Abstract
Although long interspersed element-1 (LINE-1) retrotransposons are poor prognostic indicators of non-small cell lung cancer, the genetic consequences of aberrant LINE-1 expression remain poorly understood. In this study, human bronchial epithelial cells were exposed chronically to 25 µM NiCl₂ to induce malignant transformation and compared to PBS-treated control cells or lung cancer cell lines. A 4 kb intronic LINE-1 insertion into the NACC2 (nucleus accumbens-associated protein 2) locus in nickel-transformed cells was associated with significant reductions in NACC2 mRNA and protein, along with constitutive increases in HDM2 and TP53α mRNAs, and aberrant expression of TP53β and TP53γ mRNAs. Steady-state levels of p53 and RB decreased, while EGFR and LINE-1 ORF1p increased. Silencing of NACC2 in TP53 wildtype NCI-H460 or TP53-null NCI-1299 cell lines replicated many of these changes, with profiles varying as a function of p53 status. Stable overexpression of HDM2 increased LINE-1 ORF1p in cancer cell lines. Human lung adenocarcinomas with wild-type TP53 showed sexually dimorphic profiles, with higher HDM2 levels in females than males, and shifts in LINE-1 ORF1p, p53, and RB that mirrored cancer cell lines. This study identifies the NACC2 locus as a target of LINE-1 retrotransposition and highlights critical interactions with the HDM2/TP53/RB regulatory axis.
Similar content being viewed by others
Introduction
Aberrant expression of human long interspersed element-1 (LINE-1) retrotransposons has been linked to poor clinical outcomes in non-small cell lung cancers (NSCLC)1. LINE-1 propagates autonomously through a copy-and-paste reverse transcription mechanism known as retrotransposition2. This process involves the transcription of both open reading frames (ORFs) from a common 5′-UTR promoter by RNA Polymerase II, followed by translation in the cytoplasm3. The major proteins encoded by LINE-1, ORF1p, and ORF2p, exhibit a predominant cis-preference (major) but can also act in trans (minor), binding mRNAs to form ribonucleoprotein particles (RNPs)4. While RNPs primarily localize to the cytoplasm, they can translocate to the nucleus, where the endonuclease domain of ORF2p nicks a single strand of genomic DNA to expose a 3′-OH group. This site is used by ORF2p’s reverse transcriptase domain to prime and reverse transcribe mRNAs into complementary DNA (cDNA)5,6. This process results in full-length or truncated insertions of the newly synthesized DNA into different genomic locations, leading to genomic instability and aberrant gene expression. Most LINE-1 retrotransposition events occur in gene-poor regions of the genome7,8. LINE-1 retrotransposition contributes to genetic diversity, with previous estimates indicating that any two unrelated individuals differ by approximately 300 LINE-1 insertions9,10,11.
In somatic cells, environmental carcinogens activate LINE-1 retrotransposition12,13, resulting in early stop codons14, deletions15, aberrant splicing16, polyadenylation17, and CpG island/histone methylation18. These genetic abnormalities are prevalent in lung cancer, where carcinogens found in various forms of environmental pollution and tobacco smoke activate LINE-1 via loss of DNA methylation and epigenetic disruption of the NuRD corepressor complex19,20. In fact, LINE-1 hypomethylation has been associated with the acquisition of stem cell-like characteristics by lung epithelial cells in lung cancer patients21,22,23. In vivo and in vitro experiments have demonstrated that aberrant LINE-1 expression promotes tumorigenic phenotypes24,25,26,27.
In the present study, we evaluated LINE-1 retrotransposition and its functional consequences in human lung epithelial cells challenged with NiCl2, an occupational risk factor and established human lung carcinogen associated with chronic inflammation, epigenetic deregulation, and DNA damage28,29,30. We also investigated the disruption of regulatory control at the NACC2 (nucleus accumbens-associated protein 2) locus, along with its interactions with the human double minute 1 (HDM2, also known as MDM2) and the TP53/RB axis, and their impact on LINE-1 expression in lung cancer cells. A deeper understanding of the genetic interactions between NACC2, the HDM2/TP53/RB axis, and LINE-1 retrotransposons may lead to the development of novel therapeutic strategies tailored to specific NSCLC genotypes.
Results
LINE-1 retrotransposition in human lung epithelial cells following NiCl2 exposure
Repeated exposure of BEAS2B cells to 25 mM NiCl2 for 6 months is associated with malignant transformation, as evidenced by colony formation in soft agar and tumorigenicity in nude mice31. The morphology of BEAS2B cells cultured for six months is presented in Fig. 1a. Since most LINE-1 insertions lack 5′UTR sequences due to poor processivity of reverse transcriptase and the presence of cryptic polyadenylation sites32,33, LINE-1 DNA was amplified using primers specific to the LINE-1 5′-UTR to detect full-length copies, as well as primers directed at downstream ORF2 sequences to detect both full-length and truncated copies (Fig. 1b). The relative number of full-length copies was lower than the number of truncated copies, with higher total copy numbers seen in transformed cells compared to control cells (Fig. 1b). Matched LINE-1 reads from next-generation sequencing (NGS) were queried against the hs37d5 genome build to identify novel insertions using RetroSeq software29. A total of 444 de novo insertions were identified, with 231 found in decoy genomes (not shown). The remaining insertions were plotted in a circus plot to depict patterns, distribution, and rates of retrotransposition (Fig. 1c). LINE-1 retrotransposition was directed to intronic/intergenic regions, with no exon insertions detected in this model. Most insertions went to chromosomes 20 and 1, with no insertions identified on chromosomes 15 or Y (Fig. 1c).
a In vitro malignant transformation: BEAS-2B cells, an immortalized, non-malignant human bronchial epithelial cell line, were treated with 25 mM NiCl2 at each media change for 6 months. Photomicrographs depict the morphology of control and NiCl2-treated cells. Malignant transformation was confirmed in soft agar assays as described (31). b LINE-1 Copy Number: Full-length and truncated copies of LINE-1 DNA were quantified in control (CTR) and transformed (TrF) cells using RT-PCR. c Circos Plot depicting the pattern, distribution, and rates of LINE-1 retrotransposition: The outer frame rectangles represent the 23 human chromosomes, scaled to their actual size. Individual lines within the light purple bands beneath each chromosome represent intergenic regions where LINE-1 insertions were identified. Light green lines within the beige bands denote intronic regions with LINE-1 insertions, while dark gray dots indicate all insertions and dotted bars show the total number of intronic insertions (n = 103). The light blue and white bands represent insertions into non-coding RNAs (blue) and untranslated regions (UTRs) (white). The rates of insertions within each chromosome are illustrated by the inner red histograms. BCL2L1, GRID, TMEM8B, and NACC2 were chosen as candidate genes involved in malignant transformation. d Gene ontology (GO) analysis of genes with novel LINE-1 insertions: the pie chart displays the molecular functions of the identified genes. Up to 42% of the genes affected by LINE-1 insertions are involved in protein-binding functions.
Gene ontology (GO) enrichment analysis (p < 0.05) identified protein interactions and apoptosis as the most affected pathways in nickel-transformed lung bronchial epithelial cells (Fig. 1d). PCR amplification and sequencing using primers flanking the NACC2 intron-2 sequence confirmed the presence of a 4 kb LINE-1 insertion in transformed cells (Fig. 2a). RT-PCR amplification using primers spanning exons 2 and 3, as well as Western blotting, revealed significant decreases in NACC2 mRNA and protein levels, respectively, in transformed cells compared to controls (Fig. 2b, c). A 6 kb insertion was also detected in TMEM8B (Fig. 2d), but this was not pursued further due to the functional redundancy provided by numerous TMEM8B splice variants and their low abundance in human lung34. Intronic insertions into GRID1 and BCL2L could not be validated by PCR amplification and were not investigated further. The LINE-1 insertion into the NACC2 locus was considered significant due to the oncogenic activity of an NACC2-neurotrophic receptor tyrosine kinase fusion protein in pilocytic astrocytoma and pediatric glioblastoma35, as well as our finding in the TCGA database that NACC2 interacts with TP53 to impact lung cancer mortality (not shown).
a PCR Validation of the intron-2 LINE-1 insertion: a schematic representation of NACC2 mRNA (top) with blue arrows depicting RT-PCR primers and red arrows indicating validation primers. PCR analysis confirmed a ~4 kB insertion into intron-2 of the NACC2 gene. b RT-PCR analysis of NACC2 mRNA expression in transformed and control BEAS-2B cells: data are presented as the relative quantity of NACC2 mRNA in control cells compared to transformed cells. c Western Blot analysis of NACC2 protein levels in transformed and control cells: similar results were observed in three independent experiments. d Validation of TMEM8B insertion: A ~6 kB insertion of LINE-1 into the TMEM8B gene was confirmed via PCR. Insertions into GRID1 and BCL2L were not confirmed.
LINE-1 insertion into the NACC2 locus downregulates p53 and RB through HDM2
Ingenuity pathway analysis (IPA) identified a functional NACC2 genetic regulatory network connected to lung carcinogenesis via HDM2, TP53, RB, and EGFR (Fig. 3a). Thus, subsequent studies explored the functional impact of the NACC2 insertion on HDM2, a zinc finger nuclear phosphoprotein with E3 ubiquitin ligase activity that negatively regulates p53 via deacetylation and polyubiquitination36. Previous studies showed that NACC2 represses HDM2 by recruiting the NuRD macromolecular repressor complex37. HDM2 can switch to become a positive regulator by binding to the TP53 mRNA to promote translation, a change mediated by ATM kinase-dependent phosphorylation38. HDM2 has also been shown to degrade RB through ubiquitin-dependent and ubiquitin-independent pathways37,39,40. These findings align with our observation that HDM2 mRNA levels increased in nickel-transformed cells carrying the NACC2 insertion (Fig. 3b), while p53 and RB levels decreased in transformed cells and EGFR levels slightly increased (Fig. 3c). These data suggest that LINE-1 insertion into NACC2 disrupted regulatory control in human lung bronchial epithelial cells, leading to HDM2-mediated degradation of p53 and RB.
a IPA resolution of the NACC2 gene regulatory network: the NuRD complex (genes within the complex are indicated by blue arrows) is recruited by NACC2 to the HDM2 promoter (red arrows), leading to repression of HDM2 expression. HDACs can deacetylate RB and TP53 either directly (green arrows) or through interaction with HDM2 (brown arrow), leading to their degradation. TP53 also represses EGFR expression transcriptionally (black arrow). b RT-PCR analysis of HDM2 mRNA levels in control and transformed cells: data are presented as the logarithmic relative quantity of HDM2 mRNA in control versus transformed cells. c Genes within the NACC2 genetic regulatory network: the expression levels of TP53, RB, and EGFR were examined by Western blotting. The same membrane was probed with different antibodies to assess relative expression levels. Similar results were observed in three independent experiments.
Functional disruption of the NACC2–HDM2 axis increases TP53β and TP53γ isoforms in nickel-transformed lung epithelial cells
A prominent 48 kDa band was recognized by the anti-TP53 antibody in nickel-transformed cells, but not in controls (Fig. 3c). Given that twelve TP53 isoforms can be produced via alternative splicing, promoter activity, and translation41,42, we hypothesized that the smaller 48 kDa band represented a TP53 isoform lacking the oligomerization domain, such as TP53β, TP53γ (Fig. 4a). Acetylation was assessed using an antibody targeting lysine-382 in the oligomerization domain, revealing a marked reduction in acetylation in transformed cells compared to controls (Fig. 4b). This decrease suggests reduced TP53α resistance to HDM2-mediated degradation, supporting the hypothesis that the 48 kDa band may in fact represent TP53 isoforms lacking the oligomerization domain. Expression of the TP53β and TP53γ isoforms is regulated by the proximal TP53 promoter (P1), which undergoes alternative splicing at stop codons within intron 9. Thus, RT-PCR primers targeting exon-9 and exon-10, or exon-9 and intron-9 (stop 1/2), were used to compare the relative abundance of TP53α (wild-type), TP53β, and TP53γ mRNAs in nickel-transformed and control cells. All TP53 mRNAs were significantly increased in transformed cells, with notable shifts in the TP53α to TP53β/γ ratio (Fig. 4c). These results suggest that the 48 kDa band represents TP53β/γ isoforms, which are resistant to proteasomal degradation and accumulate in nickel-transformed cells.
a Schematic diagram of TP53 gene structure and splicing patterns: TP53β and TP53γ are produced through differential splicing of intron-9, a process regulated by two distinct stop codons. TP53β and TP53γ lack exons 10 and 11 but add 10 and 15 new amino acids, respectively, to the first 306 amino acids of P53 when splicing of the exons is inhibited. The structures of TP53α (53 kDa, wildtype), TP53β (48 kDa), and TP53γ (47 kDa) show that the β and γ isoforms lack the oligomerization domain (OD domain). b TP53 acetylation in control and transformed cells: Western blot analysis of TP53α acetylation status. Similar results were observed in three independent experiments. c RT-PCR analysis of TP53 mRNA levels in control and transformed cells: similar results were observed in three independent experiments.
HDM2 overexpression increases LINE-1 mRNA and protein levels in nickel-transformed cells and human lung adenocarcinomas
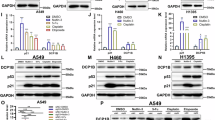
We next assessed the relative expression of LINE-1 mRNA and protein in nickel-transformed cells compared to controls. Significant increases in LINE-1 mRNA (Fig. 5a) and LINE-1 ORF1p (Fig. 5b) were observed in transformed cells. These findings are consistent with the roles of TP53 and RB as negative regulators of LINE-1 expression in human lung epithelial cells20,43. As HDM2 amplification has been reported in non-small cell lung cancers (NSCLC)44, we examined LINE-1 ORF1p, p53, and RB expression in male and female TP53 wild-type adenocarcinomas overexpressing HDM2. Sexually dimorphic profiles were found, with increased ORF1p and HDM2 and decreased p53 and RB in females compared to males (Fig. 6). These findings align with the sexually dimorphic profiles of mice carrying a synthetic human LINE-1 transgene45. Together, these results establish a functional connection between NACC2, the HDM2/TP53/RB axis, and regulatory control of LINE-1 retrotransposons.
Genetic silencing of NACC2 in NSCLC cell lines recapitulates changes in LINE-1 and the HDM2/TP53/RB axis
We next determined if the molecular changes observed in nickel-transformed lung bronchial epithelial cells were conserved in other NSCLC cells and whether TP53 status influenced these responses. NCI-H460 (TP53 wild-type) and NCI-1299 (TP53 null) cells were transfected with two distinct siRNAs targeting the NACC2 gene and compared to a control siRNA. Changes in HDM2, ORF1p, p53, RB, and EGFR expression were examined after confirming genetic silencing (Fig. 7). NACC2 knockdown reproduced many of the changes seen in nickel-transformed cells, with molecular profiles varying as a function of TP53 status (Fig. 7a, c). HDM2 and ORF1p increased in both cell lines, recapitulating the primary responses seen in nickel-transformed cells. Other genes within the network exhibited variable expression: p53 and RB increased in wild-type NCI-H460 cells, RB decreased in TP53-null NCI-1299 cells, and EGFR decreased in both (Fig. 7b, d). These findings support the concept that HDM2 modulates p53 levels depending on genetic context and TP53 mutation status, highlighting the complex interplay between HDM2, TP53, and LINE-1.
Two distinct siRNAs targeting NACC2 were used to knock down NACC2 expression in NCI-H460 cells (TP53 wild-type) and NCI-H1299 cells (TP53 mutant). qPCR was performed to confirm knockdown (panels a and c). Representative Western blots were conducted to examine protein profiles for NACC2, HDM2, ORF1p, p53, RB, and EGFR (panels b and d, respectively). Beta-actin was used as a loading control. N = 4.
Stable HDM2 overexpression confirms regulatory control of LINE-1 in NSCLC cell lines
The HDM2 sequence was inserted into a pCMV-a myc3-HDM2 plasmid via PCR. HDM2 DNA was purified and ligated into a pWPXL vector to create pWPXL-HDM2. This plasmid was co-transfected with the packaging plasmid psPAX2 and envelope plasmid pMD.2G in 293 T cells to collect pWPXL-HDM2 containing lentivirus. The latter was then used to stably overexpress HDM2 in NCI-H460 and NCI-H1299 cells (Fig. 8a). Since the pWPXL-HDM2 plasmid carries an EGFR marker, we confirmed cellular fluorescence after transfection (Fig. 8b–e). HDM2 overexpression mimicked the key changes seen after NACC2 silencing, with increased levels of ORF-1p in both NCI-H460 and NCI-1299 cells. HDM2 overexpression was associated with the appearance of multiple bands, a response that was most prominent in TP53 mutant H1299 cells, along with rebound expression of p53 in wild-type NCI-H460 cells, and decreased RB in both cell lines. Thus, induction of ORF1p in nickel-transformed lung bronchial epithelial cells was recapitulated by HDM2 overexpression in cancer cell lines.
a HDM2 overexpression was achieved by lentivirus-mediated transduction. Viral particles were collected from cells transfected with the HDM2 plasmid, while a vector plasmid lacking the HDM2 sequence was used as a control. b–e EGFR fluorescence of individual cells transfected with the pWPXL-HDM2 plasmid was monitored after lentivirus transfection. f Representative Western blots for HDM2, ORF1p, p53, and RB. Beta-actin was used as a loading control. N = 3.
Discussion
LINE-1 disrupts the structure and function of the genome through multiple mechanisms that contribute to genomic instability and oncogenesis46. This study presents evidence that repeated exposure of human lung bronchial epithelial cells to NiCl2, a known human lung carcinogen12, gave rise to clones that carried a 4-kb LINE-1 insertion in intron 2 of the NACC2 locus. This intronic insertion was associated with reduced levels of NACC2 mRNA and protein and disruption of the HDM2/TP53/RB axis. The NACC2 locus may be a preferred site for chromosomal rearrangements, as other studies have implicated an oncogenic NACC2 fusion protein that combines the BTB (Broad-complex, Tramtrack, and Bric-a-brac) domain of NACC2 with the transmembrane helix and tyrosine kinase domain of NTRK2, leading to the development of pilocytic astrocytomas and pediatric glioblastomas35. LINE-1 insertions into actively transcribed loci recruit the HUSH (human silencing hub) and MORC complexes, leading to transcriptional repression11 and induction of heterochromatin formation24,47. This epigenetic interference is consistent with NACC2 repression without the appearance of novel splice variants of this gene. Of note is that the oncogenic impact of LINE-1 and other retroelements is not limited to transcriptional silencing of target genes, but may also rely upon the unique ability of these mobile elements to reshape the architecture and function of the genome. For instance, 82 repetitive elements—including HERVs, satellite repeats, and simple repeats—were found to be differentially expressed in osteosarcomas compared to normal tissues48. Transposable elements can also exert eQTL effects that influence allele-specific gene expression directly (cis) or indirectly (trans)49. Additionally, truncated insertions of transposable elements can be associated with exonisation and splicing alterations, which contribute to oncogenesis50.
NACC2 recruits the NuRD complex to the HDM2 promoter to inhibit its oncogenic activity and repress transcription51. HDM2, in turn, regulates p53 by controlling its shuttling from the nucleus to the cytoplasm and promoting its proteasomal degradation38,44,52,53,54. HDM2 also recruits HDAC1 to mediate p53 deacetylation55,56, facilitating the ubiquitination of lysine residues and p53 degradation57. HDM2 also mediates RB protein degradation through both ubiquitin-dependent and ubiquitin-independent mechanisms37,39,40. Uchida et al. demonstrated that HDM2-mediated RB ubiquitination results in proteasomal degradation, while genetic knockdown of HDM2 stabilizes RB37. Sdek et al. showed that blocking HDM2, rather than inhibiting the proteasome, prevents RB degradation39,40. Although HDM2’s role in p53 and RB degradation is well established, other studies have shown that phosphorylation of Ser567 can trigger different regulatory dynamics, giving rise to positive regulation and increased p53 levels, as seen in our study in wild-type H460 cells58. This may explain how HDM2 shifted from negative to positive regulation upon binding to TP53 mRNA36. Multiple HDM2 bands were identified in stably transfected H460 and H1299 cells. Evans et al.59 showed that lung tumors can express several alternative splice variants carrying deletions in p53 binding domains or the absence of nuclear localization signals. One of these variants, HDM2-B, is known to be overexpressed in tumors and to interact with full-length HDM2 to inhibit its binding, leading to p53 accumulation and gain of functions60. The interplay between these two pathways likely contributes to context-specific regulation of oncogenic phenotypes, as seen in our comparative studies with TP53 wild-type and TP53 mutant cancer cell lines.
HDM2 is frequently amplified or overexpressed in a variety of sarcomas, where it drives tumorigenesis by disrupting TP53-mediated tumor suppression61,62. Beyond sarcomas, HDM2 also plays a critical role in lung cancer malignancies by dysregulating TP53 stability and function, as well as promoting aberrant expression of TP53 isoforms. TP53 has long been recognized as a critical tumor suppressor gene in lung cancer, with extensive evidence linking mutant forms of TP53, as well as its downregulation, with disruptions of apoptosis, transcriptional control, and tumor suppressive activities59. In nickel-transformed cells, HDM2-mediated degradation of TP53 may have contributed to the appearance of TP53β and TP53γ isoforms and changes in the relative abundance of isoforms with deficient activities. While complete splicing of intron 9 produces wild-type TP53 (TP53α), partial retention of intron 9 gives rise to the TP53β and TP53γ isoforms, with the oligomerization domain replaced by 10 and 15 amino acids, respectively, due to the presence of two stop codons in intron 9. Alternative translation does not likely account for the appearance of these isoforms, as one would expect isoforms that either retain the oligomerization domain or have significantly smaller molecular weights than those observed in our study.
There were certain limitations to the current study. The tumorigenicity of NiCl₂-transformed cells was not assessed in vivo, nor was the broader impact of NACC2 downregulation upon HDM2 signaling in bronchial epithelial cells. In this context, future studies should test the effects of HDM2 inhibitors such as nutilin 3a and the phosphorylation status of EGFR following disruption of the NACC2 locus in human lung bronchial epithelial cells. Nevertheless, compelling evidence is presented here that genetic silencing of NACC2 disrupts the HDM2/TP53/RB axis, with HDM2 playing a central role in the deregulation of oncogenic signaling. These findings align with the demonstration that HDM2 is overexpressed more frequently in adenocarcinomas compared to other NSCLC subtypes63,64. Loss of TP53 can induce EGFR expression, functionally mimicking EGFR-activating mutations65. Our findings suggest that tumors overexpressing HDM2 would not only lose TP53 activity, leading to induction of EGFR and increased activity, but also increased LINE-1 activity and aberrant cycles of retrotransposition that may be closely linked to the acquisition and progression of malignancy66,67,68,69. The profiles observed in NSCLC cells with different TP53 genotypes confirmed the complexity of these genetic interactions and revealed a novel role for NACC2 as a regulator of HDM2 in lung cancer. The recapitulation of key genetic alterations, namely, increased ORF1p proteins and reduced tumor suppressor levels following NACC2 knockdown or HDM2 overexpression in NCI-H460 and NCI-1299 cells, emphasizes the context-specific nature of genetic control in lung cancer and presents promising opportunities for future development of novel therapeutic interventions.
Methods
Nickel transformation of human lung epithelial cells
The human bronchial epithelial cell line BEAS-2B (NCBI Iran Cat# C561, RRID: CVCL_0168) was purchased from the American Type Culture Collection (ATCC) (Cat# CRL-3588) and cultured as described previously26. Cells were continuously exposed to either 25 µM NiCl2 or PBS at each culture media change for 6 months and maintained under identical conditions for the duration of the experiment. NiCl2-treated and control cells were assayed for soft agar malignant colony formation as described31. BEAS-2B cells are SV40-immortalized human lung bronchial epithelial cells that maintain a diploid genome in serial culture, are non-tumorigenic in nude mice, and retain functional TP53 and RB activities26.
RT-PCR, Western blotting, and LINE-1 retrotransposition
Total RNA was extracted from both transformed and control human lung epithelial cells using the RNeasy Qiagen kit. cDNA was synthesized by combining 500 ng of RNA with 1 µL of oligo-dT primers and dNTP, adjusted to 12 µL with nuclease-free H2O. After incubation at 65 °C for 5 min, 8 µL of a master mix containing Superscript Reverse Transcriptase, RNAseOUT, DTT, and 5× buffer was added to each tube. Samples were incubated at 55 °C for 1 h, and Superscript Reverse Transcriptase was inactivated at 85 °C for 5 min. All samples were treated with 1 µL of RNase H for 20 min, followed by a 1:5 dilution with nuclease-free H₂O. Five microliters of cDNA was mixed with 0.2 µL of each ORF1 or ORF2 reverse and forward primers (20 µM), 4.6 µL of nuclease-free water, and 10 µL of CyberGreen for a total of 20 µL. 18S was used as an endogenous control. LINE-1 mRNA levels were calculated using the ΔΔCT method, and statistical significance was determined by ANOVA using raw CT values followed by Tukey’s post hoc analysis. The data presented represent the average of three independent experiments. Both male and female subjects were included in the experimental design and analysis. Western blotting was performed following the extraction of total protein from nickel-transformed cells using RIPA buffer. Thirty micrograms of total lysate was used for SDS gel electrophoresis. A custom-made LINE-1 ORF1 antibody, generated by New England Peptide, was used, with specificity confirmed by peptide blockade and genetic silencing. This experiment was repeated three times, and the data shown are representative of the findings. Genomic DNA (gDNA) was extracted using the Purelink Genomic Isolation Kit (Invitrogen). gDNA concentration was measured using a Cytation3 and normalized to 7 ng of total DNA. RT-PCR amplification was performed using primers specific to LINE-1-5′ UTR to examine full-length insertions, and ORF2 to examine both full-length and truncated insertions. The retrotransposition rate was calculated using the standard curve method. The number of truncated insertions was determined by subtracting the number of full-length insertions from the total insertions. Statistical significance was determined by ANOVA followed by Tukey’s post hoc analysis. This experiment was repeated four times, with the data shown as the average of these experiments.
Next-generation sequencing (NGS), IPA analysis, and circos plot
Whole genome sequencing was conducted by Novogene (https://en.novogene.com/) following their protocol. LINE-1 insertion sites were identified using RetroSeq software70. Gene Ontology (GO) analysis was performed by Novogene Bioinformatics. Circos plots displaying LINE-1 insertion profiles were constructed using ClicO FS, an interactive web-based software71. A molecular interaction network was generated by uploading a gene carrying a LINE-1 insertion into the Ingenuity Pathway Analysis (IPA, RRID: SCR_008653) software (Qiagen, Hilden, Germany). Functional interactions between genes in the network and actionable drug targets were determined using the Ingenuity Systems Knowledge Base.
NACC2 genetic knockdown
Non-Small Cell Lung Cancer (NSCLC) cell lines NCI-H460 (ATCC Cat# HTB-177, RRID: CVCL_0459) and NCI-H1299 (ATCC Cat# CRL-5803, RRID: CVCL_0060) were purchased from the American Type Culture Collection (ATCC). NCI-H460 and NCI-H1299 cells were seeded at a density of 1 × 10⁶ cells/well and allowed to recover for 24 h. Ten microliters of Lipofectamine™ RNAiMAX transfection reagent (Invitrogen™), along with 5 µL of siRNA1 (SASI_Hs01_00073642 from Sigma-Aldrich), siRNA2 (SASI_Hs01_00073642 from Sigma-Aldrich), or control (MISSION® siRNA Universal Negative Control #1 from Sigma-Aldrich), was added to each well for 48 h. mRNA and protein were isolated and analyzed for gene expression patterns as described.
Stable transfections and protein measurements
The 293 T cell line (ATCC Cat# CRL-3216) was purchased from the American Type Culture Collection (ATCC). 293 T cells were seeded at a density of 4 × 106 cells/dish and grown to 80% confluence for 24 h. Cells were transfected using Lipofectamine 3000 with 6 µg psPAX2 (Addgene Cat# 12260, packaging plasmid), 3 µg pMD2G (Addgene Cat# 12259, envelope plasmid), and 10 µg pwpxl vector (Addgene Cat# 12257) or pwpxl-HDM2 (transfer vector). After overnight incubation with the DNA/lipid mixture, the growth media were replenished. Cells were harvested 48 h post-transfection, and viral supernatant was collected using a 0.22 µm filter. NCI-H460 and NCI-H1299 cells were seeded at 2.5 × 105 cells/well, grown to 80% confluence, and transfected with 1 mL virus and 1 mL polybrene. Plates were centrifuged at 37 °C for 90 min and incubated overnight with the virus. Stably transfected cells were harvested 48 h post-transfection and processed for protein expression analysis. Cell lysates were prepared in RIPA buffer containing protease inhibitors at a ratio of 100:1. Lysates were centrifuged at 15,400 rpm for 15 min at 4 °C, and equal amounts of protein were loaded onto 8% SDS-PAGE gels and transferred to PVDF membranes. Primary antibodies against HDM2 (Abcam Cat# ab16895, RRID: AB_2143534), LINE-1 ORF1p (Millipore Sigma, MABC1152), p53 (Abcam Cat# ab32389, RRID: AB_776981), EGFR (Abcam Cat# ab52894, RRID: AB_869579), and RB (Abcam Cat# ab181616) were used.
Immunohistochemistry (IHC)
The HDM2 antibody (SMP14) (Santa Cruz, Cat# SC-965) was optimized for IHC using lung tumor tissues positive for HDM2 expression. Human lung adenocarcinomas (n = 10) were provided by the University of Arizona Cancer Center Biorepository through TACMASR and screened for HDM2 overexpression. Tumors with HDM2 overexpression were screened for RB, P53, and LINE-1 ORF1p expression using DAB staining. Images were captured using an AxioVision inverted microscope (AxioVision Imaging System, RRID: SCR_002677) at 40× magnification. Tissue acquisition and analysis followed institutional policies.
Quantification and statistical analysis
A power analysis was performed a priori to determine the appropriate sample size for robust statistical analyses. This ensured the study had sufficient power to detect meaningful effects and minimized the likelihood of Type II errors. ImageJ (http://rsb.info.nih.gov/ij/index.html) was used to quantify signal intensities. Immuno-stained signals were extracted as greyscale, and the mean signal in areas without tissue sections was considered background. The background signal was subtracted from the mean signal in the region of interest (ROI). DAB-stained slides were converted to 8-bit greyscale images, and the darkness in the greyscale was measured as the signal in the ROI. Statistical significance was assessed using ANOVA and a two-tailed Student’s t-test. A Pearson’s test was conducted to analyze correlations between immunohistochemical signals and LINE-1, TP53, RB, and HDM2. A p-value < 0.05 was considered statistically significant. Data were shown as mean ± standard error of the mean (S.E.M.), with the number of experiments or samples indicated as n or N. Randomization was implemented during sample allocation or treatment group assignment to ensure unbiased distribution. Blinding procedures were followed during data collection and analysis to minimize potential biases. Unblinding occurred only after statistical analyses were completed.
Data availability
The data generated in this study are available within the article.
References
Rodić, N. et al. Long interspersed element-1 protein expression is a hallmark of many human cancers. Am. J. Pathol. 184, 1280–1286 (2014).
Boeke, J. D., Garfinkel, D. J., Styles, C. A. & Fink, G. R. Ty elements transpose through an RNA intermediate. Cell 40, 491–500 (1985).
Richardson, S. R. et al. The Influence of LINE-1 and SINE Retrotransposons on mammalian genomes. Microbiol. Spectr. https://doi.org/10.1128/microbiolspec.MDNA3-0061-2014 (2015).
Martin, S. L. Ribonucleoprotein particles with LINE-1 RNA in mouse embryonal carcinoma cells. Mol. Cell Biol. 11, 4804–4807 (1991).
Feng, Q., Moran, J. V., Kazazian, H. H. Jr & Boeke, J. D. Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell 87, 905–916 (1996).
Martin, S. L., Li, J. & Weisz, J. A. Deletion analysis defines distinct functional domains for protein-protein and nucleic acid interactions in the ORF1 protein of mouse LINE-1. J. Mol. Biol. 304, 11–20 (2000).
Bojang, P. Jr & Ramos, K. S. The promise and failures of epigenetic therapies for cancer treatment. Cancer Treat. Rev. 40, 153–169 (2014).
Rodić, N. et al. Retrotransposon insertions in the clonal evolution of pancreatic ductal adenocarcinoma. Nat. Med. 21, 1060–1064 (2015).
Cordaux, R. & Batzer, M. A. The impact of retrotransposons on human genome evolution. Nat. Rev. Genet. 10, 691–703 (2009).
Ewing, A. D. & Kazazian, H. H. Jr. High-throughput sequencing reveals extensive variation in human-specific L1 content in individual human genomes. Genome Res. 20, 1262–1270 (2010).
Liu, N. et al. Selective silencing of euchromatic L1s revealed by genome-wide screens for L1 regulators. Nature 553, 228–232 (2018).
El-Sawy, M. et al. Nickel stimulates L1 retrotransposition by a post-transcriptional mechanism. J. Mol. Biol. 354, 246–257 (2005).
Stribinskis, V. & Ramos, K. S. Activation of human long interspersed nuclear element 1 retrotransposition by benzo(a)pyrene, an ubiquitous environmental carcinogen. Cancer Res. 66, 2616–2620 (2006).
Schwahn, U. et al. Positional cloning of the gene for X-linked retinitis pigmentosa 2. Nat. Genet. 19, 327–332 (1998).
Narita, N. et al. Insertion of a 5’ truncated L1 element into the 3’ end of exon 44 of the dystrophin gene resulted in skipping of the exon during splicing in a case of Duchenne muscular dystrophy. J. Clin. Invest. 91, 1862–1867 (1993).
Samuelov, L., Fuchs-Telem, D., Sarig, O. & Sprecher, E. An exceptional mutational event leading to Chanarin-Dorfman syndrome in a large consanguineous family. Br. J. Dermatol. 164, 1390–1392 (2011).
Kazazian, H. H. Jr et al. Haemophilia A resulting from de novo insertion of L1 sequences represents a novel mechanism for mutation in man. Nature 332, 164–166 (1988).
Yoshida, K., Nakamura, A., Yazaki, M., Ikeda, S. & Takeda, S. Insertional mutation by transposable element, L1, in the DMD gene results in X-linked dilated cardiomyopathy. Hum. Mol. Genet. 7, 1129–1132 (1998).
Teneng, I., Montoya-Durango, D. E., Quertermous, J. L., Lacy, M. E. & Ramos, K. S. Reactivation of L1 retrotransposon by benzo(a)pyrene involves complex genetic and epigenetic regulation. Epigenetics 6, 355–367 (2011).
Bojang, P. Jr & Ramos, K. S. Epigenetic reactivation of LINE-1 retrotransposon disrupts NuRD corepressor functions and induces oncogenic transformation in human bronchial epithelial cells. Mol. Oncol. 12, 1342–1357 (2018).
Saito, K. et al. Long interspersed nuclear element 1 hypomethylation is a marker of poor prognosis in stage IA non-small cell lung cancer. Clin. Cancer Res. 16, 2418–2426 (2010).
Ikeda, K. et al. Long interspersed nucleotide element 1 hypomethylation is associated with poor prognosis of lung adenocarcinoma. Ann. Thorac. Surg. 96, 1790–1794 (2013).
Imperatori, A. et al. LINE-1 hypomethylation is associated to specific clinico-pathological features in Stage I non-small cell lung cancer. Lung Cancer 108, 83–89 (2017).
Bojang, P. Jr, Roberts, R. A., Anderton, M. J. & Ramos, K. S. Reprogramming of the HepG2 genome by long interspersed nuclear element-1. Mol. Oncol. 7, 812–825 (2013).
Apostolou, P. et al. Involvement of retrotransposon L1 in stemness and cellular plasticity. Cell Commun. Adhes. 22, 1–7 (2015).
Reyes-Reyes, E. M. et al. LINE-1 couples EMT programming with acquisition of oncogenic phenotypes in human bronchial epithelial cells. Oncotarget 8, 103828–103842 (2017). Published 2017 Oct 23.
Tristán-Ramos, P. et al. The tumor suppressor microRNA let-7 inhibits human LINE-1 retrotransposition. Nat. Commun. 11, 5712 (2020).
Lu, Y. et al. Nickel chloride promotes lung cancer invasion and metastasis by up-regulating the expression of E3 ubiquitin ligase TRIM31 through the IL-6/STAT3 signaling axis. Life Sci. 332, 122111 (2023).
Kang, Y. T., Yang, W. J., Huang, H. C., Tang, S. C. & Ko, J. L. Exposure to nickel chloride induces epigenetic modification on detoxification enzyme glutathione S-transferase M2. Environ. Toxicol. 39, 1729–1736 (2024).
Stannard, L., Doak, S. H., Doherty, A. & Jenkins, G. J. Is nickel chloride really a non-genotoxic carcinogen?. Basic Clin. Pharm. Toxicol. 121, 10–15 (2017).
Wang, L. et al. Targeting HDAC with a novel inhibitor effectively reverses paclitaxel resistance in non-small cell lung cancer via multiple mechanisms. Cell Death Dis. 7, e2063 (2016).
Lander, E. S. et al. Initial sequencing and analysis of the human genome. Nature https://doi.org/10.1038/35057062 (2001).
Larson, H. J. et al. Measuring trust in vaccination: a systematic review. Hum. Vaccin. Immunother. 14, 1599–1609 (2018).
Zhang, X. M., Wang, X. Y., Sheng, S. R., Wang, J. R. & Li, J. Expression of tumor related genes NGX6, NAG-7, BRD7 in gastric and colorectal cancer. World J. Gastroenterol. 9, 1729–1733 (2003).
Yang, W., Meyer, A. N., Jiang, Z., Jiang, X. & Donoghue, D. J. Critical domains for NACC2-NTRK2 fusion protein activation. PloS ONE 19, e0301730 (2024).
Haupt, Y., Maya, R., Kazaz, A. & Oren, M. Mdm2 promotes the rapid degradation of p53. Nature 387, 296–299 (1997).
Xuan, C. et al. RBB, a novel transcription repressor, represses the transcription of HDM2 oncogene. Oncogene 32, 3711–3721 (2013).
Uhrik, L. et al. Allosteric changes in HDM2 by the ATM phosphomimetic S395D mutation: implications on HDM2 function. Biochem. J. 476, 3401–3411 (2019).
Uchida, C. et al. Enhanced Mdm2 activity inhibits pRB function via ubiquitin-dependent degradation. EMBO J. 24, 160–169 (2005).
Sdek, P. et al. MDM2 promotes proteasome-dependent ubiquitin-independent degradation of retinoblastoma protein. Mol. Cell. 20, 699–708 (2005).
Sdek, P. et al. The central acidic domain of MDM2 is critical in inhibition of retinoblastoma-mediated suppression of E2F and cell growth. J. Biol. Chem. 279, 53317–53322 (2004).
Matlashewski, G. J. et al. Primary structure polymorphism at amino acid residue 72 of human p53. Mol. Cell Biol. 7, 961–963 (1987).
Surget, S., Khoury, M. P. & Bourdon, J. C. Uncovering the role of p53 splice variants in human malignancy: a clinical perspective. Onco Targets Ther. 7, 57–68 (2013).
Wylie, A. et al. p53 genes function to restrain mobile elements. Genes Dev. 30, 64–77 (2016).
Momand, J., Zambetti, G. P., Olson, D. C., George, D. & Levine, A. J. The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell 69, 1237–1245 (1992).
Hassanin, A. A. I., Tavera-Garcia, M., Moorthy, B., Zhou, G. D. & Ramos, K. S. Lung genotoxicity of benzo(a)pyrene in vivo involves reactivation of LINE-1 retrotransposon and early reprogramming of oncogenic regulatory networks. Am. J. Physiol. Lung Cell Mol. Physiol. 317, L816–l822 (2019).
Kazazian, H. H. Jr & Moran, J. V. Mobile DNA in health and disease. N. Engl. J. Med. 377, 361–370 (2017).
Ho, X. D. et al. Analysis of the expression of repetitive DNA elements in osteosarcoma. Front. Genet. 8, 193 (2017).
Koks, S., Pfaff, A. L., Bubb, V. J. & Quinn, J. P. Expression quantitative trait loci (eQTLs) associated with retrotransposons demonstrate their modulatory effect on the transcriptome. Int. J. Mol. Sci. 22, 6319 (2021).
Pfaff, A. L., Bubb, V. J., Quinn, J. P. & Koks, S. A genome-wide screen for the exonisation of reference SINE-VNTR-Alus and their expression in CNS tissues of individuals with amyotrophic lateral sclerosis. Int. J. Mol. Sci. 24, 11548 (2023).
Kobet, E., Zeng, X., Zhu, Y., Keller, D. & Lu, H. MDM2 inhibits p300-mediated p53 acetylation and activation by forming a ternary complex with the two proteins. Proc. Natl Acad. Sci. USA 97, 12547–12552 (2000).
Luzzatto, L. & Gartler, S. M. Switching off blocks of genes. Nature 301, 375–376 (1983).
Roth, J., Dobbelstein, M., Freedman, D. A., Shenk, T. & Levine, A. J. Nucleo-cytoplasmic shuttling of the hdm2 oncoprotein regulates the levels of the p53 protein via a pathway used by the human immunodeficiency virus rev protein. EMBO J. 17, 554–564 (1998).
Maki, C. G., Huibregtse, J. M. & Howley, P. M. In vivo ubiquitination and proteasome-mediated degradation of p53(1). Cancer Res. 56, 2649–2654 (1996).
Momand, J., Jung, D., Wilczynski, S. & Niland, J. The MDM2 gene amplification database. Nucleic Acids Res. 26, 3453–3459 (1998).
Ito, A. et al. MDM2-HDAC1-mediated deacetylation of p53 is required for its degradation. EMBO J. 21, 6236–6245 (2002).
Li, M., Luo, J., Brooks, C. L. & Gu, W. Acetylation of p53 inhibits its ubiquitination by Mdm2. J. Biol. Chem. 277, 50607–50611 (2002).
Delston, R. B., Matatall, K. A., Sun, Y., Onken, M. D. & Harbour, J. W. p38 phosphorylates Rb on Ser567 by a novel, cell cycle-independent mechanism that triggers Rb-Hdm2 interaction and apoptosis. Oncogene 30, 588–599 (2011).
Evans, S. C. et al. An alternatively splices HDM2 product increases p53 activity by inhibiting HDM2. Oncogene 20, 4041–4049 (2001).
Zheng, T. et al. Spliced MDM2 isoforms promote mutant p53 accumulation and gain-of-function in tumorigenesis. Nat. Commun. 4, 2996 (2013).
Somaiah, N. & Tap, W. MDM2-p53 in liposarcoma: The need for targeted therapies with novel mechanisms of action. Cancer Treat. Rev. 122, 102668 (2024).
Limbach, A. L. et al. The utility of MDM2 and CDK4 immunohistochemistry and MDM2 FISH in craniofacial osteosarcoma. Head. Neck Pathol. 14, 889–898 (2020).
Levine, A. J. & Oren, M. The first 30 years of p53: growing ever more complex. Nat. Rev. Cancer 9, 749–758 (2009).
Mori, S. et al. p53 apoptotic pathway molecules are frequently and simultaneously altered in nonsmall cell lung carcinoma. Cancer 100, 1673–1682 (2004).
Higashiyama, M. et al. MDM2 gene amplification and expression in non-small-cell lung cancer: immunohistochemical expression of its protein is a favourable prognostic marker in patients without p53 protein accumulation. Br. J. Cancer 75, 1302–1308 (1997).
Bheda, A., Creek, K. E. & Pirisi, L. Loss of p53 induces epidermal growth factor receptor promoter activity in normal human keratinocytes. Oncogene 27, 4315–4323 (2008).
Cruickshanks, H. A. & Tufarelli, C. Isolation of cancer-specific chimeric transcripts induced by hypomethylation of the LINE-1 antisense promoter. Genomics 94, 397–406 (2009).
Ohms, S., Lee, S. H. & Rangasamy, D. LINE-1 retrotransposons and let-7 miRNA: partners in the pathogenesis of cancer?. Front. Genet. 5, 338 (2014). Published 2014 Oct 7.
Shukla, R. et al. Endogenous retrotransposition activates oncogenic pathways in hepatocellular carcinoma. Cell 153, 101–111 (2013).
Keane, T. M., Wong, K. & Adams, D. J. RetroSeq: transposable element discovery from next-generation sequencing data. Bioinformatics 29, 389–390 (2013).
Cheong, W. H., Tan, Y. C., Yap, S. J. & Ng, K. P. ClicO FS: an interactive web-based service of Circos. Bioinformatics 31, 3685–3687 (2015).
Acknowledgements
Kenneth S. Ramos discloses support for the research of this work from the University of Arizona Health Sciences, a GURI Award from the State of Texas to KSR, a CPRIT grant RP230204 to KSR, and an NIH grant R01ES034542 to KSR.
Author information
Authors and Affiliations
Contributions
All authors contributed to the experimental design and drafting of the intellectual content and approved the final version. All authors agree to be accountable for all aspects of the work and ensure that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Bojang, P., Wang, Y. & Ramos, K.S. LINE1 insertion into the NACC2 locus disrupts the HDM2 axis and activates lung oncogenic signaling. npj Precis. Onc. 9, 255 (2025). https://doi.org/10.1038/s41698-025-01050-0
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41698-025-01050-0