Abstract
RNA-mediated toxicity, which can be controlled by alteration of gene expression, is considered a key event in Amyotrophic Lateral Sclerosis (ALS). Transcriptomic deregulation of miRNAs expression can spread via “horizontal” RNA transfer through extracellular vesicles (EVs) to act in conjunction with proteins, leading to changes in mRNA, which can provide early signals to indicate forthcoming neuropathological changes in the brain. The aim of this work is to compare expression profiles (obtained by miRNA-seq) from different tissues to highlight commonly expressed and tissue-specific miRNAs. miRNA species from plasma EVs were correlated with miRNA profiles obtained from peripheral blood mononuclear cells (PBMCs). Each tissue from ALS patients was compared to controls, revealing 159 deregulated (DE) miRNAs in Exosomes (EXOs), 247 DE miRNAs in PBMCs and 162 DE miRNAs in Microvesicles (MVs). Next, data were filtered to include only miRNAs expressed in disease samples (not in healthy subjects), to reduce the number of tissue- and ALS- specific miRNAs (EXO n = 22, MV = 11, PBMCs n = 8). We identified specific miRNAs and pathways related to each tissue. Interestingly, in PBMCs we found mainly neuro-linked pathways, such as neurotransmitters, brain and neuron development, while in EXOs, we found miRNAs implicated in MAPK and ERB signaling. In contrast, the altered pathways in MVs were not specific. This study shows that the composition of small RNA differs significantly between blood cells and its respective EVs fraction. Differentially expressed miRNAs can target definite transcripts in different cellular and molecular fractions. It is evident that, in terms of miRNAs cargo, MVs are not specific to ALS. Therefore, future studies will focus on the interaction between cells and EXOs.
Similar content being viewed by others
Introduction
RNA-mediated toxicity, which could be controlled by alteration of gene expression, is considered a key event in Amyotrophic Lateral Sclerosis (ALS). ALS is a motor neuron (MN) disease in which upper motor neurons of the brain motor cortex and lower motor neurons of the bulbar region and the anterior horn of the spinal cord are primarily, but not exclusively, affected1.
Emerging evidence suggests dysregulation of miRNAs expression profiles in ALS patients, as well as in cellular and animal models of the disease. Altered expression levels of specific miRNAs have been implicated in the modulation of key pathways involved in ALS pathogenesis, such as oxidative stress, excitotoxicity, protein aggregation, and neuroinflammation. Furthermore, several miRNAs have been identified as potential biomarkers for disease diagnosis, prognosis, and therapeutic response assessment.
Recently, different massive transcriptome profiles have been published from different tissues such as peripheral blood mononuclear cells (PBMCs) of ALS patients and matched controls2,3,4,5,6showing an important deregulation of expressed genes, both coding and non-coding RNAs, lncRNAs and miRNAs7.
Moreover, also other cellular components, such as extracellular vesicles (EVs), have been investigated for RNA cargo.
Microvesicles (MVs) are vesicles of 100–1000 nm, shed by budding of the plasma membrane of cells, both in homeostasis, but particularly during pathological conditions; Exosomes (EXOs) instead are vesicles of 30–150 nm, which formed intracellularly and released from exocytosis of multivesicular bodies8,9. EVs can transport RNA molecules that may contribute to ALS pathology. In fact, RNA carried by EVs can significantly influence cellular behavior, particularly in the nervous system. In ALS, the different aspects of EVs and their RNA content have been investigated such as propagation of pathological RNA and for biomarkers. About ALS, different studies have been published about miRNAs cargo showing that these class of RNA may be both tissue- and pathology-specific10,11,12. Moreover, it is possible that miRNA expression can spread via “horizontal” RNA transfer through EVs to act in conjunction with proteins, leading to changes in mRNA, which can provide early signals to indicate forthcoming neuropathological changes in the brain.
This study aims to investigate the miRNA landscape within EVs, and PBMCs of ALS patients and controls. Our primary objectives are to identify miRNAs that exhibit disease-specific signatures and discern their tissue specificity. Subsequently, we endeavor to unravel the gene targets of these identified miRNAs, shedding light on the molecular pathways contributing to ALS pathogenesis. The investigation into EVs and PBMCs serves as a strategic approach, taking advantage of the systemic nature of ALS pathology and the potential of these compartments to reflect disease-related alterations. Through advanced miRNA profiling techniques, we aim to pinpoint specific miRNAs that may serve as robust biomarkers, distinguishing ALS patients from healthy controls and shedding light on the underlying molecular events.
This study strives to contribute to the growing body of knowledge surrounding ALS by providing a comprehensive analysis of miRNA profiles in EVs and PBMCs. The integration of bioinformatics and experimental approaches aims to unravel the intricate web of miRNA-gene interactions.
Materials and methods
Study subjects
Recruitment resulted in 26 ALS patients and 11 age- and sex-matched healthy controls (Table 1, Table S1)). All subjects provided written informed consent (Protocol n° 20180034329). Only subjects not affected by any neurological condition or other relevant comorbidities were selected as “healthy controls”. The IRCCS Mondino Foundation (Pavia, Italy) conducted clinical and neurological checkups of ALS patients, who were diagnosed with ALS as defined by the El Escorial criteria. In addition, genetic screening was conducted to exclude patients carrying mutations in FUS, TARDBP, SOD1, VCP, ANG and C9orf72 genes. The control subjects were recruited at the Transfusional Service and Centre of Transplantation Immunology, IRCCS Foundation San Matteo (Pavia, Italy). The Ethical Committee of the IRCCS Mondino Foundation (Pavia, Italy) approved the study protocol to obtain peripheral blood from patients and controls. All the experiments were performed in accordance with relevant guidelines and regulations.
PBMCs and EVs isolation
Peripheral blood of each individual, at the time of diagnosis for sALS patients, was processed for isolation of EVs and PBMCs. For PBMCs preparation, peripheral blood was layered (density = 1.077) with Ficoll-Histopaque (Sigma-Aldrich) and centrifuged at 1800 rpm for 30 min. Cells used for RNA extraction were previously selected after assessing viability through trypan blue exclusion test. For EVs venous blood was collected in sodium citrate tubes from all patients and controls. Within 1 h it was centrifuged at 1,000 RCF for 15 min to separate plasma, followed by an additional centrifugation at 1,600 RCF for 20 min to remove platelets. Platelet-free plasma was then transferred to a new tube and snap frozen at −80°C. Prior to the analysis, platelet-free plasma was thawed on ice and it was centrifuged at 20,000 RCF for 1 h with Centrifuge 5427 R (Eppendorf). The pellet was washed with 0.22 μm filtered PBS and centrifuged for 1 h at 20,000 RCF. The pellet was then processed for MVs analysis. The supernatant of MVs was filtered through a 0.2 μm filter and spun in an Optima MAX-TL Ultracentrifuge at 100,000 RCF for 1 h at 4 °C. After ultracentrifugation, the supernatant was removed and the EXOs pellet was washed with 1 mL of filtered PBS at 100,000 RCF for 1 h at 4 °C8. The obtained EXOs pellet was processed for analysis.
Nanoparticle-tracking analysis (NTA) of MVs and exos
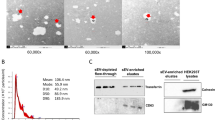
ALS patients and CTRLs EVs samples were analyzed by nanoparticle-tracking analysis (NTA) using a NS300 instrument (NanoSight, Amesbury, UK) in order to detect size and concentration of MVs and EXOs. For a more accurate detection, MVs and EXOs samples were diluted with PBS to an optimal concentration. After dilution, 1 ml of diluted sample was loaded on the machine and read in a rate of about 30 frames/sec. Particle movement videos were recorded 3 times per test and dimension and mean concentration were analysed by the NTA software (version 2.2, NanoSight). The results of NTA were presented as the mean of the 3 tests (Fig. S1).
RNA extraction
Total RNA have been extracted from PBMCs using TRIzol™ reagent (ThermoFisher Scientific), while for EVs (both EXOs and MVS) miRNeasy Serum/Plasma kit (QIAGEN) have been employed. RNA quantification and quality evaluation have been carried out using a Nanodrop ND-100 Spectrophotometer (Nanodrop Technologies) and a Tape Station 4200 (Agilent) respectively.
RNA-Seq library Preparation and bioinformatic data analysis
Libraries for miRNA sequencing have been prepared using the Small RNA-Seq Library Prep kit (Lexogen) according to the user manual. Sequencing steps have been carried out on the Illumina NextSeq 500. FastQ files were generated via Illumina bcl2fastq2 (Version 2.17.1.14 http://support.illumina.com/downloads/bcl2fastq-conversion-software-v217.html).
Quantification of miRNAs was done as described in the literature13. The workflow, including quality control filter, trimming of adapters, and reads mapping against miRNA precursors, is implemented in Docker4Seq package13,14.
Once the abundance of each miRNA was calculated and counts were available, all tissue samples were filtered, removing miRNAs with zero counts in more than 2/3 of the samples in the same tissue. Given the variability among these, different thresholds were applied for different tissues (t1 for EXO and MV, t2 for PBMCs). Based on the number of counts, different miRNA classes were defined:
-
Expressed miRNA: a miRNA for which reads count is above the threshold in at least one sample per condition (or tissue);
-
Tissue specific miRNA: a miRNA for which reads count is above the threshold for more than 50% of the samples from the same tissue;
-
Condition specific miRNA: a tissue-specific miRNA (as described above) that is expressed only in samples from patients affected by the same condition (“overall” if expressed in EXO, MV and PBMC, or specifically for each tissue otherwise) and absent from samples of all the other patients. MiRNAs have been mapped on miRBase hairpins using SHRiMP. Differential expression analysis for miRNAs has been performed with the R package DESeq. Transcripts have been considered differentially expressed (DE) and retained for further analysis with |log2(disease sample/healthy control)| ≥ 1 and a FDR ≤ 0.1.
Pathways enrichment and gene target analysis
Gene enrichment analysis was performed on coding genes with KEGG pathway analysis (Kyoto Encyclopedia of Genes and Genomes, http://www.genome.ad.jp/kegg) and enrichR web tool15,16,17. miRNA-targets analysis was done with miRWalk web tool (http://mirwalk.umm.uni-heidelberg.de)18. We have filtered the obtained results by selecting only targets reported on both TargetScan, miRDB and miRTarBase.
qPCR validation
The expression levels of miRNAs were assessed using Qiagen miRNA PCR Assays. Firstly, the total RNA from each sample was reverse transcribed into cDNA using the miRCURY LNA RT Kit (Cat. No:339340). Subsequently, the cDNAs were subjected to quantitative PCR analysis using the miRCURY LNA SYBR Green PCR Kit (Qiagen, USA, Cat. No: 339347) on a CFX 96 Real-Time PCR device (Bio-Rad Laboratories, USA). The primers used in the study are available upon request. miRNA expression was normalized against one housekeeping miRNAs (miR-16). Statistical analyses were performed using one-way ANOVA (Kruskal-Wallis) using Prism GraphPad v9 software (GraphPad Software). P-values (two tailed) with 95% confidence intervals was computed, and p < 0.05 was considered statistically significant.
Results
MiRNA enrichments in PBMCs, EVs of ALS patients and matched controls
In this work, we analyzed miRNA cargo in PBMCs and plasmatic EXOs and MVs from ALS patients and matched controls. The goal was to compare expression profiles from different tissues such as PBMCs, EXOs and MVs, in order to highlight commonly expressed miRNAs and perform functional enrichment analysis.
Differential expression analysis in EXO, MV and PBMCs of sALS patients and Healthy Controls. (A) Heatmap for top abundant miRNAs and (B) Principal component analysis (PCA) for expressed miRNAs in all tissues. The heatmap shows a clear clustering based on tissue type and differences between ALS patients and controls. The PCA plot further emphasizes these distinctions with PBMC samples forming a separate cluster along PC1, while EXO and MV show partial overlap. Notably, ALS samples shift within their respective clusters, particularly along PC2, indicating disease-specific miRNA regulation.
Starting from a total of 298 distinct miRNAs for all tissue types, in EXOs 287 miRNAs have been found deregulated, 88 are up-regulated while 199 are down-regulated. About MVs, the highest number of deregulated miRNAs has been detected (398), 183 are up-regulated while 215 are down-regulated. In PBMCs only 20 miRNAs have been found altered, mainly up-regulated (18/20) (Table 2 and Table S2). The lower dysregulation observed in PBMCs may be explained by their stability as whole cells, which are less influenced by transient changes compared to EVs, such as EXOs and MVs. EVs are constantly changing and act like messengers, carrying information about the state of the cells they come from. This could explain why the miRNA levels in EVs vary more compared to other samples. Despite the adoption of stringent measures to reduce noise in the data (miRNA filtering and different thresholds for each tissue type), the dynamic nature of EVs can still inherently bring noise in terms of the differences analyzed.
Interestingly, let-7b-5p and miRNA 10a-5p are expressed in a common way in all analyzed tissues.
Overlap and differences of MiRNAs among the three tissues
In order to understand common miRNAs of the six groups, we calculated the intersection of expressed miRNAs with http://bioinformatics.psb.ugent.be/webtools/Venn/.
We observed that 135 miRNAs were in common between PBMCs from patients and controls and MVs, 124 were in common between EXOs and MVs from patients and healthy controls and 121 between PBMCs and EXOs in both conditions (Fig. 2a, b and c). miRNAs are listed in supplementary material (Table S3).
Identification of Tissue-specific MiRNAs for ALS patients and matched controls
Next, we analyzed miRNA data to identify miRNAs that were specific for tissues and, in each tissues, for ALS disease.
All miRNAs were classified with respect to the classes described above (“expressed miRNA”, “tissue specific miRNA”, and “condition specific miRNA”), resulting in a generic expression table. The expression of a miRNA, therefore, is strictly linked to the rules and thresholds imposed on read counts during the analysis step. In this case, due to the variability of the tissues and the different number of samples per tissue, t1 = 5 and t2 = 50 were used as thresholds. These values were empirically determined by evaluating the expressed miRNAs in all classes for each threshold pair, considering an increment of 1 and an increment of 10 for t1 and t2 respectively (from t1 = 1 and t2 = 10 to t1 = 5 and t2 = 50). Once the thresholds were chosen and the expression table generated, we focused on all tissue-specific miRNAs for ALS samples, thus identifying all overexpressed or underexpressed miRNAs.
In fact, Fig. 3 showed that 247 miRNAs were detectable only in PBMCs, 162 RNAs were specific for EXOs while 159 were expressed only in MVs (Fig. 3, Table S4). From these data, we selected miRNAs that were expressed only in ALS samples to identify miRNAs disease specific and, for these three small groups, we performed pathways analysis.
In PBMCs, we identified 11 miRNAs that were expressed exclusively in ALS patients, 22 expressed only in ALS EXOs and 8 specific for MVs from ALS patients (Fig. 3). Interestingly, EXOs showed the greatest number of specific miRNAs, suggesting the importance of miRNAs cargo in EXOs in ALS patients.
Pathway analysis for miRNAs MVs-specific (Fig. 4) showed an important involvement of synaptic vesicle principally related to miR-4284.
Heatmap map calculator of pathways for selected miRNAs MVs-specific, selection is based on evidence “experimental strong” (https://mpd.bioinf.uni-sb.de/heatmap_calculator.html?organism=hsa), so the numer of rapresented miRNA is lower than reported in Fig. 3. On Y axes are reported miRNAs while in X axes are reported associated pathways.
In PBMCs, miR-331 and miR-4284 found DE have already been reported as altered in ALS patients19,20. For these three miRNAs, we investigated their possible targets (Fig. 5). Regarding miR-4284, the most interesting target genes are CXCL5 and TAF8, due to their involvement in ALS mechanisms21,22. Also miR-664a-3 has been found altered in ALS patients and has already been associate with ALS; its main target described is PPM1 A, BEND4 and ZBTB34 genes. Finally, we investigated miR-331-3p targets and identified 9 validated target genes (Fig. 5).
In EXOs, 22 miRNAs specific for this tissue and for ALS disease have been identified. Based on the literature, numerous targets of these miRNAs have not been described; in fact, only 8 miRNAs have been included in pathway analysis (Fig. 6).
Heatmap map calculator of pathways for selected miRNAs EVs-specific, selection is based on evidence “experimental strong” (https://mpd.bioinf.uni-sb.de/heatmap_calculator.html?organism=hsa). On Y axes are reported miRNAs while in X axes are reported associated pathways.
Regarding this group, mir-133-3p has been associated with degeneration and atrophy, while miR-320c and miR-449ahave been related to muscle differentiation and have already been found altered in ALS23.
Also dysregulation of miR-145-5p/3pseems to be a common feature of ALS pathology, especially in relation to TARDBP gene regulation24.
Regarding gene target searching, for miR-133a-3p 9 genes have been identified, such as Cellular Communication Network (CCN) genes (CCN1 and 2), and transcription factor Sp1 (Fig. 7A). About miR-320c, 13 genes have been identified as possible targets, such as ATG7 and ARF1 related to autophagy (Fig. 7B). More targets for miR-449a have been found (Fig. 7C), such as sirtuin and inflammatory genes like ADAM10 and transcription factors LEF1 and TSN.
In PBMCs, only 6 miRNAs were included in pathway analysis, underlining the involvement in neurological pathologies such as Huntington’s disease and pathways involving inflammation and cell cycle regulation (Fig. 8). About coding target analysis, miR-206 and miR-3204-5p were investigated (Fig. 9). For miR-206, it is known as “muscular“ miRNA25 and its targets are Syntaxin 6 and Transcriptor factors as NFAT and TBX3 (Fig. 9A). For miR-204-5p, different genes associated with immune response, such as cyclin and BDNF, have been found (Fig. 9B).
Heatmap map calculator of pathways for selected miRNAs PBMCs-specific, selection is based on evidence “experimental strong” (https://mpd.bioinf.uni-sb.de/heatmap_calculator.html?_organism=hsa). On Y axes are reported miRNAs while in X axes are reported associated pathways.
Validation microRNA-target interactions
Next, we have confirmed miRNAs data by qPCR. Validated miRNA have been selected based on relation to association with gene target and ALS disease.
About PBMC we have confirmed the up-regulation of miR-206 and we have studied the expression of Syntaxin 6 (STX6) as miRNAs targets. STX6 is part of SNARE complex, promoting movement of transport vesicles to target membranes (genecards.org). Syntaxin 6 has been found down-regulated but this difference is not significant (Fig. 10A and B). We validated the expression of miRNA miR-449a and its target ADAM10 in another cohort of ALS patients by qPCR also in EXO. This gene encodes and ADAM family member that cleaves many proteins including TNF-alpha and E-cadherin (genecards.org). It has been already related to other neurodegenerative disorders like Alzheimer Disease. We confirmed the altered expression between ALS patients and healthy subjects and the opposite trend between miRNA and gene target (Fig. 10C and D). For MV non more samples were available.
Discussion
In this study, we compared expression profiles from three different tissues of ALS patients and healthy controls to identify both commonly expressed and primarily ALS-specific miRNAs as possible biomarkers and therapeutic targets. The significance and strength of our work, lies in the possibility of identifying disease-specific biomarkers that are unique to the disease condition and that are detectable in a specific, single peripheral tissue, ensuring greater reliability of the biomarker itself. From a biological point of view, the possibility of having a deregulated miRNA panel available, which can be considered a body-fluids biomarkers of ALS pathology, represents a great challenge and opportunity. miRNAs, as well as mRNAs, can constitute a real molecular signature that is easily detectable in different patients. The next step of this study will be to evaluate the clustering of the deregulated miRNAs emerged, based on the clinical phenotypic subgroup recognizable among sporadic ALS patients. Indeed, it is known that sporadic ALS patients can be subdivided on the basis of clinical criteria26into different cluster that manifest variable site of onset, severity, and degeneration rate. Beside the purely clinical subdivision, we described in our recent work27, a specific molecular signature given by the different gene expression profiles detectable in the various phenotypes of sporadic ALS patients. No studies, based on this approach, have yet been done on miRNA deregulation profiles. Therefore, the future challenge of the work we are proposing will be to identify a molecular signature based on miRNA deregulation that is specific to the different phenotypic groups of sporadic ALS patients. The combined study of the two molecular profiles, both coding genes and miRNAs, will laid the groundwork for the inclusion of ALS patients in future clinical trials that consider phenotypic biomarkers in addition to clinical parameters, ensuring the best patient response and the most appropriate assignment of the patient into a specific therapeutic procedure.
Initially, we measured the abundance of miRNAs in each tissue, and we found that a higher amount of miRNAs has been sequenced in PBMCs. The higher amount of RNA in PBMCs compared to EVs is likely due to the fact that miRNAs are minor constituents of EVs, as previously described28. We then compared the expression profiles in each tissue between ALS patients and controls, finding that EVs were most affected by changes in miRNA expression, despite having lower miRNA amounts. This suggests that the differentially expressed miRNAs in EVs may be consistent data.
Next, we filtered for tissue-specific miRNAs expressed only in ALS samples. Regarding ALS-MVs miRNAs, we identified eight miRNAs, three of which were already associated with ALS, including hsa-miR-4284, hsa-miR-331-3p and hsa-miR-664a-3p. About hsa-miR-4284 no data are reported but it has numerous targets as TAF8 and SLC, RNA binding protein and Transporters Protein, already associated to neurodegenerative disease29,30,−31. Low expression of miR-331-3p in serum correlated with more rapid disease progression and in the extracellular matrix19. Pathway analysis supported this finding, showing that cell matrix regulation is a pathway linked to miR-331. About gene target, mitochondrial glutamyl-tRNA synthetase, EARS2 has been identified. Mutation in EARS gene has been associated to different neurological conditions (such as Leigh syndrome)32. About ALS, alteration of genes associated to RNA metabolisms, as EARS2, has been reported in C9orf72 model and in neuron33,34. Another gene target of miR-331 is FADS6 (Fatty acid desaturase domain family, member 6) that has been found altered in SODG93 A mice35.
Additionally, hsa-miR-664a-3p has been described in relation to ALS, with patients showing up-regulation of this miRNA exhibiting a longer disease course. Our analysis of PBMCs from ALS patients, which had a shorter disease course, aligns with the down-regulation of miR-664a-3p. Pathway analysis also indicated involvement in neuronal differentiation, consistent with a study demonstrating that 664a-5p promotes neuronal differentiation in neuronal cell models36. Furthermore, miR-664a-3p targets SEPT cytoskeletal GTP-binding proteins, extensively associated with neurodegeneration. Additionally, miR-664a-3p targets PPM1 A, which may play a role in neurodegenerative diseases. In fact, PPM1 A dephosphorylate AMPK and inactivate the AMPK pathway that, in ALS, is activated in abnormal way35. Also it has been found that AMPK pathway activation delayed disease progression of ALS in SOD1 mice37, 38.
Next, we examined the miRNA ALS signature in EXOs, identifying 22 miRNAs, with some already linked to ALS, notably hsa-miR-133a-3p, which has been widely reported in ALS due to its relation to muscle differentiation39. The expression profile of some miRNAs has been described in the brain or blood of some neurodegenerative diseases; for example, miR-133b is deficiently expressed in the midbrain of Parkinson’s disease patients and in animal models, and its expression in exosomes is lower compared to controls. Similarly, hsa-miR-449a has been associated with muscle differentiation and proliferation23. Moreover, one of its target is ADAM10, already associated to neurodegenerative diseases, in particular Alzheimer’s disease while not a lot is known about ALS36,40,41. About hsa-miR-320, it is already described altered in ALS patients19as possible serum biomarkers. In terms of pathway involvement, Axon guidance, ErbB, Rap1, and MAPK signaling pathways are most relevant, as previously demonstrated10. Notably, a mutation in ErbB4 has been linked to ALS42, and alterations in the ErbB pathway have been associated with ALS and FTD43.
We also analyzed PBMCs miRNA cargo specific to ALS, finding deregulation of two miRNAs already known, hsa-miR-206 and hsa-miR-320b, both identified as modulators of skeletal muscle development25,39,44. MiR-206 is the most extensively described miRNA associated with ALS. It has been found to be deregulated in ALS patients and strongly linked to muscle regeneration10,45,46,47,48. About miR-206 target, Senataxin has been confirmed to be altered in ALS compared to controls. This protein is part of SNARE complex and its inhibition or defect may be associated with neurodegeneration49. Among the involved pathways, the PI3 K-Akt pathway, which is involved in intracellular signal transduction, is associated with metabolism, proliferation, cell survival, growth, and angiogenesis in response to extracellular signals.
This study demonstrates that ALS may have a specific signature in different blood components, allowing detection of miRNAs that show disease-specific characteristics and tissue specificity. In PBMCs and exosomes, we observed significant changes in miRNAs related to muscle development, while in MVs, the main altered miRNAs are linked to the extracellular matrix. Our data suggest a more specific signature related to pathways and miRNA targets in exosomes and PBMCs. Thus, we propose that miRNA levels may serve as a starting point for identifying a specific signature for ALS, potentially leading to a future study focused on a small group of miRNAs that could serve as peripheral biomarkers for ALS. Further research is needed with larger cohorts at different stages of the disease to understand whether the deregulation of these RNAs correlates with specific clinical windows (e.g., onset, progression, or terminal stages).
Limitations of the study
The study we are proposing includes a rather homogenous cohort of sALS patients although the rarity of the condition limited recruitment and sample size. It should also be noted that from the same starting sample, material was extracted for both NTA, miRNA-seq analysis and validation by RT-qPCR. This certainly limited the possibility of confirming our data with different techniques other than those proposed, or augmenting the studies performed on this cohort of ALS patients.
Conclusions
In conclusion, our data demonstrate significant differences’ in the composition of small RNA between blood cells and its respective EVs fraction. Differentially expressed miRNAs target specific transcripts across different cellular and molecular fractions. Notably, MVs do not exhibit ALS-specific miRNA cargo, while the miRNA cargo in EXOs and the interaction with PBMCs present interesting points for further investigation in ALS research.
Data availability
The RNA-sequencing datasets for this manuscript are publicly available, as they are linked to the GEO repository (GSE155700, GSE295437).
Change history
28 May 2025
The original online version of this Article was revised: In the original version of this Article, the author names were incorrectly indexed. The original Article has been corrected.
References
Hardiman, O. et al. Amyotrophic lateral sclerosis. Nat. Rev. Dis. Prim. 3, 17071. https://doi.org/10.1038/nrdp.2017.71 (2017).
D’Erchia, A. M. et al. Massive transcriptome sequencing of human spinal cord tissues provides new insights into motor neuron degeneration in ALS. Sci. Rep. 7. https://doi.org/10.1038/s41598-017-10488-7 (2017).
Zhao, W. et al. Characterization of gene expression phenotype in amyotrophic lateral sclerosis monocytes. JAMA Neurol. 74, 677–685. https://doi.org/10.1001/jamaneurol.2017.0357 (2017).
Gagliardi, S. et al. Long non-coding and coding RNAs characterization in peripheral blood mononuclear cells and spinal cord from amyotrophic lateral sclerosis patients. Sci. Rep. 8. https://doi.org/10.1038/s41598-018-20679-5 (2018).
Zucca, S. et al. RNA-Seq profiling in peripheral blood mononuclear cells of amyotrophic lateral sclerosis patients and controls. Sci. Data. 6, 190006. https://doi.org/10.1038/sdata.2019.6 (2019).
Garofalo, M. et al. RNA molecular signature profiling in PBMCs of sporadic ALS patients: HSP70 overexpression is associated with nuclear SOD1. Cells 11 https://doi.org/10.3390/cells11020293 (2022).
Gomes, B. C. et al. Differential expression of MiRNAs in amyotrophic lateral sclerosis patients. Mol. Neurobiol. 60 (12), 7104–7117. https://doi.org/10.1007/s12035-023-03520-7 (2023).
Théry, C. et al. A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles. 7, 1535750. https://doi.org/10.1080/20013078.2018.1535750 (2018). Minimal information for studies of extracellular vesicles 2018 (MISEV2018.
Cocucci, E. & Meldolesi, J. Ectosomes and exossomes: shedding the confusion between extracellular vesicles. Trends Cell. Biol. 25, 364–372. https://doi.org/10.1016/j.tcb.2015.01.004 (2015).
Sproviero, D. et al. Different MiRNA profiles in plasma derived small and large extracellular vesicles from patients with neurodegenerative diseases. Int. J. Mol. Sci. (2021) Mar. 8, 22(5):2737. https://doi.org/10.3390/ijms22052737
Sproviero, D. et al. Extracellular vesicles derived from plasma of patients with neurodegenerative disease have common transcriptomic profiling. Front. Aging Neurosci. (2022) Feb 16, 14:785741. https://doi.org/10.3389/fnagi.2022.785741
Kim, J. A. et al. Small RNA sequencing of Circulating small extracellular vesicles MicroRNAs in patients with amyotrophic lateral sclerosis. Sci. Rep. (2023) Apr 4, 13(1):5528. https://doi.org/10.1038/s41598-023-32717-y
Cordero, F. et al. Optimizing a massive parallel sequencing workflow for quantitative MiRNA expression analysis. PLoS One. 7 (2), e31630. https://doi.org/10.1371/journal.pone.0031630 (2012).
Kulkarni, N. et al. Reproducible bioinformatics project: a community for reproducible bioinformatics analysis pipelines. BMC Bioinform. Oct 15 (Suppl 10), 349. https://doi.org/10.1186/s12859-018-2296-x (2018).
Mazin, A. V. et al. Rad54, the motor of homologous recombination, DNA repair (Amst). Mar 2, 9(3):286–302. (2010). https://doi.org/10.1016/j.dnarep.2009.12.006
Kuleshov, M. V. S. et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 44 (1), 90–97. https://doi.org/10.1093/nar/gkw377 (2016).
Bi, F. et al. Reactive astrocytes secrete lcn2 to promote neuron death. Proc. Natl. Acad. Sci. U S A. Mar 5 (10), 4069–4074. https://doi.org/10.1073/pnas.1218497110 (2013).
Sticht, C. et al. MiRWalk: an online resource for prediction of MicroRNA binding sites. PLoS One. Oct 18 (10). https://doi.org/10.1371/journal.pone.0206239 (2018).
Raheja, R. et al. Correlating serum Micrornas and clinical parameters in amyotrophic lateral sclerosis. Muscle Nerve. 58 (2), 261–269. https://doi.org/10.1002/mus.26106 (2018).
Afonso, G. J. M., Cavaleiro, C., Valero, J. & Mota, S. I. Ferreiro. Recent advances in extracellular vesicles in amyotrophic lateral sclerosis and emergent perspectives. Cells 12 (13), J1. https://doi.org/10.3390/cells12131763 (2023).
Perner, C. et al. Dysregulation of chemokine receptor expression and function in leukocytes from ALS patients. J. Neuroinflammation. Mar 28 (1), 99. https://doi.org/10.1186/s12974-018-1135-3 (2018).
Wong, K. M. et al. Mutations in TAF8 cause a neurodegenerative disorder. Brain (2022) Sep. 14, 145(9):3022–3034. https://doi.org/10.1093/brain/awac154
Pegoraro, V., Merico, A. & Angelini, C. Micro-RNAs in ALS muscle: differences in gender, age at onset and disease duration. J. Neurol. Sci. 380, 58–63. https://doi.org/10.1016/j.jns.2017.07.008 (2017). Sep 15.
Freischmidt, A. et al. Systemic dysregulation of TDP-43 binding MicroRNAs in amyotrophic lateral sclerosis. Acta Neuropathol. Commun. 1 (42). https://doi.org/10.1186/2051-5960-1-42 (2013). Jul 30.
Ma, G. et al. MiR-206, a key modulator of skeletal muscle development and disease. Int. J. Biol. Sci. Feb 5 (3), 345–352. https://doi.org/10.7150/ijbs.10921 (2015).
Chiò, A. et al. Phenotypic heterogeneity of amyotrophic lateral sclerosis: a population based study. J. Neurol. Neurosurg. Psychiatry. https://doi.org/10.1136/jnnp.2010.235952 (2011).
Dragoni, F. et al. Whole transcriptome analysis of unmutated sporadic ALS patients’ peripheral blood reveals phenotype-specific gene expression signature. Neurobiol. Dis. 206, 106823. https://doi.org/10.1016/j.nbd.2025.106823 (2025).
Albanese, M. et al. Hammerschmidt. MicroRNAs are minor constituents of extracellular vesicles that are rarely delivered to target cells. PLoS Genet. 17 (12), e1009951. https://doi.org/10.1371/journal.pgen.1009951 (2021).
Ayka, A. & Şehirli, A. Ö. The role of the SLC transporters protein in the neurodegenerative disorders. Clin. Psychopharmacol. Neurosci. (2020) May 31, 18(2):174–187. https://doi.org/10.9758/cpn.2020.18.2.174
Xue, Y. C. et al. Dysregulation of RNA-Binding proteins in amyotrophic lateral sclerosis. Front. Mol. Neurosci. (2020) May 29, 13:78. https://doi.org/10.3389/fnmol.2020.00078
Conlon, E. G., Manley, J. L. & Genes Dev. RNA-binding proteins in neurodegeneration: mechanisms in aggregate,. Aug 1, 31(15):1509–1528. (2017). https://doi.org/10.1101/gad.304055.117. Erratum in: Genes Dev. 2017;31(19):2015.
Ball, M. et al. Oct 30 [updated 2024 May 9]. In: Adam MP, Feldman J, Mirzaa GM, Pagon RA, Wallace SE, Bean LJH, Gripp KW, Amemiya A, editors. GeneReviews(R) [Internet]. Seattle (WA): University of Washington, Seattle; 1993–2024. (2003). Available from http://www.ncbi.nlm.nih.gov/books/NBK1173
Podmanicky, O. et al. Mitochondrial aminoacyl-tRNA synthetases trigger unique compensatory mechanisms in neurons. Hum. Mol. Genet. 435–447. https://doi.org/10.1093/hmg/ddad196 (2024).
Nataf, S. & Pays, L. Gene co-expression analysis unravels a link between C9orf72 and RNA metabolism in myeloid cells. Acta Neuropathol. Commun. (2015) Oct. 15, 3:64. https://doi.org/10.1186/s40478-015-0242-y
Baker, D. J. et al. Lysosomal and phagocytic activity is increased in astrocytes during disease progression in the SOD1 (G93A) mouse model of amyotrophic lateral sclerosis. Front. Cell. Neurosci. (2015) Oct. 15;9:410. https://doi.org/10.3389/fncel.2015.00410
Marcello, E., Borroni, B., Pelucchi, S., Gardoni, F. & Di Luca, M. ADAM10 as a therapeutic target for brain diseases: from developmental disorders to Alzheimer’s disease. Expert Opin Ther Targets. ;21(11):1017–1026. doi: 10.1080/14728222.2017.1386176. Epub 2017 Oct 3. PMID: 28960088. (2017).
Davies, S. P. et al. 5’-AMP inhibits dephosphorylation, as well as promoting phosphorylation, of the AMP-activated protein kinase. Studies using bacterially expressed human protein phosphatase-2 C alpha and native bovine protein phosphatase-2AC. FEBS Lett. (199) 377:421–425. https://doi.org/10.1016/0014-5793(95)01368-7
Coughlan, K. S. et al. Preconditioning with latrepirdine, an adenosine 5’-monophosphate-activated protein kinase activator, delays amyotrophic lateral sclerosis progression in SOD1(G93A) mice. Neurobiol. Aging. 36, 1140–1150. https://doi.org/10.1016/j.neurobiolaging.2014.09.022 (2015).
Foggin, S. et al. Biological significance of MicroRNA biomarkers in ALS-Innocent bystanders or disease culprits?? Front neurol. Jun 11, 10:578. https://doi.org/10.3389/fneur.2019.00578 (2019).
Sogorb-Esteve, A. et al. Levels of ADAM10 are reduced in Alzheimer’s disease CSF. J. Neuroinflammation. 15, 213. https://doi.org/10.1186/s12974-018-1255-9 (2018).
Song, F. et al. Cleavage site-directed antibodies reveal the prion protein in humans is shed by ADAM10 at Y226 and associates with misfolded protein deposits in neurodegenerative diseases. Acta Neuropathol. 148, 2. https://doi.org/10.1007/s00401-024-02763-5 (2024).
Chen, E. Y. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinform. 14, 128. https://doi.org/10.1186/1471-2105-14-128 (2013).
Ferrari, R. et al. FTD and ALS: a Tale of two diseases. Curr. Alzheimer Res. (2011) May, 8(3):273–294. https://doi.org/10.2174/156720511795563700
Przanowska, R. K. et al. miR-206 family is important for mitochondrial and muscle function, but not essential for myogenesis in vitro. FASEB J. (2020) Jun, 34(6):7687–7702. https://doi.org/10.1096/fj.201902855RR
Liscic, R. M. Als and Ftd: insights into the disease mechanisms and therapeutic targets. Eur. J. Pharmacol. 15, 817: 2–6. https://doi.org/10.1016/j.ejphar.2017.10.012 (2017).
Takahashi, Y. et al. ERBB4 mutations that disrupt the neuregulin-ErbB4 pathway cause amyotrophic lateral sclerosis type 19. Am. J. Hum. Genet. (2013) Nov 7, 93(5):900–905. https://doi.org/10.1016/j.ajhg.2013.09.008
Arbour, D., Vande Velde, C. & Robitaille, R. New perspectives on amyotrophic lateral sclerosis: the role of glial cells at the neuromuscular junction. J. Physiol. Feb 1 (3), 647–661. https://doi.org/10.1113/JP270213 (2017).
Tan, Y. et al. Downregulated miR-204 promotes skeletal muscle regeneration. Biomed. Res. Int. Nov 17, 2020 (3183296). https://doi.org/10.1155/2020/3183296 (2020).
Margiotta, A. Role of snares in neurodegenerative diseases. Cells 10 (5), 991. https://doi.org/10.3390/cells10050991 (2021). PMID: 33922505; PMCID: PMC8146804.
Acknowledgements
We would like to thank patients and their families who participated in the study without whom this work would have not been possible.
Funding
This study was funded by Ricerca Corrente 2025-2027, Italian Ministry of Health.
Author information
Authors and Affiliations
Contributions
DF: Investigation, Writing - original draft; CV: Validation; DL: Funding acquisition; Investigation; MB, ES, RB: Methodology; DGR: data curation; OP: Supervision, Writing – original draft and Writing - review & editing, SZ: Conceptualization, Supervision, Writing – original draft; GS: Supervision, Writing - original draft, review & editing, Funding acquisition.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the Helsinki declaration and its later amendments or comparable ethical standards. The study design was examined by the IRBs of the enrolling institutions. The study protocol to obtain blood from patients and healthy controls was approved by the Ethical Committee of the IRCCS Mondino Foundation (Pavia, Italy) (Code: p-20180034329). Informed consent was obtained from all subjects involved in the study.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Dragoni, F., Gerlando, R.D., Diamanti, L. et al. Cross-tissue MiRNA profiling of extracellular vesicles and PBMCs from amyotrophic lateral sclerosis patients. Sci Rep 15, 14976 (2025). https://doi.org/10.1038/s41598-025-99206-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-025-99206-2