Abstract
Fluorescent or luminescent labeling of biomolecules, as biosensors, with high sensitive and spatiotemporal resolution enable it an outstanding imaging technique for detecting and tracking biomolecular dynamics in many areas of life sciences and biomedical research. Ir(III) complex IrCN with solvent ligands could selectively recognize His via covalent attachment to the His imidazole group, and serve as a reaction-based “turn-on” fluorescent probe for the detection of His and His containing proteins. Fibroblast growth factor 21 (FGF21) was an essential glucose and lipid metabolic regulator and a promising therapeutic target for metabolic disorder syndromes. In this study, a non-emissive cyclometalated Ir(III) solvent complex IrCN was synthesized for FGF21 protein labeling. Binding test showed that the optimal binding ratio of IrCN and FGF21 protein was 1:100 (W/W). The binding between IrCN and FGF21 protein was very rapid, and the reaction could be completed in 10 min. IrCN probe no longer bound to other proteins after it specifically bound to the FGF21 protein. Biocompatibility studies shown that IrCN exhibited low cytotoxicity and tissue toxicity when the concentration was not higher than 50 μg/mL. Whereas, high concentration of IrCN caused organ-specific toxicity, with notable effects observed in both the spleen and skeletal muscle. Cell imaging experiments showed that revealed that unbound IrCN exhibits significant potential as a versatile cytoplasmic labeling agent, while its protein-conjugated form demonstrates effective protein tracing capabilities in cellular systems. Functional validation experiments by quantitative analysis of FGF21-mediated downstream pathway markers demonstrated that IrCN labeling preserves the native biological activity of FGF21 protein. This study demonstrated that IrCN served as a highly sensitive and stable probe for cell imaging and protein fluorescent labeling applications, which established a solid foundation for further exploration of its potential applications in diverse areas of biomolecular research, particularly in protein tracking and live-cell imaging studies.
Similar content being viewed by others
Introduction
Fluorescent or luminescent labeling of biomolecules, as biosensors, with high sensitive and spatiotemporal resolution enable it an outstanding imaging technique for detecting and tracking biomolecular dynamics in many areas of life sciences and biomedical research1. Metal complexes, due to their excellent photophysical properties, such as large Stokes shift, enhanced photostability, high damage threshold, long optical lifetime, high quantum yield and easy adjustment of the luminescence band, have been widely used in in many fields, such as sensing, organic light-emitting diodes, photodynamic therapy and anti-tumor2,3,4,5,6. Cyclometalated Iridium (Ir) complexes with favorable photophysical properties have been developed as a type of phosphorescent probes for amino acid detection, including cysteine (Cys), homocysteine and histidine (His)7,8,9. In particular, the nonemissive Ir(III) complex IrCN with solvent ligands could selectively recognize His via covalent attachment to the His imidazole group, and serve as a reaction-based “turn-on” fluorescent probe for the detection of His in solution9,10, which provided broad prospects for the study of His and His containing proteins. However, the knowledge about the biocompatibility of IrCN and its impact on protein function after labeling is still limited.
Fibroblast growth factor 21 (FGF21) was identified as an essential metabolic regulator involved in enhancing insulin sensitivity, lowing blood glucose and triglyceride (TG) levels, increasing brown adipocyte numbers, preserving β-cell function, inducing sustained weight loss, ameliorating hepatic steatosis and reducing cardiovascular disease risk11,12,13,14, which makes it a potential diagnostic marker and a promising therapeutic target for metabolic disorder syndromes, such as type 2 diabetes (T2D), nonalcoholic fatty liver, hyperlipidemia, and cardiovascular diseases15,16. FGF21 exerts regulation effects mainly via FGF receptor 1 (FGFR1) and a cofactor, β-Klotho (KLB)-mediated downstream cascades17, including sirtuin 1 (SIRT1), phosphatidylinositol 3-kinase (PI3K)/Akt signaling, AMPK/mTOR signaling, and ERK1/2 signaling18,19. FGF21 is mainly regulated by the nuclear receptor peroxisome proliferator‑activated receptor α (PPARα), PPARγ20 and transcriptional factor ATF4. PPAR and ATF4 can synergize to activate FGF21 transcription21,22. These findings allow a better understanding of the basic molecular mechanism of FGF21 and provided critical insights for the development of FGF21-based therapies. Efficient fluorescent labeling will help broaden the research progress and application field of FGF21 factor.
Camel can adapt to arid and semi-arid desert climates in the world mainly due to an efficient mechanism of lipid reserve and utilization in hump23. Our previous studies have claimed that camel FGF21 has distinct expression patterns and regulatory mechanisms compared to human and rodent13,23. In this work, a non-emissive cyclometalated Ir(III) solvent complex [Ir(ppy)2(CH3CN)2] OTf− (IrCN) is prepared for labeling FGF21 protein containing His tag in carboxy-terminal. After determining the binding conditions between the IrCN probe and FGF21 protein, the biological function of FGF21 is monitored via detecting the expression of FGF21 downstream signaling pathway genes. This study attempts to reveal the effect of IrCN probe labeling on protein function, providing a basis for the research and application of IrCN probe for protein labeling and cell imaging.
Results
Preparation and characterization of IrCN
To explore the protein labeling function of IrCN, this study synthesized IrCN in the laboratory and their property was characterized by 1H nuclear magnetic resonance (NMR) NMR and high-resolution mass spectrometry (HR-MS). As shown in Fig. 1, the cyclometalated IrCN complex were synthesized with following parameters, 1H NMR (400 MHz, DMSO-d6, δ ppm): 9.49 (d, J = 5.7 Hz, 2H), 8.40 (d, J = 8.1 Hz, 2H), 8.26 (dd, J = 11.4, 4.3 Hz, 2H), 7.89 (d, J = 7.6 Hz, 2H), 7.72 (dd, J = 9.7, 3.7 Hz, 3H), 7.64 (s, 1H), 7.05 (t, J = 7.5 Hz, 2H), 6.91 (t, J = 7.5 Hz, 3H), 5.91 (d, J = 7.6 Hz, 2H), 2.07 (s, 6H). In addition, the HR-MS spectrum data is in good agreement with the theoretical simulated result. These results suggested that high purity IrCN were obtained in this study.
Investigation of the binding kinetics between IrCN and FGF21 protein
To determine the optimal binding conditions of IrCN and FGF21 protein with His tag, including binding ratio, binding time, kinetic analysis, photostability and binding stability under different temperature, this study co-incubated IrCN probe and FGF21 protein with different concentrations for different durations, respectively. Fluorescence intensity was captured in a fluorescence spectrometer and analyzed using ImageJ software. As shown in Fig. 2, the IrCN probe showed binding affinity for FGF21 protein, manifesting in a distinct absorbance peak at 280 nm and a corresponding emission maximum at 515 nm. The binding of IrCN to FGF21 resulted in a 39-fold enhancement of fluorescence intensity at 515 nm and the fluorescence quantum yield exhibited a 30.67-fold enhancement (Fig. 2A and B). The fluorescence intensity resulting from IrCN-FGF21 interactions exhibited a monotonic increment with increasing IrCN concentration, reaching a saturation plateau at 20 μg/mL, and the optimal stoichiometric interaction ratio of IrCN-FGF21 protein was 1:100 (W/W, Fig. 2C). Time-resolved fluorescence spectroscopy showed that the IrCN probe exhibits rapid and specific binding kinetics with FGF21 protein. Significant fluorescence enhancement was observed within 2 min post-incubation and saturation of fluorescence intensity achieved at 10 min (Fig. 2D and E), indicating that the optimal binding time of IrCN and FGF21 protein was 10 min. Photostability assessment revealed that IrCN-FGF21 complex showed exceptional resistance to photodegradation under visible light irradiation of different wavelengths and continuous light exposure. Fluorescence intensity showed no attenuation after 10 min illumination (Fig. 2F and G). The binding stability assays showed that the IrCN-FGF21 conjugate exhibits exceptional thermal stability, maintaining persistent fluorescence signals exceeding 20 d under diverse temperature conditions, including room temperature (25 °C), 4 °C and − 20 °C (Fig. 2H), indicating that IrCN-labeled conjugates possess good stability and a long half-life. These results demonstrated that IrCN is a sensitive and stable probe for labeling proteins.
Determination of the binding kinetics of IrCN and FGF21 proteins. (A) Determination of absorption spectra of IrCN binding to FGF21 protein. (B) Determination of emission spectra of IrCN binding to FGF21 protein. (C) Determination of the optimal binding ratio of IrCN and FGF21 protein. (D) Investigation of binding kinetics between IrCN and FGF21 protein. (E) Determination of the optimal binding time between IrCN and FGF21 protein. (F) Photostability testing of IrCN binding to FGF21 protein under visible light irradiation of different wavelengths. (G) Photostability testing of IrCN binding to FGF21 protein under irradiation at 515 nm wavelength for different durations. (H) Determination of binding stability of IrCN and FGF21 protein under different temperature conditions, including room temperature (25 °C), 4 °C and − 20 °C. Fluorescence signals were captured in a fluorescence spectrometer and UV spectrometer system. The fluorescence intensity analyzed using ImageJ software.
Binding specificity of IrCN and FGF21 protein with His tag
Ir(III) complex IrCN could selectively bind to His via covalent attachment to imidazole group and “turn-on” fluorescent9,10. The His tandem tag is also a commonly used tool for protein purification. To understand the binding characteristics of IrCN with His-tagged proteins, FGF21 proteins purification used in this study were performed via His tag mediated column chromatography. Bovine serum albumin (BSA) was introduced as control. FGF21 proteins were incubated with IrCN of different concentrations for 15 min firstly, then BSA was added and maintained for 15 min. The mixture was subjected for gel electrophoresis separation and fluorescence analysis. As shown in Fig. 3, FGF21 contains five discontinuous His within the protein and six continuous His at the C-terminus. BSA contains ten discontinuous His within the protein (Fig. 3A). As IrCN probe concentration decreases, the fluorescence brightness of FGF21 protein gradually decreases. When the ratio of IrCN to FGF21 protein reaches below 1:200 (W/W), the fluorescence bands of BSA protein disappeared (Fig. 3B), suggesting that IrCN probe no longer binds to other proteins after it specifically binds to the FGF21 protein. The results indicated that IrCN probes can specifically bind to FGF21 proteins with His tag. When the concentration ratio between IrCN probe and FGF21 protein is below 1:200, there are no free IrCN binding to other proteins.
Binding specificity of IrCN and FGF21 protein with His tag. (A) Schematic diagram of protein molecules FGF21 with His tag and bovine serum albumin (BSA). The molecular weight of FGF21 and BSA is 22 kD and 66 kD, respectively. The blue line represents the position of his in the polypeptide chain. (B) Each sample tube with addition of IrCN probe, FGF21 and BSA protein in turn. The mixture after reaction for 15 min were subjected for gel electrophoresis and fluorescence detection, respectively.
Biocompatibility of the IrCN
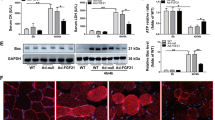
For a labeling probe, low cytotoxicity and good biocompatibility are usually prerequisites for their clinical applications. To ensure their potential for clinical application, the biocompatibility of IrCN was systematically assessed through a cytotoxicity assay, hemolysis experiment, and histopathological analysis, respectively. As shown in Fig. 4, two different cell lines BHK-21 and MAC-T cells, were exposed to varying concentrations of IrCN for 24 h at 37 °C. Addition of IrCN did not cause a significant decrease in cell survival rate until the concentration of IrCN reached 50 μg/mL (Fig. 4A), indicating that the working concentration of IrCN used for probe labeling should be maintained below 50 μg/mL to ensure proper cellular function.
Biocompatibility of the IrCN. (A) Cytotoxicity of BHK-21 and MAC-T cells was assessed by CCK8 method after 24 h exposure to IrCN at different concentrations. (B) Hemolytic effect of the IrCN. (C) Principal organs morphology was examined using H&E staining after IrCN treatment for 3 d in mice. The arrows indicated the location of the lesion. Scale bar: 100 μm. *P < 0.05; ** P < 0.01.
The blood compatibility of IrCN was determined by a hemolysis assay. As shown in Fig. 4B, the hemolysis effect showed a concentration-dependent manner for IrCN. When the concentration of IrCN exceeds 100 μg/mL, it will cause significant hemolysis of red blood cells, indicating that the concentration of IrCN in the circulatory system should be below 100 μg/mL when used for in vivo labeling.
Organ toxicity potentially induced by IrCN was evaluated through a histological examination base on H&E staining. As shown in Fig. 4C, the major organs including heart, liver, lung and kidney showed no marked pathological abnormalities or adverse effects after treating mice weighing 40 g with 50 or 200 μg IrCN. However, the mouse exposed to IrCN exhibited severe spleen lesions, including splenic corpuscle degeneration and atrophy, with blurred border boundaries as indicated by the black arrow in Fig. 4C, and severe infiltration of red blood cells and inflammatory cells in the red pulp as indicated by the red arrow in Fig. 4C. Additionally, high dose IrCN treatment (200 μg) caused diffuse necrosis and rupture of muscle fibers as indicated by the blue arrow in Fig. 4C, suggesting that high concentration IrCN demonstrated potential organ-specific toxicity, with notable effects observed in both the spleen and skeletal muscle..
IrCN probe has potential for cell imaging and protein tracing
To understand whether the IrCN probe can be used for cell fluorescence labeling and imaging, IrCN were subjected to co-culture with living cells and fixed cells, respectively. Fluorescence imaging was generated by a fluorescence microscope. As shown in Fig. 5, in living cells, IrCN treatment elicited prominent green fluorescence signals localized specifically in the cytoplasm. The fluorescence intensity exhibited a concentration-dependent relationship, with elevated IrCN concentrations yielding more intense cytoplasmic green fluorescence (Fig. 5A). In fixed cells, IrCN treatment resulted in intense green fluorescence specifically localized within the cytoplasmic compartment. (Fig. 5B). To precisely determine the subcellular localization of IrCN-induced fluorescence, confocal microscopy analysis was conducted, revealing that the fluorescence signals were predominantly localized in the perinuclear region of the cytoplasm (Fig. 5C). These results indicated that IrCN probe has potential for versatile cytoplasmic labeling both in living cells and fixed cells.
IrCN probe can be used for cell imaging. (A) Fluorescence imaging of live cells treated with IrCN of different concentrations. (B) Fluorescence imaging of fixed cells treated with IrCN. (C) Confocal imaging of cell after treatment with IrCN. (D) IrCN-labeled FGF21-His protein complex was introduced into cultured cells. IrCN signal was detected as green fluorescence through its intrinsic emission properties and FGF21-His protein was specifically labeled with anti-His tag antibodies and visualized as red fluorescence using TRITC-conjugated secondary antibodies in immunofluorescence staining. Scale bar: 50 μm.
To further investigate the potential of the IrCN probe for protein tracing in cellular environments, we introduced IrCN-labeled FGF21-His protein into cultured cells. After 4 h incubation, the cells were fixed with 4% paraformaldehyde and subjected to immunofluorescence staining with the use of anti-His tag IgG as the primary antibody and TRITC-conjugated secondary antibody. Fluorescence microscopy was then employed to capture the resulting signals. As illustrated in Fig. 5D, a significant colocalization was observed between the green fluorescence signal from the IrCN probe and the red fluorescence signal representing the FGF21 protein, particularly in the cell membrane and cytoplasmic regions, indicating a stable association between the IrCN probe and FGF21 protein within the cellular context. These findings demonstrated the potential utility of the IrCN probe as an effective tool for protein tracing applications in cells.
IrCN labeling did not affect FGF21 protein function
To determine whether IrCN labeling affect protein functionality, the study generated FGF21 proteins with C-terminal His tags from multiple species, including camel, mouse and human. BHK-21 cells were treated with either IrCN-labeled or unlabeled FGF21 proteins, followed by analysis of downstream pathway components. Gene expression levels were quantified using qPCR, while protein expression was assessed through western blot analysis. As demonstrated in Fig. 6, the expression level of FGF21 downstream pathway genes, including PI3K, Akt, and mTOR showed significantly increased both in mRNA levels and protein levels after FGF21 proteins treatment with or without IrCN labeling. Comparative analysis revealed no statistically significant differences in the expression patterns of these downstream genes between cells treated with IrCN-labeled and unlabeled FGF21 proteins (Fig. 6). The result collectively indicated that the IrCN labeling process does not compromise the biological activity or functional integrity of FGF21 proteins.
IrCN labeling did not affect FGF21 protein function. (A) The mRNA expression levels of PI3K, AKT and mTOR after FGF21 protein treatment from different species with or without IrCN labeling. (B) The protein expression levels of PI3K, AKT and mTOR after FGF21 protein treatment from different species with or without IrCN labeling. Relative intensity of protein bands was captured and analyzed using ImageJ software. cFGF21: camel FGF21. mFGF21: mouse FGF21. hFGF21: human FGF21. * P < 0.05; ** P < 0.01.
Discussion
Recently, Ir(III) complexes has gained widespread attention due to their excellent photophysical properties as a specific biomarker probes for biomolecule imaging and their great potential in the life sciences and biomedical field24,25,26. Intriguingly, Ir(III) complexes could selectively recognize His via covalent attachment to the imidazole group, and serve as a reaction-based “turn-on” fluorescent probe for the detection of His in solution, which provided broad prospects for the study of proteins containing His27,28. However, the knowledge about the biocompatibility of Ir(III) complexes and whether Ir(III) probes labeling will affect protein function is still limited. FGF21 was a novel metabolic regulator and showed great prospects for treatment of metabolic disorders, such as type 2 diabetes mellitus (T2DM), obesity and cardiovascular diseases (CVDs), which made FGF21 a promising therapeutic target and valuable diagnostic marker for metabolic disease15,16,29. Developing a specific labeling method for FGF21 will provide support for further in-depth research on FGF21 function.
In this study, Ir(III) complex IrCN was synthesized as a fluorescent probe to label FGF21 protein containing His tag, and was proven to be an effective protein labeling probe. The IrCN-mediated labeling of FGF21 proteins exhibits several advantageous characteristics, including high sensitivity, rapid reaction kinetics, exceptional stability, and remarkable specificity. Remarkably, the labeling process requires only 1% (w/w) of IrCN relative to the target FGF21 protein and can be completed within 10 min. The IrCN-induced fluorescence in labeled FGF21 proteins demonstrated extraordinary stability, maintaining detectable signals for over 20 d across a wide range of temperature conditions. Furthermore, the binding specificity analysis revealed that once IrCN forms a complex with FGF21, it exhibits exclusive binding characteristics, demonstrating no detectable affinity for other proteins. These findings demonstrated that that IrCN is an efficient protein specific probe, especially for proteins containing His tag, which may be attributed to IrCN’s ability to recognize and covalently bind imidazole groups of His, which then turn on fluorescence27,28. However, there are still some issues that need to be addressed, such as whether IrCN has better binding affinity with continuous His-tag compared to dispersed His in proteins.
Consequently, the IrCN-labeled FGF21 protein maintains remarkable stability within cellular environments, as evidenced by our experimental observations. This stability profile, combined with the probe’s specific binding characteristics, underscores the significant potential of the IrCN probe as a robust and reliable tool for real-time protein tracing and visualization in cell systems. Furthermore, this study demonstrated that unbound IrCN functions as an effective and versatile cytoplasmic staining probe in both live and fixed cells, which provided an alternative approach for comprehensive cell imaging applications, especially for real-time visualization of living cell behavior and intracellular dynamics in biomedical research. The IrCN probe showed good cell compatibility when its concentration is below 50 μg/mL, which is consistent with previous report30,31. However, IrCN with a concentration higher than 100 μg/mL will cause an increase in cell mortality and hemolysis. Actually, various nanomaterials can produce cytotoxicity and hemolysis when the concentration is too high32,33,34. IrCN exposure showed no marked damage to major organs including heart, liver, lung and kidney, even at concentrations of 200 μg per 40 g body weight in mice. The result is consistent with previous report that Ir(III) complex is a promising avenue for the treatment of liver damage35. Notably, administration of IrCN in vivo caused severe spleen lesions in mouse, including splenic corpuscle degeneration and atrophy, with blurred border boundaries. And high dose IrCN (200 μg per 40 g body weight) caused diffuse necrosis and rupture of muscle fibers in mouse, which casted a shadow over the in vivo application of IrCN probe, although the possible toxicological impacts of Ir(III) complex-based nanomaterials warrant further consideration and investigation.
FGF21 exerts metabolic regulatory activities mainly via the PI3K/AKT/mTOR signaling cascade36. This study evaluated the expression levels of PI3K, Akt, and mTOR, and found that adding exogenous FGF21 protein, whether from camels, mice, or humans, can activate PI3K, Akt, and mTOR expression. IrCN labeling did not affect FGF21-stimulated downstream gene expression levels, including PI3K, Akt, and mTOR, indicating that IrCN labeling did not impact the biological function of FGF21 protein, which may be due to that His is not the key residue for FGF21 protein to bind to receptor.
In summary, this study synthesized Ir(III) complex IrCN for labeling FGF21 protein and found that IrCN is a sensitive, rapid, stable, and specific FGF21 protein probe. The IrCN probe showed good biocompatibility when its concentration is below 50 μg/mL. IrCN labeling did not affect the biological function of FGF21 protein. In addition, IrCN is also a sensitive and stable probe for cell imaging and protein tracing. These findings established a solid foundation for further exploration and potential widespread application of the IrCN probe in various biomedical research fields, particularly in protein tracking and live-cell imaging studies.
Materials and methods
Synthesis and characterization of IrCN
IrCN fluorescent probe was synthesized using phenylpyridine and IrCl3·3H2O as raw materials. Briefly, phenylpyridine, IrCl3·3H2O, 2-Ethoxyethanol and H2O were sequentially added to a 100 mL three necked flask. The reaction was maintained under N2 atmosphere, and at 110 °C in the dark for 24 h. After the reaction was completed and cooled to room temperature, a yellow solid was observed to precipitate. After vacuum filtration and ethanol washing, a yellow solid M2 was obtained. M2, silver nitrate, and acetonitrile were weighed and sequentially placed into a 100 mL three necked flask. The reaction was refluxed under N2 atmosphere for 24 h. After the reaction was completed and cooled to room temperature, concentrated filtrate was obtained via vacuum filtration. After adding a small amount of CH2Cl2, filter under reduced pressure to remove excess silver nitrate, concentrating the filtrate to obtain a purple yellow solid, iridium complex IrCN. The synthesized IrCN was characterized by 1H nuclear magnetic resonance (NMR) and high-resolution mass spectrometry (HR-MS), respectively.
FGF21 proteins and cells
Camel, mouse and human FGF21 proteins containing His tag in carboxy-terminal were expressed and purified using prokaryotic expression system and preserved in our laboratory. Bovine serum albumin (BSA) was purchased from Solarbio (Beijing, China). BHK-21 and MAC-T cells were purchased from the ATCC corporation (Beijing, China), and cultured in DMEM medium (BasalMedia, Shanghai, China) containing 10% fetal bovine serum (FBS, Gibco, Carlsbad, CA, USA) and maintained in dishes or plates in a cell culture incubator with 5% CO2 at 37 °C.
Exploring the optimal binding conditions of IrCN and FGF21 protein
The IrCN was dissolved in dimethyl sulfoxide (DMSO) to dilute into different concentration gradients, ranging from 0.05 to 1 mg/mL. The optimal binding ratio between IrCN probe and FGF21 protein was determined by co-incubating IrCN and FGF21 protein at different concentrations. The binding time and stability was determined by co-incubating probes with proteins for different durations. Fluorescence signals were captured using a fluorescence spectrometer (Horiba, Shizuoka, Japan) and UV imaging system (Tannon, Shanghai, China). The fluorescence intensity was analyzed using ImageJ software. The fold-change in fluorescence quantum yields at room temperature was measured using the following equation.
where Ф is the quantum yield, D is the integrated area of the emission spectrum, A is the absorbance at the excitation wavelength, respectively.
Biocompatibility evaluation of the IrCN
Biocompatibility of the IrCN was systematically assessed through a cytotoxicity assay, hemolysis test, and histopathological analysis after using different concentrations of IrCN to treat cells, red blood cells and mice, respectively. Cytotoxicity effect was performed by a standard CCK8 method according to the manufacture’s instruction (Beyotime, Beijing, China). For histopathological analysis, mouse tissues were collected 3 d after intramuscular injection of IrCN to prepare paraffin sections, which were then stained with haematoxylin and eosin (H&E) staining as previously described37. The animal use protocol listed in this study was reviewed and approved by the Animal Ethical and Welfare Committee of Gansu Agricultural University, and mice were euthanized by injection of pentobarbital sodium in accordance with American Veterinary Medical Association (AVMA) guidelines. For hemolysis test, fresh blood was collected from healthy mouse. After adding 10 times the volume of 0.9% NaCl solution, centrifuging and discarding the supernatant, a 4% red blood cell suspension was prepared via adding 0.9% NaCl solution. Hemolysis phenomenon was observed by mixture of IrCN of different concentrations into 200 μL of red blood cell suspension for 15 min in room temperature. The absorbance value at 540 nm was recorded by Spectrophotometer (BioTek, Winooski, VT, USA). Hemolysis rate = (ODsample test group − ODnegative group)/(ODpositive group − ODnegative group) × 100%.
Fluorescence imaging analysis
For live cell imaging, BHK-21 cells were seeded into small dishes. When the cells reached a 70% confluence, different concentrations of IrCN were added into the dishes and co-cultured with the cells for 10 min. After washing twice with phosphate buffer saline (PBS), fluorescence signals were captured using a fluorescence microscope (Echo-Labs, Englewood, CO, USA) and the fluorescence intensity was quantified by ImageJ software.
For fixed cell imaging, BHK-21 cells were seeded into dishes. After reaching a 70% confluence, the cells were fixed with 4% paraformaldehyde at room temperature for 30 min. After washing three times with PBS, the cells were permeabilized in 0.2% Triton X-100 (Beyotime). Different concentrations of IrCN were added into the dishes and co-cultured with the cells for 10 min. Fluorescence was captured using a fluorescence microscope (Echo-Labs) and the fluorescence intensity was quantified by ImageJ software.
Immunofluorescence assay
For immunofluorescence staining, cells after IrCN-FGF21 conjugate treatment for 2 h were collected and fixed with 4% paraformaldehyde (PFA) for 15 min at room temperature. After permeabilization with 0.1% Triton X-100 for 10 min and blocking with 5% BSA for 1 h, cells were incubated with rabbit anti-His tag IgG (Immunoway, San Jose, CA, USA) overnight at 4 °C. Subsequently, cells were washed three times with PBS and incubated with TRITC-conjugated goat anti-rabbit IgG (H + L) (Proteintech, Chicago, IL, USA) for 1 h at room temperature in the dark. Nuclei were counterstained with DAPI (Beyotime) for 5 min, and coverslips were mounted onto glass slides using antifade mounting medium for imaging under a fluorescence microscope (Echo-Labs).
Real-time quantitative PCR (qPCR) assay
Total RNA was extracted from BHK-21 using the TRIzol reagent (Beyotime). cDNA was synthesized according to the instructions of the PrimeScript RT Reagent Kit (Takara, Dalian, China). The primers sequences for specific genes detection are listed as follow. β-actin forward primer: 5′-GGCTGTATTCCCCTCCATCG-3′ and reverse primer: 5′-CCAGTTGGTAACAATGCCATGT-3′. PI3K forward primer: 5′-CAAAGCCGAGAACCTATTGC-3′ and reverse primer: 5′-GGTGGCAGTCTTGTTGATGA-3′. AKT forward primer: 5′-AGATGGACTCAAGAGGCAGGAAG-3′ and reverse primer: 5′-TACTCAAACTCGTTCATGGTCACAC-3′. mTOR forward primer: 5′-TTGAGGTTGCTATGACCAGAGAGAA-3′ and reverse primer: 5′-TTACCAGAAAGGACACCAGCCAATG-3′. The primers were synthesized by Sangon Biotech (Shanghai, China). The qPCR reaction system was prepared according to the protocol of the SYBR Premix Ex Taq II Kit (Takara), and the reaction was performed on the LightCycler instrument (Roche, Indianapolis, IN, USA). Relative gene expression quantity was analyzed using the 2−ΔΔCt method and the amount of transcript in each sample was normalized using Actin as the internal control.
Western blot assay
Cellular total protein was extracted using RIPA lysis buffer (Beyotime) and protein concentration was measured using the BCA protein assay kit (Beyotime). Equal total protein was subjected to 10% SDS-PAGE gel for electrophoresis separation, and then electrotransferred onto a 0.45 µm polyvinylidene difluoride (PVDF) membrane (Biosharp, Beijing, China). The membrane was blocked for 2 h in 5% skim milk, followed by overnight incubation at 4 °C with primary antibodies, including anti-β-tubulin antibody (1:10,000, Proteintech, Chicago, IL, USA), anti-PI3K antibody (1:1000, Immunoway), anti-AKT antibody (1:1000, Immunoway, Plano, TX, USA), and anti-mTOR antibody (1:1000, Immunoway). After washing with PBS for 3 times, the membrane was co-incubated with the HRP-conjugated goat anti-rabbit IgG (1:5000, Proteintech) for 2 h at room temperature. Finally, protein bands were visualized using the ECL chemiluminescence detection kit (NeoBioscience, Suzhou, China) and captured by the Gel imaging system (Tannon, Shanghai, China).
Statistical analysis
Data were shown as mean ± SD, and the GraphPad Prism software (version 9.3.1, San Diego, CA, USA) was used for analysis of results. The significance of the differences between two groups and multiple groups were analyzed by two-tailed Student’s t-test and one-way ANOVA, respectively. P-value < 0.05 was considered to be statistically significant.
Data availability
All data generated or analysed during this study are included in this published article.
References
Wang, X., Jia, J., Huang, Z., Zhou, M. & Fei, H. Luminescent peptide labeling based on a histidine-binding iridium(III) complex for cell penetration and intracellular targeting studies. Chemistry 17, 8028–8032. https://doi.org/10.1002/chem.201100568 (2011).
Chen, Y. et al. Two-photon luminescent metal complexes for bioimaging and cancer phototherapy. Coord. Chem. Rev. 310, 16–40. https://doi.org/10.1016/j.ccr.2015.09.010 (2016).
Imberti, C., Zhang, P., Huang, H. & Sadler, P. J. New designs for phototherapeutic transition metal complexes. Angew. Chem. Int. Ed. Engl. 59, 61–73. https://doi.org/10.1002/anie.201905171 (2020).
Zhao, Z., Gao, P., Ma, L. & Chen, T. A highly X-ray sensitive iridium prodrug for visualized tumor radiochemotherapy. Chem. Sci. 11, 3780–3789. https://doi.org/10.1039/d0sc00862a (2020).
Wang, S., Wang, B., Zhu, L., Hou, J. T. & Yu, K. K. A ratiometric fluorescent probe for monitoring pH fluctuations during autophagy in living cells. Chem. Commun. 57, 1510–1513. https://doi.org/10.1039/d0cc07788g (2021).
Liang, X. et al. Fast synthesis of iridium(III) complexes incorporating a bis(diphenylphorothioyl)amide ligand for efficient pure green OLEDs. ACS Appl. Mater. Interfaces 11, 7184–7191. https://doi.org/10.1021/acsami.8b19318 (2019).
Li, C. et al. A nonemissive iridium(III) complex that specifically lights-up the nuclei of living cells. J. Am. Chem. Soc. 133, 11231–11239. https://doi.org/10.1021/ja202344c (2011).
Wu, Y. et al. A near-infrared phosphorescent iridium(iii) complex for imaging of cysteine and homocysteine in living cells and in vivo. RSC Adv. 7, 52621–52625. https://doi.org/10.1039/c7ra09798k (2017).
Wang, H. et al. A fluorescent probe based on Ir(III) solvent complex for specific recognition of histidine in aqueous solution and the application in cell imaging. Inorg. Chim. Acta 511, 119799. https://doi.org/10.1016/j.ica.2020.119799 (2020).
Chen, T. et al. Highly selective “off-on” fluorescent probe for histidine and its imaging in living cells. Biosens. Bioelectron. 66, 259–265. https://doi.org/10.1016/j.bios.2014.11.005 (2015).
Li, X. The FGF metabolic axis. Front. Med. 13, 511–530. https://doi.org/10.1007/s11684-019-0711-y (2019).
Szczepanska, E. & Gietka-Czernel, M. FGF21: A novel regulator of glucose and lipid metabolism and whole-body energy balance. Hormone and metabolic research = Hormon- und Stoffwechselforschung = Hormones et metabolisme 54, 203–211. https://doi.org/10.1055/a-1778-4159 (2022).
Yong, F. et al. Analysis of functional promoter of camel FGF21 gene and identification of small compounds targeting FGF21 protein. Vet. Sci. 10, 452. https://doi.org/10.3390/vetsci10070452 (2023).
Tan, H. et al. Targeting FGF21 in cardiovascular and metabolic diseases: From mechanism to medicine. Int. J. Biol. Sci. 19, 66–88. https://doi.org/10.7150/ijbs.73936 (2023).
Post, A. et al. Circulating FGF21 concentration, fasting plasma glucose, and the risk of type 2 diabetes: Results from the PREVEND study. J. Clin. Endocrinol. Metab. 108, 1387–1393. https://doi.org/10.1210/clinem/dgac729 (2023).
Kliewer, S. A. & Mangelsdorf, D. J. A dozen years of discovery: Insights into the physiology and pharmacology of FGF21. Cell Metab. 29, 246–253. https://doi.org/10.1016/j.cmet.2019.01.004 (2019).
Ogawa, Y. et al. BetaKlotho is required for metabolic activity of fibroblast growth factor 21. Proc. Natl. Acad. Sci. U. S. A. 104, 7432–7437. https://doi.org/10.1073/pnas.0701600104 (2007).
Flippo, K. H. & Potthoff, M. J. Metabolic messengers: FGF21. Nat. Metab. 3, 309–317. https://doi.org/10.1038/s42255-021-00354-2 (2021).
Kilkenny, D. M. & Rocheleau, J. V. The FGF21 receptor signaling complex: Klothobeta, FGFR1c, and other regulatory interactions. Vitam. Horm. 101, 17–58. https://doi.org/10.1016/bs.vh.2016.02.008 (2016).
So, W. Y. et al. High glucose represses beta-klotho expression and impairs fibroblast growth factor 21 action in mouse pancreatic islets: involvement of peroxisome proliferator-activated receptor gamma signaling. Diabetes 62, 3751–3759. https://doi.org/10.2337/db13-0645 (2013).
Schaap, F. G., Kremer, A. E., Lamers, W. H., Jansen, P. L. & Gaemers, I. C. Fibroblast growth factor 21 is induced by endoplasmic reticulum stress. Biochimie 95, 692–699. https://doi.org/10.1016/j.biochi.2012.10.019 (2013).
Ord, T., Ord, D. & Ord, T. TRIB3 limits FGF21 induction during in vitro and in vivo nutrient deficiencies by inhibiting C/EBP-ATF response elements in the Fgf21 promoter. Biochim. Biophys. Acta Gene Regul. Mech. 1861, 271–281. https://doi.org/10.1016/j.bbagrm.2018.01.014 (2018).
Gao, Y. et al. Localization of FGF21 protein and lipid metabolism-related genes in camels. Life 13, 432. https://doi.org/10.3390/life13020432 (2023).
Caporale, C. & Massi, M. Cyclometalated iridium(III) complexes for life science. Coordin. Chem. Rev. 363, 71–91. https://doi.org/10.1016/j.ccr.2018.02.006 (2018).
Aoki, S. et al. Post-complexation functionalization of cyclometalated iridium(III) complexes and applications to biomedical and material sciences. Top. Curr. Chem. 380, 36. https://doi.org/10.1007/s41061-022-00401-w (2022).
Jhun, B. H., Song, D., Park, S. Y. & You, Y. Phosphorescent Ir(III) complexes for biolabeling and biosensing. Top. Curr. Chem. 380, 35. https://doi.org/10.1007/s41061-022-00389-3 (2022).
Szymaszek, P., Tyszka-Czochara, M. & Ortyl, J. Iridium(III) complexes as novel theranostic small molecules for medical diagnostics, precise imaging at a single cell level and targeted anticancer therapy. Eur. J. Med. Chem. 276, 116648. https://doi.org/10.1016/j.ejmech.2024.116648 (2024).
Shi, H., Wang, Y., Lin, S., Lou, J. & Zhang, Q. Recent development and application of cyclometalated iridium(III) complexes as chemical and biological probes. Dalton Trans. 50, 6410–6417. https://doi.org/10.1039/d1dt00592h (2021).
Xiao, F. et al. Dose-response relationship between serum fibroblast growth factor 21 and liver fat content in non-alcoholic fatty liver disease. Diabetes Metab. 47, 101221. https://doi.org/10.1016/j.diabet.2020.101221 (2021).
Lau, J. S. et al. Luminescent cyclometalated iridium(III) polypyridine indole complexes–synthesis, photophysics, electrochemistry, protein-binding properties, cytotoxicity, and cellular uptake. Inorg. Chem. 48, 708–718. https://doi.org/10.1021/ic801818x (2009).
Lee, P. K., Law, W. H., Liu, H. W. & Lo, K. K. Luminescent cyclometalated iridium(III) polypyridine di-2-picolylamine complexes: synthesis, photophysics, electrochemistry, cation binding, cellular internalization, and cytotoxic activity. Inorg. Chem. 50, 8570–8579. https://doi.org/10.1021/ic201153d (2011).
Golubeva, O. Y., Alikina, Y. A. & Brazovskaya, E. Y. Particles morphology impact on cytotoxicity, hemolytic activity and sorption properties of porous aluminosilicates of kaolinite group. Nanomaterials https://doi.org/10.3390/nano12152559 (2022).
Huang, H. et al. An evaluation of blood compatibility of silver nanoparticles. Sci. Rep. 6, 25518. https://doi.org/10.1038/srep25518 (2016).
Ru, J. et al. Exploring hollow mesoporous silica nanoparticles as a nanocarrier in the delivery of foot-and-mouth disease virus-like particle vaccines. ACS Appl. Bio Mater. 7, 1064–1072. https://doi.org/10.1021/acsabm.3c01015 (2024).
Li, G. et al. A bioactive ligand-conjugated iridium(III) metal-based complex as a Keap1-Nrf2 protein-protein interaction inhibitor against acetaminophen-induced acute liver injury. Redox Biol. 48, 102129. https://doi.org/10.1016/j.redox.2021.102129 (2021).
Arias-Calderon, M. et al. Fibroblast growth factor 21 is expressed and secreted from skeletal muscle following electrical stimulation via extracellular ATP activation of the PI3K/Akt/mTOR signaling pathway. Front. Endocrinol. 14, 1059020. https://doi.org/10.3389/fendo.2023.1059020 (2023).
Zhao, S. Q., Gao, Y., Zhang, Y., Yang, X. P. & Yang, Z. cAMP/PKA/CREB signaling pathway-mediated effects of melatonin receptor genes on clock gene expression in Bactrian camel ovarian granulosa cells. Domest. Anim. Endocrinol. 76, 106609. https://doi.org/10.1016/j.domaniend.2021.106609 (2021).
Acknowledgements
This research was funded by the Longyuan Youth Talent Project (LYYC-2024-01), the National Natural Science Foundation of China (32060783, 03122071), and the Scientific Startup Foundation for Doctors of Gansu Agricultural University (GAU-KYQD-2021-18).
Author information
Authors and Affiliations
Contributions
LZ, WJ, RM and DY performed the experiments; JL and YN analyzed data; SZ and YG provide essential reagents and materials; SD and YG supervised the study; JR and YG designed the experiments and wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
The animal use protocol listed in this study was reviewed and approved by the Animal Ethical and Welfare Committee of Gansu Agricultural University (Approval No. GAU-Eth-LST-2021-004). This study is reported in accordance with ARRIVE guidelines.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zhang, L., Du, S., Ji, W. et al. Development of an iridium complex fluorescent probe for FGF21 protein labeling and tracking. Sci Rep 15, 12365 (2025). https://doi.org/10.1038/s41598-025-97470-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-025-97470-w