammation. In preclinical studies, compounds that inhibit the inflammasome or antagonize IL-1β have shown promising results in reducing inflammatory cytokines and tissue destruction in experimental models of periodontitis. Activation of the NLRP3 inflammasome produces IL-1β and IL-18, exacerbating the inflammatory response and tissue destruction in periodontal tissues.
The AIM-2 inflammasome, a multimeric protein complex, regulates the inflammatory response in periodontal tissues. Activated by cytosolic DNA, AIM-2 recruits an adaptor protein ASC, which then recruits and activates caspase-1, producing proinflammatory cytokines. AIM2 consists of an N-terminal pyrin domain (PYD), a central DNA-binding domain (HIN200), and a C-terminal regulatory domain (CARD)9. Once activated, AIM2 recruits ASC (apoptosis-associated speck-like protein containing a caspase recruitment domain), which recruits and activates caspase-1, producing proinflammatory cytokines. The upregulation of AIM-2 and associated inflammasome components in patients with periodontal disease is attributed to the recognition of bacterial DNA released during infection6. The AIM-2 inflammasome also interacts with other innate immune pathways, such as toll-like receptors (TLRs) and NOD-like receptors (NLRs), to modulate the inflammatory response in periodontal tissues. Targeting AIM2 may represent a promising therapeutic strategy to control the inflammatory response and mitigate tissue damage. Further research is needed to explore the efficacy and safety of targeting AIM-2 for managing periodontal diseases. One previous study found that increased expression of inflammasome components, including Aim2, Ifi204, and Nlrp3, increased expression of proinflammatory Il1b in gingival tissues of murine experimental periodontitis patients7.
Periodontal inflammation, caused by dental plaque accumulation, is a significant health issue requiring effective therapeutic interventions. The AIM-2 inflammasome plays a crucial role in regulating the immune response during inflammation, influencing the extent and persistence of inflammation. Advanced computational models like transformers and graph attention networks (GAT) are used to analyze vast datasets to identify peptide sequences associated with AIM-2, a key factor in periodontal inflammation severity and treatment response10. This leads to targeted therapeutics, predictive models, patient outcomes, educational campaigns, and clinical practice integration. Sequence prediction is crucial in understanding genetic mutation dynamics, particularly in periodontal inflammation. This approach can help identify novel therapeutic agents and polymorphisms that may influence disease progression and contribute to personalized diagnostic tools for periodontal conditions.
Peptide sequence prediction using Language Models (LLM) is a method that leverages deep learning architectures, such as GPT and BERT, to predict peptide sequences with high accuracy11,12,13. Peptides are crucial in biological processes, including signaling, enzyme activity, and immune responses. LLM models learn from vast amounts of text data through unsupervised pre-training, enabling them to recognize sequence patterns. These models can then be fine-tuned using labeled data for specific tasks like peptide sequence prediction12,13. CAMP, a deep learning framework, outperforms current methods for binary peptide-protein interaction prediction and binding residue identification, facilitating peptide drug discovery based on attention mechanisms14 and another recent study showed that AlphaPeptDeep offers modular APIs for N.N. architectures like LSTM, CNN, and transformers, with a HuggingFace transformer library. It offers universal training and transfer learning steps and saves learned parameters, source code, and hyperparameters15. These studies inspire us to explore peptide sequence prediction using advanced algorithms.
LLM models employ a multi-layer architecture with self-attention mechanisms to capture long-range dependencies and complex relationships between protein sequences. Despite the challenges of requiring large amounts of labeled training data and significant computational resources11,12,13. LLM-Attention Networks, particularly Transformers, are advanced deep-learning models that predict peptide sequences. These models have shown potential in drug discovery, biomarker identification, and vaccine design, accurately predicting peptide binding affinities to receptors or major histocompatibility complex molecules. They have also been used to identify antimicrobial peptides and toxin sequences, aiding in developing new antibiotics and toxin inhibitors10. However, LLM-Attention Networks demand substantial training data and computational resources, limiting their applicability in data-scarce tasks. Further research is necessary to optimize these models and address their limitations. Nonetheless, with ongoing advancements, LLM-Attention Networks hold significant promise for revolutionizing peptide sequence prediction and biomedical applications13,16,17.
Limited studies have focused on using LLM-based prediction for the AIM-2 inflammasome, a crucial molecular complex involved in the inflammatory response. By understanding and accurately predicting the peptide sequences associated with AIM-2, we can potentially develop targeted therapeutics to modulate its activity and mitigate the inflammatory response in periodontal disease. This study aims to explore the prediction of AIM-2 inflammasome sequences using transformers and graph attention networks to treat periodontal inflammation.
Materials and methods
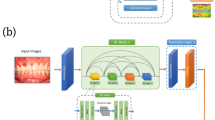
Using UniProt18, the AIM-2 inflammasome proteins and their corresponding FASTA sequences were downloaded, including I.D.s with 100%, 90%, and 50% similarity: O14862, A0A8Q3WLZ2, Q5T3W0, and Q5T3W0. These sequences were identified, downloaded, and checked for quality. The FASTA sequences were then processed using DeepBIO, a tool for analyzing large language models and deep attention networks (Fig. 1).
The protein sequence analysis process involves several preprocessing steps, including deduplication, similarity filtering, data splitting, and regularization. Deduplication removes duplicated protein sequences, while similarity filtering ensures diversity and minimizes redundancy. Data splitting divides the deduplicated dataset into three parts: the training set, validation set, and test set. Early stopping prevents overfitting by monitoring the model’s performance on the validation set. Cross-validation, such as k-fold cross-validation, maximizes available data while providing a robust evaluation. These steps enhance the dataset quality and model performance, yielding more reliable biological insights from protein sequence analysis.
DeepBIO
DeepBIO19 is a web service that allows researchers to create deep-learning architectures for biological problems, visualize sequencing data, compare and enhance models, and provide in-depth interpretations and visualizations. It uses sequence-based datasets to offer conservation motif analysis and well-trained architectures for over 20 tasks.
Transformers architecture
Introduced in 2017, the Transformers architecture has revolutionized natural language processing and computer vision. Its self-attention mechanism allows the model to weigh the importance of input tokens, capture dependencies, and learn contextual representations, making it applicable across various domains. The architecture comprises an encoder and decoder, each equipped with a multi-head self-attention mechanism and a feed-forward neural network. The self-attention mechanism transforms the input sequence into queries, keys, and values vectors, while the feed-forward neural network refines these representations. The encoder-decoder structure stacks multiple layers to produce contextual representations and generate output sequences.
Key components of the transformers architecture:
-
1.
Encoder-Decoder Structure: The model consists of an encoder and a decoder. The encoder processes the input sequence into hidden representations while the decoder generates the output sequence.
-
2.
Self-Attention Mechanism: This allows the model to focus on different parts of the input sequence during encoding and decoding, capturing dependencies between sequence points. It assigns weights to each position in the sequence based on their relevance.
-
3.
Multi-Head Attention: Transformers use parallel self-attention layers, where each head learns different attention weights and representations. The outputs from all heads are concatenated and linearly transformed to capture diverse relationships.
-
4.
Positional Encoding: To incorporate word order and position information, positional encoding vectors are added to the input embeddings, indicating each word’s position in the sequence.
-
5.
Position-wise Feed-Forward Networks: Each position in the sequence receives a fully connected feed-forward network after the self-attention layers, capturing complex non-linear relationships between sequence positions.
-
6.
Layer Normalization and Residual Connections: Residual connections are used around self-attention and feed-forward layers to facilitate gradient flow and optimization in deep networks. Layer normalization is applied to the output of each sublayer.
-
7.
Masking: During training, transformers use masking to prevent the model from looking ahead in the input sequences. In the encoder, masking restricts attention to earlier positions, while in the decoder, it focuses on preceding output positions.
Graph attention network architecture
The Graph Attention Networks (GAT) architecture is a neural network model tailored for graph-structured data, integrating attention mechanisms to assess the significance of neighboring nodes and learn node representations effectively. It incorporates multiple attention heads, each responsible for computing attention weights that capture different relationships between nodes. The model utilizes a self-attention mechanism to compute these weights, which are then aggregated to form comprehensive node representations, capturing diverse and intricate relationships within the graph structure. The multi-head attention scheme facilitates enhanced information exchange and captures higher-order dependencies in the graph.
Overview of graph attention networks architecture:
-
1.
Input Layer: The model takes in a graph with nodes and edges, where each node possesses specific features, and edges connect pairs of nodes.
-
2.
Node Embedding Layer: Node features are transformed into embeddings, linearly and non-linearly activating node characteristics.
-
3.
Attention Mechanism: The architecture relies on attention mechanisms to determine the relevance and compute attention weights for nodes and edges. These weights, learned through parameters, signify the importance of one node or edge relative to others.
-
4.
Graph Convolution: Attention convolutions utilize the computed attention weights on edges connecting neighboring nodes, influencing how each node aggregates information. This process enables comprehensive circulation and integration of information throughout the graph.
-
5.
Pooling and Aggregation: Following multiple graph convolution layers, pooling and aggregation techniques summarize graph information. This can involve pooling nodes or edges based on attention weights or aggregating data using sum and mean operations.
-
6.
Output Layers: Fully connected layers, often including an activation function, process the aggregated data to solve specific tasks such as node classification, edge prediction, or graph classification. The structure of these output layers varies depending on the nature of the task.
GAT has proven effective in various graph-related tasks, making it a popular choice in graph representation learning.
RNN-CNN architecture
The RNN-CNN architecture integrates the strengths of Recurrent Neural Networks (RNNs) and Convolutional Neural Networks (CNNs), offering a robust framework for processing sequential data. It begins with an input layer that handles data sequences, such as text or time-series information, where vectors like word embeddings in text data represent each element.
Overview of RNN-CNN architecture:
-
1.
Input Layer: The model takes in data sequences, with each element represented as vectors (e.g., word embeddings for text).
-
2.
Convolutional Layer: A 1D convolutional layer filters the input sequences to capture local patterns or features. Sliding window filters analyze sequences at different positions to extract relevant features.
-
3.
Pooling Layer: Max or average pooling layers follow the convolutional layer to reduce the dimensionality of the extracted features while preserving key patterns.
-
4.
Recurrent Layer: Pooled features are fed into a recurrent layer such as LSTM (Long Short-Term Memory) or GRU (Gated Recurrent Unit). This layer maintains an internal state to process sequences and capture temporal dependencies and contextual information.
-
5.
Output Layers: The final output from the recurrent layer is processed through task-specific layers. For tasks like sentiment analysis, a fully connected layer with a softmax activation function can estimate sentiment based on the processed sequence data.
The RNN-CNN architecture excels in tasks that require both local pattern extraction and understanding of contextual dependencies, such as sentiment analysis, text classification, and speech recognition. Its effectiveness in handling sequence-based data has made it popular in natural language processing and sequence modeling applications.
Table 1 shows the hyperparameter tuning of three models.
This table presents hyperparameters and configurations for three machine learning models: TRANSFORMERS, GAT, and RNN-CNN. Each model has CUDA enabled for GPU acceleration, a random seed of 43, and 4 worker threads for data loading. The models have two classes in the classification task, a k-mer size of 3, and no specific sequence for heatmap generation. The mode of operation is “train-test,” and the data type is “prot” (likely protein data). The model architecture is specified by TRANSFORMERS, GAT, and RNN-CNN, with a data type of “userprovide.” The learning rate is 1.00E-05, while GAT and RNN-CNN use 0.0001. The regularization parameter is set to 0.0025, and the maximum sequence length is 35, GAT 207, and RNN-CNN. The embedding layer dimension is 32, and the mini mode mode suggests a model comparison.
Results
Transformer, RNN-CNN, and GAT architectures were employed to extract latent features and weights from FASTA-formatted protein sequences. Subsequently, the model was fine-tuned using backpropagation algorithms with the ADAM optimizer over 50 iterations.
Table 2 presents accuracy results for the Transformer, RNN-CNN, and GAT models, achieving 84%, 64%, and 86% accuracy, respectively, along with class accuracy values of 85%, 76%, and 80%. These metrics evaluate peptide sequence prediction methods, showing the Transformer model with the highest overall accuracy at 84%, followed by GAT at 86% and RNN-CNN at 64%, indicating superior prediction capabilities of Transformer and GAT over RNN-CNN. Class accuracy values reveal the Transformer model’s dominance across peptide classes with an average of 85%, followed by GAT at 80% and RNN-CNN at 76%. Sensitivity (T.P. / (T.P. + F.N.)) and specificity (T.N. / (T.N. + F.P.)) metrics further demonstrate model performance: Transformer has a sensitivity of 0.85 and specificity of 0.83, RNN-CNN has a sensitivity of 0.76 and specificity of 0.52. At the same time, GAT shows a sensitivity of 0.80 and a specificity of 0.80. These findings underscore the Transformer model’s effectiveness in peptide sequence prediction tasks, both overall and across specific peptide classes.
Figure 2 compares True Positive Rate (TPR) and False Positive Rate (FPR) across various thresholds. The Precision-Recall Curve graphically illustrates the balance between precision and recall as classification thresholds vary. Starting at a precision of 1 and a recall of 0, indicating all predictions are correct. Still, no true positives. The curve shows that lowering the threshold identifies more true positives and increases false positives, thereby reducing precision. The curve progresses to a precision of 0 and a recall of 1, indicating all true positives are identified with numerous false positives. Higher precision values across different recall levels signify a model’s ability to detect true positives while minimizing false positives accurately. The Area Under the ROC Curve (AUC-ROC) summarizes overall model performance. The Transformer model achieves an AUC of 0.891, indicating good performance, whereas the GAT model slightly outperforms with an AUC of 0.923, suggesting superior classification capability for the dataset. Additionally, the Transformer model exhibits an Average Precision (A.P.) of 0.851, while the GAT model achieves 0.922, indicating a stronger balance between precision and recall. Both metrics in Fig. 1 highlight the GAT model’s superiority over the Transformer model in terms of ROC AUC and Precision-Recall AP, underscoring its potential suitability for the task.
Figure 3 displays three SHAP value plots corresponding to the Transformer, RNN-CNN, and GAT models. SHAP (Shapley Additive explanations) values utilize cooperative game theory to explain predictions made by machine learning models. They assign importance to individual features, accounting for their interactions and dependencies. Known for their properties, such as local accuracy, consistency, and handling of missing data, SHAP values decompose model predictions into the contributions of each feature, offering insights into key drivers and specific instances. Analyzing SHAP values aids in understanding the decision-making processes of models and validating their predictions. Higher SHAP values positively influence the Transformer model’s outputs, whereas lower values exert a negative impact. The SHAP values for the RNN-CNN model illustrate how each feature influences predictions, with the spread indicating variability. Color coding indicates the correlation between feature values and their impact on predictions. These SHAP plots are essential for interpreting model behavior, ensuring fairness, and enhancing transparency in machine learning applications.
SHAP values evaluate the predictive power of three machine learning models: Transformer, RNN-CNN, and GAT. These values use cooperative game theory to assign importance scores to individual features, allowing for a comprehensive understanding of their impact on predictions. They are characterized by local accuracy, consistency, and the ability to manage missing data. Higher SHAP values indicate a greater positive influence on the model’s outputs, while lower values indicate negative impacts. SHAP plots enhance interpretation and transparency, ensuring fairness and accountability in machine learning applications. Exploring SHAP values can help uncover the biological significance of high-importance features, particularly in inflammatory responses.
Figure 4 shows an epoch plot, a graphical representation of a machine learning model’s training progress over multiple iterations known as epochs. It shows how the model’s performance changes, indicating improvement or degradation. Epoch plots are commonly used in deep learning to understand the model’s learning process, make informed decisions about further training, and track overfitting, underfitting, and other issues during the training process by comparing the performance of three different algorithms: Transformer, RNN_CNN, and GAT. Transformer starts with lower accuracy but increases steadily, surpassing RNN_CNN and nearly approaching GAT’s performance. GAT exhibits the highest accuracy, while RNN_CNN starts higher but doesn’t improve much. The test loss curve shows lower loss values, with Transformer showing better performance and GAT showing the lowest loss.
Figure 5 presents a Venn diagram depicting the classification outcomes of a model, where overlapping circles illustrate instances classified as both positive and negative. The size of each circle indicates the number of predictions falling into each category, while the intersection reveals the proportion of ambiguous or overlapping predictions. This diagram visually represents the model’s classification performance, highlighting its ability to distinguish between positive and negative classes and identifying cases where predictions overlap or are uncertain.
Figure 6 depicts a UMAP plot, a visualization technique used in data analysis and machine learning to project high-dimensional data onto a lower-dimensional space while preserving local and global structures. This plot visualizes relationships and patterns in the data by representing each data point as a dot or marker. UMAP plots are valuable for visualizing complex datasets, identifying clusters, exploring data structure, and providing insights for further analysis or modelling. The figure compares the clustering of data points generated by three algorithms: Transformer, RNN-CNN, and GAT. It illustrates how each algorithm groups data points, revealing similarities or differences in their interpretations. Transformer clusters may exhibit distinct patterns, whereas RNN-CNN clusters might vary in density or separation. GAT’s clusters could display unique patterns, with overlaps indicating shared interpretations. The UMAP plot aids in comprehending each algorithm’s strengths and weaknesses in handling the dataset effectively.
Discussion
Inflammasomes are intracellular complexes crucial for activating caspase-1 in response to various signals, producing interleukin-1β and interleukin-181,20,21. Their role in periodontal disease and potential therapeutic applications are actively under investigation. Evidence supports their involvement in inflammatory disorders, including periodontitis, with clinical and preclinical data linking inflammasomes to periodontal and comorbid diseases and ongoing research exploring potential therapies targeting inflammasomes. Multiple proteins and processes regulate inflammasomes, such as PYD-only proteins, CARD-only proteins, TRIMs, autophagy, and interferons9. Studies indicate that NLRP3 and AIM2 inflammasomes play roles in periodontal disease, with observed upregulation of inflammasomes and downregulation of inflammasome regulator proteins in this context. IL-1β, produced as pro-IL-1β in response to PAMPs and DAMPs binding to PRRs on cell membranes, is cleaved into its active form by the inflammasome complex, comprising a PRR, an adaptor protein (ASC), and active caspase-1. Various types of inflammasomes, including NLRP1, NLRP2, NLRP3, NLRP6, NLRP12, NLRC4, IPAF, NLRC5, PYHINS, AIM2, and Ifi-16, have been identified. For instance, NLRP1 is activated by a lethal toxin from Bacillus anthracis. Previous studies have examined the expression of NLRP3 and AIM2 inflammasomes, caspase-1, and IL-1β in peri-implantitis biopsies, revealing associations with increased inflammation, probing depth, biofilm presence, and bleeding on probing9. Moreover, another study measured salivary levels of DNA sensing inflammasomes (AIM2, IFI16, IL18) in individuals with periodontitis, diabetes, and healthy controls, showing correlations with periodontal clinical parameters and predictors such as glycated hemoglobin, gingival index, PISA, and CAL6,22,23,24.
Furthermore, genetic studies have identified frequent genotypes in AIM2 and Pycard genes in patients with periodontitis and atherosclerosis coronary heart disease, highlighting their pivotal roles in inflammatory diseases8. Exploring peptide sequences is crucial for targeting inflammasome proteins like AIM-2 in periodontal diseases. AI-driven peptide sequence prediction enhances research efficiency across drug discovery, immunology, protein engineering, diagnostics, and peptide synthesis, advancing therapeutics and personalized medicine. This study utilizes advanced algorithms such as transformers, GAT, and RNN-CNN to predict AIM-2 peptide sequences, aiming to enhance the understanding and treatment of periodontal diseases through precision medicine approaches.
Transformers are renowned for their effectiveness in natural language processing and ability to capture intricate dependencies over long distances23,24,25. Given that peptide sequences can be seen as analogous to linguistic information, the Transformer’s ability to grasp complex relationships across sequences likely contributes to its superior accuracy in peptide sequence prediction, akin to machine learning models predicting TCR-pMHC interactions using amino acid sequences of TCR CDR3 and peptides, achieving competitive performance on benchmark and external datasets26,27. The Transformer model associates neural network weights with protein structural properties, aiding in molecular recognition, as exemplified by PiTE, a two-step pipeline for predicting TCR-epitope binding affinity using a pre-trained amino acid embedding model and a Transformer-like sequence encoder to enhance prediction accuracy by capturing contextual information between amino acids. Graph Attention Networks (GAT), incorporating attention mechanisms28,29, also demonstrate high accuracy due to their ability to capture intricate amino acid interactions in peptide sequences through graph-based models. In contrast, the RNN-CNN model exhibits lower accuracy and class accuracy, indicating limitations in capturing complex peptide sequence patterns and correlations despite combining RNNs and CNNs to capture temporal and local information. Overall, Transformers and GAT outperform RNN-CNN in peptide sequence prediction and class accuracy, highlighting their adeptness in capturing peptide sequence connections and dependencies, as supported by experimental results demonstrating sAMPpred-GAT’s superiority in AUC and other metrics across multiple test datasets30. Specifically, the Transformer model excels in predicting AIM-2 peptide sequences with 84% overall accuracy and 85% class accuracy, while the RNN-CNN model shows lower overall accuracy (64%) but comparatively higher-class accuracy (76%). The GAT model achieves the highest overall accuracy (86%) and class accuracy (80%) by effectively capturing the graph structure and linkages in AIM-2 peptide sequences, leveraging its attention mechanism to prioritize relevant amino acids for improved prediction. It’s important to note that model performance depends upon dataset quality and size, as detailed in Table 2; Figs. 2, 3, 4, 5 and 6.
The Transformer, RNN-CNN, and GAT models showed high accuracy in peptide sequence prediction, with the GAT model achieving the highest accuracy. The transformer model maintained consistent performance across different peptide classes, effectively identifying specific peptide sequence types. The Transformer model’s high accuracy and class performance demonstrate its ability to identify complex biological sequences, particularly those related to the AIM-2 inflammasome’s function in periodontal inflammation, which is crucial for understanding immune response interactions. With an impressive 86% accuracy, the GAT model captures structural information within peptide sequences, effectively modelling amino acid relationships and spatial configurations. This performance is particularly useful in understanding the spatial arrangement of peptide sequences contributing to immune responses. The RNN-CNN model has lower accuracy (64%), sensitivity (0.76), and specificity (0.52) due to sequential data processing and vanishing gradient issues, which may hinder its ability to predict peptide sequences and capture long-range dependencies accurately. The Transformer and GAT models offer valuable insights into peptide sequences related to the AIM-2 inflammasome. In contrast, the limitations of the RNN-CNN model highlight the need for improved architectures to better capture biological sequence complexities.
The AIM-2 inflammasome, a crucial part of the innate immune system, plays a role in recognizing cytosolic DNA from pathogens and cellular stress signals. In periodontal inflammation, the AIM-2 inflammasome can be activated by microbial DNA from oral pathogens, contributing to inflammatory responses and tissue destruction. The evaluation of predictive models, Transformers, Graph Attention Networks (GAT), and RNN-CNN provides significant insights into identifying and characterizing AIM-2 inflammasome sequences associated with periodontal inflammation. The Transformer model has the highest predictive accuracy and class performance, suggesting it can effectively identify AIM-2-related sequences that play a role in periodontal inflammation. GAT models highlight the importance of relational and structural information in peptide sequences, which can be particularly relevant for AIM-2 inflammasome sequences, which often interact with other molecular players within the inflammatory pathways. The insights gathered from these models can guide future research toward enhancing predictive capabilities.
Protein language model embeddings can significantly improve the Transformer and GAT models in peptide sequence prediction. This is due to enhanced feature representation, improved predictive performance, generalization, and interpretability. These embeddings are trained on extensive protein datasets, allowing them to capture intricate patterns and contextual relationships in amino acid sequences. Experimental model performance comparisons before and after embeddings provide quantitative evidence of their impact. Including embeddings can also bridge the gap between training scenarios and practical applications, making the models more robust. SHAP analysis aids in model decision-making, providing biological insights like AIM-2 inflammasome activation and facilitating further research, feature selection, and personalized medicine.
While the Transformer and GAT models demonstrate high accuracy in AIM-2 peptide sequence prediction, critical areas remain for enhancement and consideration. Factors such as dataset quality and size, training methodologies, feature engineering, interpretability, and model optimization significantly influence performance. Improving accuracy entails gathering a more extensive and varied dataset, refining hyperparameters, employing robust data preprocessing methods, and enhancing training protocols.
Transformer and GAT models, promising for AIM-2 peptide sequence prediction, face several limitations that could impact their clinical implications. Data quality, size, training methods, feature engineering, interpretability, model optimization, regulatory and ethical considerations, and integration into clinical workflows are key factors in successfully translating medical models into clinical practice. These factors can lead to overfitting, biased predictions, and challenges in data privacy and health disparities. Moreover, exploring domain-specific feature engineering strategies and integrating external biological insights could further elevate model effectiveness. Future research efforts should prioritize advancements in these areas to achieve enhanced predictive outcomes and deepen understanding of peptide sequence prediction.
Conclusions
Inflammasomes, particularly AIM-2, play a significant role in periodontal disease and other inflammatory conditions. AI-driven peptide sequence prediction utilizing advanced algorithms like transformers, GAT, and RNN-CNN shows considerable potential in accurately predicting AIM-2 peptide sequences. Transformers and GAT models demonstrate superior accuracy and class accuracy compared to the RNN-CNN model, highlighting their effectiveness in capturing peptide sequence relationships and dependencies. Nevertheless, further enhancements can be achieved by addressing dataset quality and size, refining training methodologies, optimizing feature engineering approaches, and improving model optimization. Ongoing research efforts hold promise for advancing peptide sequence prediction capabilities and exploring therapeutic applications in the context of periodontal diseases.
Data availability
The datasets used and/or analysed during the current study available from the corresponding author on reasonable request.
References
Sordi, M. B., Magini, R., de Panahipour, S. & Gruber, R. L. Pyroptosis-Mediated periodontal disease. Int. J. Mol. Sci. 23(1), 372 (2021).
Marchesan, J. T. et al. Role of inflammasomes in the pathogenesis of periodontal disease and therapeutics. Periodontology 82, 93–114 (2020).
Thilagar, S. et al. Periodontal treatment for chronic periodontitis with rheumatoid arthritis. Int. Dent. J. 72(6), 832–838 (2022).
Yadalam, P. K. et al. Future drug targets in periodontal personalised Medicine—A narrative review. J. Pers. Med. 12(3), 371 (2022).
Aral, K., Milward, M. R., Kapila, Y., Berdeli, A. & Cooper, P. R. Inflammasomes and their regulation in periodontal disease: A review. J. Periodontal Res. 55, 473–487 (2020).
Arunachalam, L. T. et al. Association of salivary levels of DNA sensing inflammasomes AIM2, IFI16, and cytokine IL18 with periodontitis and diabetes. J. Periodontol. 95, 114–124 (2024).
Marchesan, J. T. Inflammasomes as contributors to periodontal disease. J. Periodontol. 91, S6–11 (2020).
Ali Daily, Z., Al-Ghurabi, B. H., Al-Qarakhli, A. M. A. & Hussein, H. M. Association between AIM2 and pycard genes polymorphisms and susceptibility to periodontitis with coronary heart disease. Clin. Cosmet. Investig Dent. 15, 307–320 (2023).
Galindo-Moreno, P. et al. Inflammasomes NLRP3 and AIM2 in peri-implantitis: A cross-sectional study. Clin. Oral Implants Res. 34, 1342–1353 (2023).
Park, E. et al. Activation of NLRP3 and AIM2 inflammasomes by Porphyromonas gingivalis infection. Infect. Immun. 82, 112–123 (2014).
Marks, D. S., Hopf, T. A. & Sander, C. Protein structure prediction from sequence variation. Nat. Biotechnol. 30, 1072–1080 (2012).
Liu, J. & Gong, X. Attention mechanism enhanced LSTM with residual architecture and its application for protein-protein interaction residue pairs prediction. BMC Bioinform. 20, 609 (2019).
Lupo, U., Sgarbossa, D. & Bitbol, A. F. Protein Language models trained on multiple sequence alignments learn phylogenetic relationships. Nat. Commun. 13, 6298 (2022).
Lei, Y. et al. A deep-learning framework for multi-level peptide-protein interaction prediction. Nat. Commun. 12, 5465 (2021).
Zeng, W. F. et al. AlphaPeptDeep: a modular deep learning framework to predict peptide properties for proteomics. Nat. Commun. 13, 7238 (2022).
Ferruz, N., Schmidt, S. & Höcker, B. ProtGPT2 is a deep unsupervised Language model for protein design. Nat. Commun. 13, 4348 (2022).
Brandes, N., Ofer, D., Peleg, Y., Rappoport, N. & Linial, M. ProteinBERT: a universal deep-learning model of protein sequence and function. Bioinformatics 38, 2102–2110 (2022).
UniProt. The universal protein knowledgebase in 2023. Nucleic Acids Res. 51, D523–531 (2023).
Wang, R. et al. DeepBIO: an automated and interpretable deep-learning platform for high-throughput biological sequence prediction, functional annotation and visualization analysis. Nucleic Acids Res. 51, 3017–3029 (2023).
Li, Y., Ling, J. & Jiang, Q. Inflammasomes in alveolar bone loss. Front. Immunol. 12, 691013 (2021).
Wu, X., Zeng, W., Lin, F., Xu, P. & Li, X. Anticancer peptide prediction via Multi-Kernel CNN and attention model. Front. Genet. 13, 887894 (2022).
Aral, K., Milward, M. R. & Cooper, P. R. Inflammasome dysregulation in human gingival fibroblasts in response to periodontal pathogens. Oral Dis. 28, 16–24 (2022).
Ding, P. H. et al. Porphyromonas gingivalis-Induced NLRP3 inflammasome activation and its downstream Interleukin-1β release depend on Caspase-4. Front. Microbiol. 11, 1881 (2020).
Barnett, K. C. et al. An epithelial-immune circuit amplifies inflammasome and IL-6 responses to SARS-CoV-2. Cell. Host Microbe. 31, 243–259 (2023).
Yadalam, P. K. et al. Machine learning predicts patient tangible outcomes after dental implant surgery. IEEE Access 10 (2022).
Yadalam, P. K. et al. Gene and protein interaction network analysis in the epithelial-mesenchymal transition of Hertwig’s epithelial root sheath reveals periodontal regenerative drug targets – An in silico study. Saudi J. Biol. Sci. 29 (2022).
Yadalam, P. K. et al. Gene interaction network analysis reveals IFI44L as a drug target in rheumatoid arthritis and periodontitis. Molecules 27 (2022).
Koyama, K., Hashimoto, K., Nagao, C. & Mizuguchi, K. Attention network for predicting T-cell receptor-peptide binding can associate attention with interpretable protein structural properties. Front. Bioinform. 3, 1274599 (2023).
Zhang, P., Bang, S. & Lee, H. PiTE: TCR-epitope binding affinity prediction pipeline using Transformer-based sequence encoder. Pac. Symp. Biocomput. 28, 347–358 (2023).
Yan, K., Lv, H., Guo, Y., Peng, W. & Liu, B. sAMPpred-GAT: Prediction of antimicrobial peptide by graph attention network and predicted peptide structure. Bioinformatics 39 (2023).
Acknowledgements
We would like to thank the Center of Medical and Bioallied Health Sciences and Research, Ajman University, Ajman, UAE.
Author information
Authors and Affiliations
Contributions
Conceptualization, P. Y., D.A., P. N., and C. A.; Data curation, P. Y., D.A., P. N., and C. A.; Formal analysis, P. Y., D.A., P. N., and C. A.; Funding acquisition, P. N.; Investigation, P. Y., D.A., P. N., and C. A.; Methodology, P. Y., D.A., P. N., and C. A.; Project administration, P. Y.; Resources, P. Y. and P. N.; Software, P. Y.; Supervision, P. Y. and C.A.; Validation, P. Y., D.A., P. N., and C. A.; Visualization, P. Y., D.A., P. N., and C. A.; Writing – original draft, P. Y., D.A., P. N., and C. A.; Writing – review & editing, P. Y., D.A., P. N., and C. A. All authors have read and agreed to the published version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Yadalam, P.K., Arumuganainar, D., Natarajan, P.M. et al. Artificial intelligence-powered prediction of AIM-2 inflammasome sequences using transformers and graph attention networks in periodontal inflammation. Sci Rep 15, 8733 (2025). https://doi.org/10.1038/s41598-025-93409-3
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-93409-3
Keywords
This article is cited by
-
Identification and analysis of neutrophil extracellular trap-related genes in periodontitis via bioinformatics and experimental verification
BMC Oral Health (2025)
-
Comprehensive single-cell transcriptome analysis of autologous platelet-rich plasma therapy on human thin endometrium
Scientific Reports (2025)