Abstract
In the domain of medical image segmentation, while convolutional neural networks (CNNs) and Transformer-based architectures have attained notable success, they continue to face substantial challenges. CNNs are often limited in their ability to capture long-range dependencies, while Transformer models are frequently constrained by significant computational overhead. Recently, the Vision Mamba model, combined with KAN linear attention, has emerged as a highly promising alternative. In this study, we propose a novel model for medical image segmentation, termed VMKLA-UNet. The encoder of this architecture harnesses the VMamba framework, which employs a bidirectional state-space model for global visual context modeling and positional embedding, thus enabling efficient feature extraction and representation learning. For the decoder, we introduce the MKCSA architecture, which incorporates KAN linear attention—rooted in the Mamba framework—alongside a channel-spatial attention mechanism. KAN linear attention substantially mitigates computational complexity while enhancing the model’s capacity to focus on salient regions of interest, thereby facilitating efficient global context comprehension. The channel attention mechanism dynamically modulates the importance of each feature channel, accentuating critical features and bolstering the model’s ability to differentiate between various tissue types or lesion areas. Concurrently, the spatial attention mechanism refines the model’s focus on key regions within the image, enhancing segmentation boundary accuracy and detail resolution. This synergistic integration of channel and spatial attention mechanisms augments the model’s adaptability, leading to superior segmentation performance across diverse lesion types. Extensive experiments on public datasets, including Polyp, ISIC 2017, ISIC 2018, PH2, and Synapse, demonstrate that VMKLA-UNet consistently achieves high segmentation accuracy and robustness, establishing it as a highly effective solution for medical image segmentation tasks.
Similar content being viewed by others
Introduction
Medical image segmentation is a pivotal technology in medical image processing and computer vision, widely applied in areas such as diagnosis, surgical planning, and treatment evaluation. Its primary goal is to delineate structures or regions of interest—e.g., organs, tumors, and blood vessels—from complex medical images. With the rapid advancement of imaging technologies, the volume of medical data has grown exponentially, increasing the demands on segmentation techniques. In recent years, the advent of deep learning has revolutionized the field, driving significant progress in medical image segmentation.
In medical image segmentation using deep learning, the encoder-decoder architecture is a prevalent framework. In this design, the encoder extracts feature from the input image, progressively compressing high-dimensional data into low-dimensional representations to capture global context. The decoder then recovers these representations, gradually restoring them to the original input size to produce refined segmentation results. Numerous studies have demonstrated that this architecture significantly enhances segmentation performance by effectively integrating global information and enriching multiscale feature representation.
U-Net1 is one of the most widely used frameworks, known for its balanced and symmetrical encoder-decoder design and the integration of skip connections. The hierarchical structure of the encoder and decoder allows the model to extract and process features at varying depths, enabling it to capture the multi-scale details of the image. Additionally, skip connections facilitate the effective transfer of feature information. Numerous studies on U-Net focus on several key areas: the encoder, by replacing the backbone networks to obtain feature maps at different levels; the skip connections, by incorporating various channel attention mechanisms and adjusting them at different points in the network; and the decoder, by exploring different sampling methods and feature fusion strategies.
Models based on convolutional neural networks (CNNs) have difficulty capturing long-distance information due to the limitations of their local receptive field. This limitation may lead to poor feature extraction and thus affect the quality of segmentation results. Models based on Transformer2 perform well in global modeling, but the quadratic complexity of their self-attention mechanism leads to high computational costs, especially in tasks that require dense predictions, such as medical image segmentation. These limitations have prompted us to develop a new architecture for medical image segmentation that can not only effectively capture long-distance information but also maintain linear computational complexity. Recently, advances in state-space models (SSMs), especially structured space models S4, have provided an effective solution because they perform well in processing long sequences, e.g., the Mamba model3. The Mamba model enhances S4 through selection mechanisms and hardware optimizations, and performs well in dense data areas. By using the visual state-space model (VMamba)4.The addition of the Cross Scan Module (CSM) further improves the applicability of Mamba in computer vision tasks. The three frame structures are shown in Fig. 1, which fully demonstrates the process of three mainstream models processing image data. CNN focuses on local context information, while transformer and SSM focus on global context information.
Inspired by the great success of VMamba4 in image classification tasks and VM-UNet5 in medical segmentation tasks, this paper introduces a new medical segme ntation model Vision Mamba with KAN Linear Attention UNet (VMKLA-UNet). The model is based on the U-Shape structure, and the encoder adopts the VMamba structure, which enables the encoding stage of the model to selectively focus on the key features of the input data, and this selective mechanism allows the model to more effectively extract and represent the key information of the image in the encoding stage, especially when dealing with complex medical images, it can better capture subtle structural differences. In the decoder, in order to improve its efficiency and robustness, we first replaced SSM with KAN’s linear attention. Although SSM performs well in selective feature extraction in the encoder, it has high computational complexity in the decoding stage and because of its special selective mechanism, it usually ignores some important features, resulting in information loss and affecting the final segmentation results. At this time, the advantages of SSM in the encoder become the disadvantages of the decoder. However, by integrating KAN linear attention, the decoder not only reduces the computational overhead, but also improves global feature integration and enhances generalization between different datasets. The linear attention mechanism not only speeds up the decoding process, but also ensures more comprehensive utilization of the encoded features, thereby achieving excellent segmentation accuracy. These improvements make the KAN-based linear attention module a more suitable choice for the decoder. Specially, the combination of Vision Mamba (SS2D), KAN, and Linear Attention is driven by the strengths each component contributes to medical image segmentation. The core of the VSS Block in Vision Mamba, SS2D, excels at capturing global and local image structures through multi-directional scanning. This technique is particularly effective for extracting features in medical images with complex geometries and multi-scale information. By selectively focusing on relevant regions, SS2D dynamically adapts to intricate edge patterns, textures, and specific regions of interest like tumors or organ boundaries. Additionally, its design for 2D medical images, such as CT or MRI slices, ensures efficient processing without unnecessary computational overhead. As an encoder, SS2D generates rich multi-scale representations that form a robust foundation for downstream tasks. However, SS2D has limitations when used in the decoder, where global information integration is paramount. Its reliance on multi-directional scanning primarily models local features and struggles with capturing nonlinear relationships or integrating complex global context. This limitation becomes evident in scenarios with blurred boundaries or diverse feature distributions, making SS2D suboptimal for decoding tasks. To address this, we integrate KAN Linear Attention into the decoder. While traditional linear attention is computationally efficient, it often fails to model complex high-dimensional interactions adequately. KAN compensates for this by decomposing high-dimensional features into low-dimensional representations, capturing deeper relationships through mathematical decomposition6. This allows KAN Linear Attention to enhance interaction modeling while retaining the efficiency of linear attention. Furthermore, KAN enriches feature diversity during dimensional mapping6, enabling the attention mechanism to better represent both global and local structures. This is particularly important in medical image segmentation, where accurate modeling of discontinuities, regional boundaries, and subtle features is critical. By combining KAN with Linear Attention, we achieve a balance of computational efficiency, expressiveness, and robustness, ensuring superior performance in extracting meaningful features from complex high-dimensional data. This thoughtful integration ensures the model remains lightweight and efficient, making it ideal for real-world medical applications that demand high accuracy and resource-conscious solutions. Besides this, we also added channel attention blocks and spatial attention blocks to the decoder to further enhance the model’s ability to segment objects in complex areas.
We conducted extensive experiments on multiple segmentation-related tasks to demonstrate the capabilities of the SSM combined with linear attention model in the field of medical image segmentation. In particular, we conducted extensive tests on ISIC17, ISIC18, PH2, Polyp, Synapse, and other public datasets. The results show that VMKLA-UNet can provide competitive results.
The main contributions of this paper can be summarized as follows:
-
We proposed VMKLA-UNet, which is the first to introduce SSM combined with KAN linear attention into the field of medical image segmentation. KAN is a neural network architecture different from the traditional multi-layer perceptron, namely MLP.
-
We designed the MKCSA module, which is based on Mamba-Shape and first proposed the KAN linear attention block. We also added channel attention and spatial attention to the structure to extract global context information to improve the semantic segmentation ability.
-
We conducted a large number of experiments on three skin lesion datasets ISIC17, ISIC18, PH2, a colon polyp dataset Polyp, and a multi-organ segmentation task dataset Synapse, verifying the effectiveness of MKCSA on multiple different modality medical image segmentation datasets, which not only improved the segmentation accuracy but also reduced the model calculation complexity.
Related work
With the significant advancements in computational power, the field of computer vision has emerged as one of the most critical areas in modern computer science. The development of deep learning has led fully convolutional models (FCN)7 to achieve remarkable performance in image segmentation. Soon after, another fully convolutional model, U-Net, gained widespread attention1. The skip connections in U-Net al.low for effective integration of high-level and low-level features, which is particularly crucial for image segmentation tasks, especially in cases requiring fine-grained segmentation, such as in medical imaging. In this section, we provide a concise overview of prevalent medical image segmentation methods, focusing on their strategies for effectively modeling contextual information. These methods can be broadly categorized into three groups: convolutional neural network (CNN)-based approaches, Transformer-based approaches, and state-space model (SSM)-based approaches.
CNN-based models
Since the introduction of U-Net, algorithms for medical image segmentation—represented by skin lesion segmentation—have seen rapid development. The MHorUNet model8 proposed a high-order spatial interaction U-Net for skin lesion segmentation. Although the high-order spatial interaction module is introduced to enhance context modeling, its manually designed interaction rules have bottlenecks in generalization. In Wu et al.‘s study, an adaptive high-order U-Net model was introduced for sequential interactions in skin lesion segmentation, which optimized the interaction efficiency to a certain extent, but sacrificed the computational efficiency. Attention-UNet9 leverages attention gates to dynamically modulate the importance of features, enabling the model to focus more precisely on target areas. However, this added mechanism also increases computational overhead and model complexity, which can lead to longer training and inference times, greater sensitivity to hyperparameter tuning, and a heightened risk of overfitting, especially when training data is scarce. For medical image segmentation tasks, modeling global context is an important test for the model, but it is obvious that CNN-based models cannot capture long-distance features.
Transformer-based models
Inspired by the breakthrough success of Vision Transformers (ViTs)10 in vision tasks, Chen et al. introduced TransUNet11, marking the first use of a Transformer-based architecture in the encoding phase instead of convolutional networks in U-Net. Aghdam et al. proposed a cascaded attention suppression mechanism for skin lesion segmentation based on Swin U-Net12. Additionally, Xu et al.13 introduced a segmentation algorithm combining Transformers with CNNs, which demonstrated strong performance on skin lesion datasets. Other U-Net-based improvements for skin lesion segmentation include models such as Attn-Swin UNet14 which integrates cross-attention in the decoder, further enhancing Swin U-Net’s segmentation capabilities. Although Transformers excel at capturing long-range dependencies, the quadratic complexity of their self-attention mechanism with respect to input size presents challenges, particularly in pixel-level inferences required for medical image segmentation. This computational burden limits the practical applicability of Transformer-based methods.
SSM-based models
Recent advances in state-space models (SSMs), particularly the Mamba model, have shown the ability to model long-range dependencies with linear complexity, while also demonstrating superior performance across various vision tasks. U-Mamba15 introduced a novel hybrid model combining CNN with SSM, effectively capturing both fine-grained local features and long-range contextual information. In this architecture, features extracted from CNNs are flattened into 1D sequences and processed by Mamba to extract global features. Unlike natural language data, images lack a fixed causal relationship. Thus, Hao et al. proposed T-Mamba16, which improved image modeling by introducing both forward and backward feature scanning, achieving state-of-the-art results in tooth segmentation. Currently, the most successful SSM-based vision model is VMamba4. Its most significant contribution is the introduction of a cross-scanning module called SS2D4, which employs a four-directional scanning strategy. Although as an encoder, SS2D can generate rich multi-scale representations and lay a solid foundation for downstream tasks, it has difficulty capturing nonlinear relationships or integrating complex global contexts. This limitation becomes apparent in scenarios with fuzzy boundaries or diverse feature distributions, making SS2D less suitable for decoding tasks. To address the limitations of SS2D as a decoder, we proposed an effective method called VMKLA-UNet. This method is based on the structure of VMamba4, but replaces the core functional module SS2D in the decoder with the KAN linear attention mechanism we first proposed, and combines channel and spatial attention mechanisms, which performs well in local and long-distance dependency modeling and computational efficiency.
Methods
Overall framework
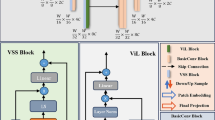
The model VMKLA-UNet proposed in our paper is shown in Fig. 2.a.
Overall framework of the model. (a) VMKLA-UNet. Both the encoder and decoder consist of 4 stages, where each stage of the encoder contains a down-sampling operation and a VSS block, and each stage of the decoder contains an up-sampling operation and an MKLA-CA block. (b) the specific architecture of MKCSA. (c) the VSS Block.
Encoder structure
State space model (SSM)
In modern state-space (SSM) based models, namely structured state-space sequence models (S4) and Mamba3, both rely on a traditional continuous system that maps a one-dimensional input function or sequence \(\:x\left(t\right)\in\:\mathbb{R}\:\)to an output \(\:y\left(t\right)\in\:\mathbb{R}\mathbb{\:}\)through an intermediate hidden state \(\:h\left(t\right)\in\:{\mathbb{R}}^{N}\). This process can be described as a linear ordinary differential equation (ODE):
where A is the state matrix, B and C are the input matrix and output matrix respectively. S4 and Mamba extended this continuous time dynamic modeling to discrete time series data by introducing a time scale parameter Δ and converting A and B into discrete parameters\(\:\:\widehat{\user2{A}\:}\)and using a fixed discretization rule\(\:\:\widehat{\user2{B}}\), as shown in Eq. (2):
After discretization, the SSM-based model can be calculated in two ways: (1) linear recursion, (2) global convolution, as shown in Eqs. (3) and (4).
where\(\:\:\widehat{\user2{K}}\:\)represents a structured convolution kernel and L represents the length of the input sequence x.
VSS block
The VSS module proposed in VMamba4 serves as the backbone of the VMKLA-UNet encoder, and its structure is shown in Fig. 2.c. The input first passes through an initial linear embedding layer and is then split into two independent information streams. One stream flows through a 3 × 3 depth-wise convolution layer, followed by a SiLU activation function, before entering the main 2D-Selective Scan Module (SS2D). The output of SS2D is then processed by a layer normalization layer and combined with the output from the other stream, which has also been activated by SiLU. The combined output forms the final result of the VSS module.
where \(\:\user2{E}\:\)is the output of the initial linear embedding, \(\:{\user2{S}}_{1}\:\)is the first information stream after processing by the SS2D module, and \(\:{\user2{S}}_{2}\:\)is the output of the second information stream. The final VSS module output \(\:\user2{Y}\:\)is the combined result of the two information streams.
2D selective scan (SS2D)
The 2D-Selective-Scan (SS2D)4 is the core component of the VSS block, designed to efficiently extract features from two-dimensional images. The main idea behind SS2D is to capture long-range dependencies and complex spatial structures through a multi-directional scanning strategy. Specifically, it employs a selective scanning approach to traverse the image from various directions (e.g., horizontal, vertical, diagonal), extracting features along these paths. This strategy selectively focuses on specific scanning directions, i.e., those that are most relevant for capturing key image patterns. As a result, it enables more effective modeling of both global and local image structures.
SS2D includes three operations: (1) a scan expanding operation, (2) S6 operation, which adds a selectable mechanism based on S4 to achieve linear time variability of the model, and (3) a scan merging operation. The visualization process of the SS2D algorithm is shown in Fig. 3, which tells us that the process of the SS2D algorithm is to scan from four different directions and finally merge them.
Specifically, the input data is flattened into 1D vectors along four different directions (e.g., upper left, lower right, lower left, and upper right) using a Scan Expanding operation. These 1D vectors are then processed by the S6 operation within the S6 Block. Finally, the vectors are fused into a 2D feature map via Scan Merging. SS2D ensures that the VSS block achieves a global receptive field, i.e., it captures information across the entire image, while maintaining linear computational complexity.
Decoder structure
The decoder is structured differently from the encoder. To enhance its feature representation capability, we designed a unique MKCSA structure, as illustrated in Fig. 2.b. In this work, we first proposed KAN linear attention in the decoder design, combining it with a channel-space attention mechanism. This novel approach significantly improves medical image segmentation performance while also reducing the model’s computational complexity.
Kolmogorov–Arnold networks (KAN)
Kolmogorov-Arnold Networks (KAN)6 is a neural network architecture based on the Kolmogorov-Arnold theorem, which is specifically designed to approximate arbitrary multi-dimensional continuous functions. The theorem was proposed by Andrey Kolmogorov and Vladimir Arnold and describes that any n-dimensional continuous function can be represented as a combination of a series of single-variable functions, as shown in Eq. (6):
where\(\:\:{\varphi\:}_{i\:}\)and\(\:\:{\psi\:}_{ij}\:\)are some single variable continuous functions. This formula means that any n-dimensional continuous function can be approximated by linear combination and nonlinear transformation of a finite number of one-dimensional functions.
Based on the Kolmogorov-Arnold theorem, KAN designs a three-layer neural network architecture to approximate complex functions in high-dimensional input space, including input layer, mapping layer, combination layer, nonlinear activation layer and output layer, as shown in Eq. (7).
where\(\:\:\mathcal{X}\mathcal{\:}\)is the multidimensional vector of input, \(\:{z}_{ij}\:\)is the one-dimensional features after mapping, \(\:{\psi\:}_{ij}\:\)is the mapping function, \(\:{h}_{i}\:\)is the new feature representation obtained by linearly combining the output of the mapping layer, \(\:{\varphi\:}_{i}\left({h}_{i}\right)\:\)is the nonlinear activation function applied to the output of the combination layer, and\(\:\:f\left(x\right)\:\)is the final output after superposition of all nonlinearly activated features.
The design of KAN is rooted in well-established mathematical theorems, providing a strong theoretical foundation for its expressive power. This architecture allows the model to handle highly complex, high-dimensional input data without significantly increasing computational complexity. Furthermore, its network structure offers a distinct advantage in interpretability, as the computations at each layer have precise mathematical meanings—i.e., they correspond to specific single-variable functions. This makes KAN particularly well-suited for applications where clarity and theoretical rigor are essential.
KAN with linear attention
Linear attention17 is an approach designed to optimize traditional self-attention mechanisms (e.g., the attention mechanism in Transformers) by reducing computational complexity. The complexity of traditional self-attention is O(N²), where N represents the length of the input sequence. This quadratic complexity results in a significant increase in computational resource consumption when processing long sequences. Linear attention addresses this by reducing the complexity to O(N) through specific optimizations, making it more suitable for modeling long sequences. It achieves this by first projecting queries and keys into a lower-dimensional feature space, followed by computing the weighted sum.
where\(\:\:Q=X{W}_{Q},\:\:K=X{W}_{K},\:\:V=X{W}_{V}\), \(\varphi (\cdot)\)is a point-to-point nonlinear mapping function, and 1 represents a vector whose elements are all ones.
Medical images often contain intricate patterns and structures, e.g., the boundaries of lesions or subtle anatomical variations, which are high-dimensional in nature. Linear attention mechanisms, while efficient, may struggle to represent such complex patterns effectively. However, KAN can model these complex relationships more effectively by decomposing high-dimensional functions into a combination of simpler one-dimensional functions. The KAN paper points out the application of the Kolmogorov-Arnold theorem, which mathematically proves that any continuous multivariable function can be decomposed into a finite combination of univariate functions. This formula has been mentioned in the previous introduction of KAN. We can explain how KAN captures deeper relationships from two aspects. The first aspect is the hierarchical nature of mathematical decomposition. The paper6 mentions that each layer of KAN performs two steps. The first step is local feature extraction, that is, the univariate function\(\:\:F\:\)independently processes each input feature to extract low-dimensional local patterns; the second step is global interaction combination, that is, the outer function\(\:\:S\:\)sums and combines the low-dimensional representation, gradually constructing high-order interactions, and hierarchical stacking can model nonlinearities of arbitrary depth (Theorem 2.1 in the paper6). The second aspect is the dynamic depth adjustment of KAN. KAN automatically expands the network depth through the “pruning-growth” mechanism (Sect. 2.5.1 of the paper6), prioritizes modeling low-order interactions, and then gradually introduces high-order terms to avoid falling into complex noise too early. In practice, this decomposition allows the model to capture intricate dependencies between input features while maintaining computational efficiency. As for the capture mechanism of complex dependencies, the paper6 has mentioned that KAN uses L1 regularization to sparse single-variable functions and automatically identifies key feature interactions. E.g., in the real world, there is a high-dimensional function\(\:\:f\left({x}_{1},\dots\:{,x}_{100}\right)={x}_{1}{x}_{2}+sin\left({x}_{3}\right)\). KAN can remove irrelevant terms\(\:\:{x}_{4},\dots\:,{x}_{100}\:\)through pruning. In addition, the spline curve of the single-variable function can also intuitively display the feature contribution. E.g.,\(\:\:{\varphi\:}_{\text{1,3}}{x}_{3}\:\)presents a sinusoidal state, indicating that\(\:\:{x}_{3}\:\)participates in the interaction through\(\:\:sin\left({x}_{3}\right)\). As for the guarantee of computational efficiency, Sect. 4.1 of the paper6 also mentioned that for\(\:\:n\)-dimensional input, the number of parameters of a single KAN layer is\(\:\:n\times\:k\times\:(2n+1)\), where\(\:\:k\:\)is the number of\(\:\:B\)-spline basis functions, which is much smaller than\(\:\:O\left({n}^{2}\right)\:\)of MLP.
By integrating KAN into linear attention, we achieve the following improvements:
Feature decomposition and nonlinear mapping
KAN decomposes the input feature space into simpler components, the simple components here refer to the decomposition of multivariable functions into combinations of univariate functions\(\:\:{\varphi\:}_{q,p}\left({x}_{p}\right)\:\)by KAN. Each\(\:\:{\varphi\:}_{q,p}\:\)only processes a single input feature, and the complexity is much lower than the multidimensional weight matrix. The\(\:\:B\)-spline function\(\:\:{B}_{i}\left(x\right)\:\)has local support (non-zero only in the interval\(\:\:[{t}_{i},\:{t}_{i+1}]\), so that each univariate function can be interpreted as a piecewise local response to the input feature, and the symbolic regression and physical law discovery mentioned in the paper also demonstrate the ability of KAN to recover real components6, and applies nonlinear mappings to each. E.g., if the input features\(\:\:X\:\)consist of multiple channels or modalities (e.g., grayscale, texture, or gradient information), KAN transforms\(\:\:X\:\)into a representation that highlights relationships between channels as shown in Eq. (9).
where\(\:\:{\varphi\:}_{i}\:\)and\(\:{\:\psi\:}_{ij}\:\)are nonlinear functions designed to capture local and global dependencies. This ensures that even subtle interactions between features are preserved.
Capturing global context
In medical images, the relationship between local regions (e.g., tumor boundaries) and global structures (e.g., organ shapes) is crucial. KAN enhances linear attention by embedding these relationships into the attention mechanism. For instance, after KAN processes the features, the attention scores calculated in linear attention better reflect the interplay between different regions.
Improved boundary and detail recognition
Boundary regions in medical images are often challenging to model due to their fine-grained details. By leveraging KAN’s ability to capture high-dimensional interactions, the attention mechanism can focus more accurately on these critical regions, improving segmentation precision.
Therefore, we consider combining KAN with linear attention to make up for the shortcomings of linear attention in expressing high-dimensional features, increase the expressiveness of the attention mechanism, and thus improve the performance of the overall model.
MKCSA block
As illustrated in Fig. 2.b, the MKCSA architecture is primarily comprised of two key components: KAN linear attention and a channel-spatial attention mechanism. The KAN linear attention is designed to capture the intricate relationships within high-dimensional inputs, enabling the network to effectively discern subtle features in medical images. The channel attention mechanism selectively emphasizes the most salient feature channels, such as the distinct tissue distributions present in medical images, while the spatial attention mechanism enhances the spatial representation of the image, ensuring precise segmentation of complex anatomical structures. By integrating both channel and spatial attention, the model is able to more comprehensively capture and represent the multidimensional information embedded within the image, leading to improved performance in fine-grained segmentation tasks.
In medical images, it is crucial to accurately identify lesion areas, subtle tissues, or organ boundaries. After deep convolution filtering of local pixels, the response to details is enhanced, while KAN linear attention captures the contextual information of local areas to ensure that subtle features are not missed when processing complex structures. This is especially important when processing high-resolution medical images and helps improve the accuracy of lesion identification.
where\(\:\:{x}_{i,j,c}\:\text{i}\text{s}\:\)the value of channel\(\:\:c\:\)at position\(\:\:\left(i,j\right)\:\)on the input feature map, and\(\:\:{w}_{m,n,c}\:\)is the weight of the corresponding filter. In this way, deep convolution can capture local features while maintaining channel independence, and can capture subtle structures and edges more accurately, especially in high-resolution medical images.\(\:\:Q,\:K\:\)and\(\:\:V\:\)represent query, key, and value matrices, respectively, where\(\:\:Q\) and\(\:\:K\:\)are generated by KAN and\(\:\:{\upsigma\:}\:\)are activation functions. This linear attention mechanism can effectively integrate global context information through matrix multiplication and reduce the computational complexity of traditional self-attention while maintaining sensitivity to local information.
Tissue structures in medical images often exhibit complex global relationships. The KAN linear attention mechanism establishes long-range dependencies between features on a global scale, ensuring the model retains critical details when interpreting the overall structure. This integration of global information is essential for accurately segmenting intricate anatomical structures and lesion regions, thereby enhancing both the precision and consistency of segmentation. Furthermore, the channel attention mechanism dynamically adjusts the weights of each feature channel, allowing the model to emphasize critical information and improve its ability to distinguish between different tissues or lesion areas. The spatial attention mechanism further refines this process by directing the model’s focus to key regions within the image, optimizing boundary delineation and enhancing detail accuracy. By combining channel and spatial attention, the model achieves greater adaptability and improved segmentation performance across various lesion types.
when\(\:\:GAP\left(X\right)\:\)is the global average pooling, \(\:{W}_{1}\:\)and \(\:{W}_{2}\:\)is the weight matrix of the fully connected layer, \(\:\sigma\:\:\)is the activation function, \(\:W\:\)is the weight of the convolutional layer, \(\:{F}^{{\prime\:}}\:\)is the output of channel attention. The channel-spatial attention mechanism18 is shown in Fig. 4.
Compared with traditional SSM models, e.g., SS2D4, the incorporation of KAN linear attention significantly reduces computational complexity. This reduction is particularly advantageous for datasets that require processing large-scale 3D medical images, i.e., MRI and CT scans. The use of KAN linear attention accelerates the model’s inference process, while also reducing training time and resource consumption. As can be seen from Table 1, the model complexity of the KAN linear attention module is much smaller than that of the SS2D model. Therefore, using the KAN linear attention module can significantly improve the lightweight of the model.
The specific process of this MKCSA module. First input features\(\:\:\mathcal{X}\in\:{\mathbb{R}}^{\user2{C}\times\:\user2{H}\times\:\user2{W}}\), where C represents the number of channels, H and W represent the height and width respectively. For the main branch, it first undergoes a linear transformation:
Then\(\:\:{Y}_{1}\:\)it goes through a 3 × 3 depth-wise separable convolution and an activation function:
The final result of the main branch is obtained by feeding \(\:{Y}_{2}\:\)it into the KAN linear attention module:
For the secondary branch, first go through the channel attention module:
Then \(\:{Y}_{1}^{{\prime\:}}\)the final result of the sub-branch is obtained through the spatial attention module:
Finally, the results of the two branches are multiplied element by element and then subjected to a linear transformation:
Skip connections
In order to effectively fuse the features of the encoder and decoder, we introduced skip connections between the corresponding encoder and decoder stages. This connection strategy enables the decoder to utilize feature information at different levels in the encoder, thereby improving the accuracy and robustness of segmentation. In each skip connection, features are fused by element-by-element addition to ensure full integration of local and global information.
Loss function
The proposed VMKLA-UNet is designed to tackle medical image segmentation tasks. For binary classification, we employ the binary cross-entropy (BCE) function combined with the Dice function as the loss functions. In the case of multi-class classification, we use the cross-entropy (CE) function along with the Dice function, as .
where N represents the total number of samples, and C represents the sample category.\(\:\:{y}_{i}\:\)and\(\:\:{\widehat{y}}_{i}\:\)represent the true label and predicted value, respectively. When\(\:\:{y}_{i,c}\:\)is equal to 1, it means that sample i belongs to class c, otherwise, it is equal to 0. \(\:{\widehat{y}}_{i,c}\:\)is the probability that the model predicts that sample i belongs to class c. |X| and |Y| represent the true label and predicted value respectively.\(\:\:{\lambda\:}_{1},\:\:{\lambda\:}_{2}\:\)represent the weight of the loss function, and the default value is 1.
Experiments
Dataset
We utilized five datasets across three categories to validate the effectiveness of the proposed model. The first category comprises open-source skin disease datasets, including ISIC201719,20, ISIC201820, and PH2 21, which were used to evaluate the model’s performance on 2D image segmentation. The ISIC 2017 dataset, part of the ISIC Challenge, aims to advance melanoma diagnosis using dermoscopic images, with a focus on lesion segmentation, i.e., accurately delineating skin lesion boundaries in dermoscopic images. The training set contains 2,000 images with corresponding segmentation labels, while the test set includes 600 images for model evaluation. ISIC2018 features a training set of 2,594 images and a test set of 1,000 images. The PH2 dataset, focused on skin cancer (primarily melanoma) detection, consists of 200 dermoscopic images depicting both benign lesions (e.g., moles) and malignant ones (melanomas). It serves as a benchmark for skin lesion classification, segmentation, and diagnosis tasks. Following previous successful models5, we split the ISIC skin lesion dataset into training and test sets at a 7:3 ratio and the PH2 dataset into a 1:1 ratio. The second category includes open-source polyp segmentation datasets, primarily used for polyp segmentation tasks. This category comprises subsets such as Kvasir-SEG, CVC-ClinicDB, CVC-ColonDB, EndoScene, and ETIS. We utilized four of these datasets: Kvasir-SEG22, ClinicDB23, ColonDB24, and ETIS25. The third category consists of the 3D medical image dataset Synapse, a multi-organ CT dataset for medical image segmentation. It contains 30 abdominal CT scans, totaling 3,779 axial enhanced abdominal CT images, with annotations for eight abdominal organs: the aorta, gallbladder, spleen, left kidney, right kidney, liver, pancreas, and stomach. This dataset is widely used to assess medical image segmentation algorithms, with evaluation metrics such as the Dice similarity coefficient (DSC) and Hausdorff distance (HD). For training, we applied the BceDice loss function on the ISIC, PH2, and Polyp datasets, and the CeDice loss function on the Synapse dataset.
Experimental environment
We resized the image resolution of all datasets to 256 × 256 and customized data augmentation methods, such as random flipping, random rotation, and center cropping. In the hyperparameter setting, we set the batch size to 32, used the AdamW optimizer, and the initial learning rate was 0.0001. The learning scheduling strategy used the classic CosineAnnealingLR, whose operation spanned a maximum of 50 iterations and the learning rate was as low as 1e-5. The epoch of the entire training process was set to 300. The implementation environment and hyperparameter settings for this experiment is presented in Table 2.
Analysis of experimental results
We compared VMKLA-UNet with some SOTA models, and the specific results are shown in Tables 3, 4, 5 and 6. For ISIC, Polyp and PH2 datasets, we compared mean intersection over union(mIoU), Dice coefficient(DSC), Accuracy(Acc), Specificity(Spe) and Sensitivity(Sen). Among them, mIoU is used to measure the overlap between the predicted area and the true area, Dsc is used to measure the overlap between the predicted segmentation results and the true segmentation results, Acc is used to measure the classification accuracy of the model on all pixels, Spe is used to measure the model’s ability to identify negative classes (background) and Sen is used to measure the model’s ability to identify the positive class (target area). And for the Synapse dataset, we mainly compared the DSC and HD95 indices as well as the DSC on each individual class.
In addition to computing the standard evaluation metrics, we also calculated the standard deviation of certain metrics on the ISIC, Polyp, and PH2 datasets. The standard deviation measures the variability or dispersion of a set of values. In the context of evaluation metrics, it quantifies the variation in performance indicators (e.g., mIoU, DSC) across different samples, providing deeper insights into the model’s stability and robustness. A smaller standard deviation indicates more consistent performance, while a larger standard deviation suggests greater variability.
By comparing VMKLA-UNet with other state-of-the-art (SOTA) models, we observe that our proposed model exhibits notable advantages across multiple datasets, including ISIC17, ISIC18, PH2, Polyp, and Synapse, as illustrated in Figs. 5, 6, 7, 8 and 9. These advantages are particularly evident in terms of edge completeness and lesion area detection accuracy. While many existing models either fail to fully perceive the lesion boundary or primarily focus on its most prominent regions, our model effectively captures the entire lesion area with high precision. Specifically, on the ISIC17 and ISIC18 datasets, our model achieves mIoU scores of 84.51% and 84.16%, Dice scores of 91.60% and 91.40%, and accuracy (Acc) values of 97.39% and 96.14%, respectively. Additionally, specificity (Spe) and sensitivity (Sen) are significantly improved, reaching 98.13% and 93.24% for ISIC17, and 97.56% and 91.26% for ISIC18. Compared to the SOTA models, our method increases mIoU, Dice, Acc, and Sen by 2.33%, 1.38%, 0.61%, and 3.34% on ISIC17, while on ISIC18, it improves mIoU, Dice, Acc, and Spe by 2.81%, 1.69%, 1.23%, and 0.62%, respectively. Similarly, on the PH2 dataset, our model improves Dice, Acc, and Spe by 0.15%, 0.25%, and 0.34%, respectively, over SOTA methods.
Additionally, we evaluate our model on the Polyp dataset, specifically on the Kvasir-SEG, ClinicDB, ColonDB, and ETIS benchmarks, where it consistently achieves strong performance. The experimental results demonstrate that our model is particularly effective in detecting polyp regions with high completeness and precision, even in cases where the polyps have indistinct or irregular boundaries. The improved segmentation quality in these datasets further highlights the robustness of our model in medical image analysis. Moreover, on the Synapse dataset, our model achieves a significant increase in total mDice and demonstrates superior segmentation accuracy for six out of eight organs. These improvements can be attributed to the unique combination of KAN linear attention and channel-spatial attention mechanisms within our model, which are built upon the Mamba architecture. This design enhances the model’s capability to capture both global and local spatial dependencies, leading to more complete segmentation contours and better differentiation between lesion areas and background regions. These results underscore the effectiveness of our model’s attention mechanism in refining segmentation quality and highlight its robustness across diverse medical imaging tasks.
To further prove that our designed KAN Linear Attention is superior to SS2D in the decoder, we compare and analyze the heat maps generated by SS2D and KAN Linear Attention from the perspective of interpretability, as shown in Fig. 10, and find that there is a significant difference in performance between the two. For the overall lesion area, SS2D mainly focuses on the “directly visible part” of the lesion in the original image, and has limited perception of the potential lesion area. In contrast, KAN Linear Attention has a more comprehensive understanding of the lesion area, and the edge depiction is clearer and more complete, which can be seen from the clear boundaries in its heat map. In addition, in terms of heat distribution, the hot spots generated by KAN Linear Attention are more concentrated and comprehensive, closely fitting the actual target area, while the hot spots of SS2D are more scattered or irrelevant. Importantly, for medical image segmentation tasks, accurate and complete coverage of the lesion area is crucial. The heatmaps generated by KAN Linear Attention not only better reflect the actual shape of the target region in terms of color and intensity, but also demonstrate excellent detection ability and consistency with the ground truth.
Ablation study
To demonstrate the effectiveness of MKCSA, we conducted relevant ablation experiments on ISIC17, ISIC18 and Polyp. In the ablation experiments, the encoder remained unchanged and all changes were made to the decoder.
The baseline model was VM-UNet5. We called the model in which the SS2D block was changed to KAN linear attention MKLA; the model in which only spatial and channel attention was added was called Only-CSA; the model in which SS2D was replaced with a normal linear attention module was called MLLA26; the model in which channel and spatial attention were added to the decoder of MLLA was called MLCSA. The results are shown in Tables 9 and 10.
In addition, we also conducted comparative experiments on the ISIC dataset and Polyp with encoders of different depths. As shown in Tables 7 and 8, as the encoder depth increases, the model is able to extract richer hierarchical features, thereby better capturing the edges and details of the target area, leading to a gradual improvement in performance.
In Table 9 show that after adding the new components, the mIoU of ISIC17 increased from 80.23 to 84.51%, and the Dice coefficient increased from 89.03 to 91.60%; the mIoU of ISIC18 increased from 81.35 to 84.16%, and the Dice coefficient increased from 89.71 to 91.40%. And in Table 10, by adding new components, the mIoU of the Kvair-SEG dataset increased from 80.32 to 86.43%, and the Dice coefficient increased from 89.09 to 92.72%; the mIoU of the ClinicDB dataset increased from 81.95 to 90.64%, and the Dice coefficient increased from 90.08 to 95.09%; the mIoU of the ColonDB dataset increased from 55.28 to 64.90%, and the Dice coefficient increased from 71.20 to 78.72%; the mIoU of the ETIS dataset increased from 66.41 to 73.53%, and the Dice coefficient increased from 79.81 to 84.74%. It is not difficult to see from the table that each component contributes to the improvement of model performance.
Through ablation experiments, we not only confirmed the key role of the new components in improving model performance and the effectiveness of model design, but also proved that the model encoder can learn more abstract and high-level features in a deeper network structure, which is consistent with the performance increase phenomenon observed in our experiments.
Conclusion
In this paper, we present a medical image segmentation model, VMKLA-UNet, which integrates KAN linear attention with channel-spatial attention and the Vision Mamba architecture. To the best of our knowledge, this is the first work to explore the combination of KAN-based linear attention with Vision Mamba and channel-spatial attention. To validate the model’s effectiveness in segmentation tasks, we conducted extensive experiments on the ISIC17, ISIC18, PH2, Polyp, and Synapse datasets. The results demonstrate that VMKLA-UNet offers notable advantages in medical image segmentation and shows promise for future exploration. However, there is still room for improvement, such as reducing the number of model parameters, incorporating a dedicated edge feature processing module, and further optimizing the encoder.
For future work, we plan to: (1) continue refining the model architecture, particularly by exploring more suitable SSM-based structures (e.g., encoder, decoder, and skip connections) for medical image segmentation; (2) further investigate the intersection of Mamba and KAN to develop a more lightweight model that reduces overall complexity; and (3) leverage the strengths of the Mamba structure to explore other downstream tasks in medical imaging, aiming to create a scalable, shareable, and unified multi-task model.
Data availability
The datasets we used in our experiments are all public datasets. The ISIC series datasets can be accessed at https://challenge.isic-archive.com/data/, the PH2 dataset is available at https://www.fc.up.pt/addi/ph2%20database.html, and the Polyp dataset originates from https://github.com/yaoppeng/U-Net_v2. Lastly, the Synapse dataset can be found at https://github.com/HuCaoFighting/Swin-Unet.
Change history
02 August 2025
The original online version of this Article was revised: In the original version of this Article the Acknowledgements section contained an error. The Acknowledgements section now reads: “This work was supported by the Innovation Team Funds of China West Normal University under Grant KCXTD2022-3, and the Chinese Government Guidance Fund on Local Science and Technology Development of Sichuan Province (2024ZYD0272)”.
References
Ronneberger, O., Fischer, P., Brox, T. & U-Net Convolutional networks for biomedical image segmentation. International Conference on Medical Image Computing and Computer-Assisted Intervention, 234–241 (2015).
Vaswani, A. et al. Attention is All You Need. Advances in Neural Information Processing Systems (NeurIPS), 5998–6008 (2017).
Gu, A., Dao, T. & Mamba Linear-time sequence modeling with selective state spaces. ArXiv Preprint (2023). arXiv:2312.00752.
Yue Liu, Tian, Y. et al. VMamba: Visual state space model. arXiv preprint arXiv:2401.10166 (2024).
Ruan, J. & Xiang, S. VM-UNet: vision Mamba UNet for medical image segmentation. ArXiv Preprint (2024). arXiv:2402.02491.
Liu, Z. et al. KAN: Kolmogorov-Arnold networks. ArXiv Preprint (2024). arXiv:2404.19756.
Long, J., Shelhamer, E. & Darrell, T. Fully convolutional networks for semantic segmentation. ArXiv Preprint (2015). arXiv:1411.4038.
Wu, R. et al. High-order Spatial interaction UNet for skin lesion segmentation. Biomed. Signal Process. Control. 88, 105517 (2024).
Oktay, O. et al. Attention U-Net: learning where to look for the pancreas. ArXiv Preprint (2018). arXiv:1804.03999.
Dosovitskiy, A. et al. An Image is Worth 16x16 Words: Transformers for Image Recognition at Scale. arXiv preprint arXiv:2010.11929(2020).
Chen, J. et al. TransUNet: Transformers make strong encoders for medical image segmentation. arXiv preprint arXiv:2102.04306 (2021).
Cao, H. et al. SwinUNet: UNet-like pure transformer for medical image segmentation. ArXiv Preprint (2021). arXiv:2105.05537.
Xu, Z., Guo, X. & Wang, J. Enhancing skin lesion segmentation with a fusion of convolutional neural networks and transformer models. Heliyon 10 (5), 101234 (2024).
Aghdam, E. K., Azad, R., Zarvani, M. & Merhof, D. Attention Swin U-Net: Cross-contextual attention mechanism for skin lesion segmentation. ArXiv Preprint (2022). arXiv:2210.16898.
Ma, J., Li, F. & Wang, B. U-Mamba: enhancing Long-Range dependency for biomedical image segmentation. ArXiv Preprint (2024). arXiv:2401.04722.
Hao, J. et al. T-Mamba: A unified framework with Long-Range dependency in dual-domain for 2D&3D tooth segmentation. ArXiv Preprint (2024). arXiv:2404.01065.
Li, R. et al. Linear attention mechanism: an efficient attention for semantic segmentation. ArXiv Preprint (2020). arXiv:2007.14902.
Woo, S. et al. CBAM: convolutional block attention module. ArXiv Preprint (2018). arXiv:1807.06521.
Matt Berseth ISIC 2017-Skin lesion analysis towards melanoma detection. ArXiv Preprint (2017). arXiv:1703.00523.
Noel Codella, V. et al. Skin lesion analysis toward melanoma detection 2018: A challenge hosted by the international skin imaging collaboration (ISIC). ArXiv Preprint (2019). arXiv:1902.0336.
Teresa Mendonc¸a, Pedro, M., Ferreira, J. S., Marques, A. R. S., Marcal & Rozeira, J. PH2-A Dermoscopic Image Database for Research and Benchmarking. Proceedings of the Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), 5437–5440 (2013). (2013).
Debesh Jha, Pia, H. et al. Kvasir-SEG: A Segmented Polyp Dataset. Proceedings of the 26th International Conference on Multimedia Modeling (MMM), 451–462 (2020).
Jorge Bernal, F., Javier Sánchez, C., Rodríguez & Vilarino, F. WM-DOVA Maps for Accurate Polyp Highlighting in Colonoscopy: Validation vs. Saliency Maps from Physicians. Computerized Medical Imaging and Graphics 43, 99–111 (2015).
Nima Tajbakhsh, Suryakanth, R., Gurudu & Liang, J. Automated polyp detection in colonoscopy videos using shape and context information. IEEE Trans. Med. Imaging. 35 (2), 630–644 (2015).
Juan Silva, A., Histace, O., Romain, X., Dray & Granado, B. Toward embedded detection of polyps in WCE images for early diagnosis of colorectal cancer. J. Comput. Assist. Radiol. Surg. 9, 283–293 (2014).
Dongchen et al. Demystify Mamba in vision: A linear attention perspective. ArXiv Preprint (2024). arXiv:2405.16605.
Zhang, Y., Liu, H., Hu, Q. & TransFuse Fusing transformers and CNNs for medical image segmentation. International Conference on Medical Image Computing and Computer-Assisted Intervention, 14–24 (2021).
Ruan, J., Xiang, S., Xie, M., Liu, T. & Fu, Y. MalUNet: A multi-attention and lightweight UNet for skin lesion segmentation. 2022 IEEE Int. Conf. Bioinf. Biomed. (BIBM). 1150, 1156 (2022).
Peng, Y., Sonka, M. & Chen, D. Z. U-Net v2: rethinking the skip connections of U-Net for medical image segmentation. ArXiv Preprint (2023). arXiv:2311.17791.
Zhou, Z., Siddiquee, M. M. R., Tajbakhsh, N., Liang, J. & UNet++: A nested U-Net architecture for medical image segmentation. Deep Learn. Med. Image Anal. Multimodal Learn. Clin. Decis. Support, 3–11 (2018).
Wei, J. et al. Shallow attention network for polyp segmentation. International Conference on Medical Image Computing and Computer-Assisted Intervention, 699–708 (2021).
Milletari, F., Navab, N., Ahmadi, S. A. & V-Net Fully convolutional neural networks for volumetric medical image segmentation. Fourth International Conference on 3D Vision (3DV), 565–571 (2016). (2016).
Alom, M. Z., Yakopcic, C., Hasan, M., Taha, T. M. & Asari, V. K. Recurrent residual U-Net for medical image segmentation. J. Med. Imaging. 6 (1), 014006–014006 (2019).
Azad, R., Al-Antary, M. T., Heidari, M., Merhof, D. & TransNorm Transformer provides a strong Spatial normalization mechanism for a deep segmentation model. IEEE Access. 10, 108205–108215 (2022).
Azad, R. et al. Convolution-free transformer-based DeepLab v3 + for medical image segmentation. International Workshop on Predictive Intelligence in Medicine, 91–102 (2022).
Wang, H., Cao, P., Wang, J., Zaiane, O. R. & UCTransNet Rethinking the skip connections in U-Net from a channel-wise perspective with transformer. Proceedings of the AAAI Conference on Artificial Intelligence, 2441–2449 (2022).
Wang, H. et al. Mixed Transformer U-Net for medical image segmentation. ICASSP IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), 2390–2394 (2022). (2022).
Ruan, J., Xie, M., Xiang, S., Liu, T. & Fu, Y. MeW-UNet: Multi-axis representation learning in frequency domain for medical image segmentation. ArXiv Preprint (2022). arXiv:2210.14007.
Gao, Y. et al. UTNet: A hybrid transformer architecture for medical image segmentation. ArXiv Preprint (2021). arXiv:2107.00781.
Chao et al. SliceMamba with neural architecture search for medical image segmentation. ArXiv Preprint. arXiv, 240708481 (2024).
Zhang, M. et al. SCRNet: A retinex Structure-based Low-light enhancement model guided by Spatial consistency. ArXiv Preprint (2023). arXiv:2305.08053.
Renkai, W. et al. UltraLight VM-UNet: parallel vision Mamba significantly reduces parameters for skin lesion segmentation. ArXiv Preprint (2024). arXiv:2403.20035.
Acknowledgements
This work was supported by the Innovation Team Funds of China West Normal University under Grant KCXTD2022-3, and the Chinese Government Guidance Fund on Local Science and Technology Development of Sichuan Province (2024ZYD0272).
Author information
Authors and Affiliations
Contributions
C.S. conceived the study, contributed to data collection, data analysis, data interpretation, manuscript preparation. S.L., and X.L. contributed to data collection. L.C., and J.W. contributed data analysis and interpretation. C.S. wrote the manuscript. J.W. contributed to funding acquisition. All authors edited and reviewed the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Su, C., Luo, X., Li, S. et al. VMKLA-UNet: vision Mamba with KAN linear attention U-Net. Sci Rep 15, 13258 (2025). https://doi.org/10.1038/s41598-025-97397-2
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-97397-2