Abstract
Broadly neutralizing antibodies (bNAbs) targeting the HIV-1 envelope glycoprotein (Env) have the capacity to delay viral rebound when administered to people with HIV-1 (PWH) during anti-retroviral therapy (ART) interruption. To further enhance the performance of bNAbs through their Fc effector functions, in particular NK cell-mediated killing of HIV-1 infected cells, we have produced a panel of glyco-engineered (afucosylated) bNAbs with enhanced affinity for Fc gamma receptor IIIa. These afucosylated anti-HIV-1 bNAbs enhance NK cell activation and degranulation compared to fucosylated counterparts even at low antigen density. NK cells from PWH expressing exhaustion markers PD-1 and TIGIT are activated in a similar fashion by afucosylated bNAbs as NK cell from HIV-1 negative individuals. Killing of HIV-1 infected cells is most effective with afucosylated bNAbs 2G12, N6, PGT151 and PGDM1400, whereas afucosylated PGT121 and non-neutralizing antibody A32 only induce minor NK cell-mediated killing. These data indicate that the approach angle and affinity of Abs influence the capacity to induce antibody-dependent cellular cytotoxicity. Thus, afucosylated bNAbs have the capacity to induce NK cell-mediated killing of infected cells, which warrants further investigation of afucosylated bNAb administration in vivo, aiming for reduction of the viral reservoir and ART free durable control.
Similar content being viewed by others
Introduction
Although anti-retroviral therapy (ART) effectively suppresses HIV-1 replication and prevents progression toward acquired immunodeficiency syndrome (AIDS), it is not able to clear the persistent viral reservoir and life-long adherence to ART is necessary to prevent viral rebound1,2. Therefore, alternative therapeutic strategies are required to efficiently target the viral reservoir and achieve durable control or complete eradication of the viral reservoir from people with HIV-1 (PWH)2.
Engaging natural killer (NK) cells to eliminate HIV-1 infected cells has gained attention since the level of NK cell activity was found to be inversely correlated with HIV-1 DNA levels in HIV-1 controllers and with HIV-1 DNA levels in PWH during analytical treatment interruption (ATI)3,4,5,6,7,8. Furthermore, NK cell-mediated elimination of HIV-1 infected cells has been observed in humanized mice models9,10. NK cells are part of the innate immune system and contribute to control and elimination of virally infected cells11,12. Interaction of activating receptors on NK cells, such as KIR, NKG2A, NKG2D and CD16, with ligands presented on target cells triggers degranulation of lytic granules and production of pro-inflammatory cytokines such as interferon gamma (IFNγ)11. Antibody opsonized HIV-1 infected cells are recognized by NK cells through multivalent interaction with Fc gamma receptor IIIa (FcγRIIIa = CD16a), which induces NK cell-mediated antibody-dependent cellular cytotoxicity (ADCC)11,13,14.
Broadly neutralizing antibodies (bNAbs) targeting the HIV-1 envelope glycoprotein (Env) trimer were found to delay viral rebound in ATI studies and have the capacity to induce CD8-mediated control when administered at the start of ART7,15,16,17,18,19. Fc-mediated effector functions contribute to the efficacy of bNAbs20 in vivo and have the potential to eliminate latently infected cells and reduce the size of the viral reservoir through ADCC, complement dependent cytotoxicity (CDC) and antibody-dependent cellular phagocytosis (ADCP). The efficacy of bNAb therapy in ATI studies was found to be correlated with the activity of NK cells, indicating that efficacy of bNAbs depends on the recruitment and activation of NK cells through the interaction of antibodies with Fc gamma receptors (FcγR)6,7,21.
Efforts to increase the affinity of bNAbs for FcγRIIIa are of considerable interest to boost NK cell-mediated elimination of HIV-1 infected cells22. The N-linked glycan in the IgG1 Fc domain (N297) is involved in the interaction with FcγRs and the absence of the fucose moiety has been shown to increase the affinity of the antibody for FcγRIIIa and enhance NK cell-mediated ADCC23,24,25,26,27. Afucosylated IgG responses predominantly occur in the context of membrane-expressed antigen through regulatory pathways in B cells that orchestrate the expression of glycosyltransferases28. HIV-1 specific afucosylated IgG responses are found more prominent in elite controllers and have been shown to correlate with enhanced NK cell-mediated ADCC in vitro29. Interestingly, afucosylated antibodies induce ADCC at lower antigen density, which is a clear advantage in the context of the sparse antigen expression of Env on HIV-1 infected cells23,26. In addition, interaction of NK cells with afucosylated antibodies induces a stronger intracellular signaling pathway that results in CD16 shedding and faster detachment of NK cells from target cells30,31. This allows for sequential engagement of multiple target cells, resulting in serial killing of infected cells by NK cells30,31,32.
Glyco-engineering techniques facilitate the production of afucosylated variants of monoclonal anti-HIV-1 antibodies33,34. Afucosylated anti-HIV-1 Abs have been studied in the context of viral protection (monoclonal antibodies (mAbs) 2G12, b12) and elimination of HIV-1 infected cells (mAbs LSEVh and PGT121)35,36,37,38,39. In a simian HIV-1 challenge model, the afucosylated b12 antibody did not improve protection from infection compared to the wild-type b12 antibody, indicating that the contribution of ADCC activity in the context of HIV-1 protection is minimal40. Interestingly, afucosylated versions of LSEVh and PGT121 demonstrated enhanced ADCC activity and clearance of HIV-1 infected cells38,39, arguing that glyco-engineering is a powerful tool to enhance in vivo activity of bNAbs.
To boost NK cell cytolytic activity and enhance the elimination of HIV-1 infected cells by bNAbs, we produced a panel of afucosylated bNAbs targeting various sites of the HIV-1 Env trimer. While Env binding and neutralization capacity of afucosylated bNAbs was preserved, NK cell activation and NK cell-mediated ADCC was significantly improved for the afucosylated antibodies. These afucosylated antibodies have the potential to advance bNAb therapeutic strategies and may contribute to combinatorial HIV-1 cure strategies that aim for reservoir reduction and durable control.
Results
Afucosylated anti-HIV-1 bNAbs demonstrate enhanced binding to FcγRIIIa
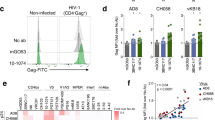
To study the influence of fucosylation on antibody effector function in the context of HIV-1, a panel of afucosylated bNAbs (2G12, N6, PGT121, PGT151, PGDM140041,42,43,44,45) was produced in vitro using a decoy substrate of the fucosyl transferase, 2-Deoxy-2-fluoro-L-fucose (2FF) (Fig. 1a)33. These bNAbs are very distinct in the epitope they target on the HIV-1 Env trimer, the angle of approach they use and the stoichiometry of interaction (1, 2 or 3 antibodies binding to one HIV-1 Env trimer). In addition to the various bNAbs, we also produced an afucosylated variant of the non-neutralizing antibody A32, which has been described to induce strong ADCC46,47. Similar yields were obtained for antibodies produced in the absence and presence of 2FF, and antibodies were validated using a reduced SDS-PAGE (Supplementary Fig. 1a). Then, we used mass-spectrometry analysis to determine the N-linked glycosylation profile of the antibodies produced in the presence or absence of 2FF. Depending on the antibody, we found that the degree of fucosylation was reduced to 5–20% for the Abs produced in the presence of 2FF, compared to >95% fucosylation for the Abs produced conventionally (Fig. 1b). Other glycosylation features were largely comparable between the antibodies except for higher galactosylation and sialylation levels of the afucosylated A32 antibody (Supplementary Fig. 1b). In general, neutralization capacity of afucosylated bNAbs was comparable to that of their fucosylated counterparts, (Supplementary Fig. 2a, b). In line with preserved neutralization capacity, afucosylated Abs showed similar binding to monomeric BG505 gp12048 and trimeric BG505 SOSIP.v4.249 compared to their fucosylated counterparts (Supplementary Fig. 1c and Fig. 1c) indicating that the glyco-engineering strategy did not influence antigen binding. Next, we studied binding of the afucosylated antibodies to FcγRIIa and FcγRIIIa in the context of HIV-1 Env using dimeric FcγR proteins50. All afucosylated antibodies except A32, bound with increased capacity to FcγRIIIa compared to their fucosylated counterparts, whereas binding to FcγRIIa remained unchanged, confirming that only FcγRIIIa binding is sensitive to Ab fucosylation degree (Fig. 1c and Supplementary Fig. 1c, d). FcγRIIIa binding to afucosylated antibodies was also tested in a gp120-bound context, allowing better binding of the A32 antibody. However, gp120-bound afucosylated antibody A32 did not show enhanced binding compared to its fucosylated counterpart, in contrast to all other afucosylated Abs (Supplementary Fig. 1c). A SARS-CoV-2 specific afucosylated antibody (afucosylated COVA1-1851) and an Fc dead PGT121 antibody (PG-LALA mutations52;) did not bind to HIV-1 Env or FcγR respectively (Fig. 1c). In summary, we produced a panel of afucosylated anti-HIV-1 Abs that significantly improved binding to FcγRIIIa, while preserving HIV-1 Env binding.
a Schematic representation of an IgG1 monoclonal antibody with the N-linked 297 glycan depicted in the Fc domain on both Ab heavy chains. The fucosylated complex glycan represents one of the major glycoforms (G0F) found in serum and in in vitro produced monoclonal Ab. Glyco-engineering techniques facilitate the production of antibodies with an afucosylated Fc-glycan. b Mass spectrometry based glyco-peptide analysis of anti-HIV-1 Abs produced in HEK-293F cells in the absence or presence of a decoy substrate for the fucosyltransferase (2FF). Fucosylated Ab variants (produced under standard conditions) and afucosylated variants (produced in the presence of 2FF) are depicted in dark gray and light gray respectively. c Env, FcγRIIa and FcγRIIIa binding of fucosylated (dark gray) and afucosylated (light gray) anti-HIV-1 Abs in the context of BG505 SOSIP.v4.2 as determined with ELISA in triplo. Mean OD450 values + standard deviation are plotted at 5 µg/ml Ab concentration. Antibody titrations can be found in Supplementary Fig. 1E. Controls (PGT121 PG-LALA, afucosylated anti-SARS CoV-2 Ab COVA1-18 and absence of Ab) are depicted in white bars. Statistical differences between fucosylated and afucosylated bNAbs were determined using an ordinary one way Anova with a Šídák's multiple comparisons test and significant differences are indicated with asterisks. *p < 0.05, **p < 0.01, ***p < 0.001.
Afucosylated anti-HIV-1 bNAbs enhance activation and degranulation of NK cells
Next, we isolated NK cells from healthy donors and determined their activation (intracellular IFNγ), receptor shedding (CD16) and degranulation (CD107) upon interaction with fucosylated or afucosylated antibodies in the context of HIV-1 Env (Fig. 2a and Supplementary Fig. 3a). Strong activation through antibody interaction with CD16 is known to result in ADAM17-mediated CD16 shedding and acts as a feedback loop to prevent hyperinflammation32,53. Of the conventional fucosylated antibodies, N6, PGT121, A32 and particularly 2G12 were found to induce antibody-dependent activation of NK cells, illustrated by the increase in CD107+IFNγ+ double positive cells and the induction of CD16 shedding (Fig. 2b). Fucosylated PGDM1400 and PGT151 induced only minor Ab mediated CD16 shedding and no activation of NK cells. All afucosylated Abs significantly enhanced NK cell activation and CD16 shedding in comparison to their fucosylated counterparts in an antibody concentration-dependent manner up to >10 fold (Fig. 2b). NK cell activation and CD16 shedding were strongly correlated, confirming the presence of the feedback loop upon activation through CD16 (Supplementary Fig. 3b). NK cell activation by afucosylated Abs was also enhanced compared to the fucosylated variant in the context of membrane-expressed Env instead of plate-coated Env protein (Supplementary Fig. 3c).
a Schematic representation of the plate-based NK cell activation assay. Expression of CD107, CD16 and IFNγ was analyzed after incubating NK cells with HIV-1 Env bound antibodies. NK cells were isolated from healthy donors using a negative selection kit (b) CD107+IFNγ+ double-positive NK cells, and % CD16 shedding is plotted for NK cells after incubation with a serial dilution of fucosylated or afucosylated anti-HIV-1 Abs 2G12, N6, PGT151 and PGDM1400. The dotted line indicates the Ab concentration (5μg/ml) that was used to analyze NK cell activation of all other anti-HIV-1 Abs. These triplicate measurements + standard deviation at 5μg/ml are plotted in the bar graphs next to the titration curves in which fucosylated Abs are depicted in dark gray and afucosylated Abs in light gray. Statistical differences between fucosylated and afucosylated bNAbs were determined using an ordinary one way Anova with a Šídák's multiple comparisons test and significant differences are indicated with asterisks. *p < 0.05, **p < 0.01, ***p < 0.001. c Expression of TIGIT and PD-1 on NK cells from PWH (red triangles) and HIV-1 negative donors (black dots) prior to stimulation and percentage CD107+IFNγ+CD3− cells in response to fucosylated or afucosylated N6 Ab for five PWH and five uninfected donors. Statistical differences between fucosylated and afucosylated bNAbs were determined using a Wilcoxon matched-pairs signed rank test and significant differences are indicated with asterisks. *p < 0.05, **p < 0.01, ***p < 0.001.
Next, we compared NK cell activation by N6 and afucosylated N6 in PBMCs from 5 uninfected donors and 5 PWH on ART, since the presence of dysfunctional NK cell subsets in PWH might prevent a strong antibody-mediated NK cell activation through afucosylated Abs54. In line with our expectation, we observed an elevated expression of exhaustion markers TIGIT and PD-1 on NK cells from PWH compared to uninfected individuals (Fig. 2c). Nonetheless, NK cells from PWH were still potently activated through the afucosylated N6 antibody (Fig. 2c, Supplementary Fig. 3d). Thus, afucosylated Abs strongly activated NK cells from PWH, supporting the supposition that they can contribute to eradicating HIV-1 infected cells through NK cell-mediated ADCC.
ADCC of HIV-1 infected cells
To determine killing by activated NK cells (i.e. ADCC), we infected CEM.NKr-CCR5-luc cells with viruses with Env’s from three different strains (LAI, BG505.T332N55 and AMC01656) and used the infected cells as target cells (Supplementary Fig. 4a). ADCC capacity of our panel of fucosylated and afucosylated bNAbs was measured by a loss of luciferase activity in the presence of antibody and PBMCs (Effector to target cell ratio (E:T) = 10:1). We performed a serial dilution of 2G12 and N6 antibodies and found a strong enhancement of ADCC capacity by afucosylated antibodies in comparison to their fucosylated counterparts (Fig. 3a). ADCC-mediated killing by afucosylated bNAbs was even observed at low antibody concentrations, which is in line with the NK cell activation data (Fig. 2b). ADCC activity was significantly enhanced for afucosylated 2G12, N6, PGT151 and PGDM1400 antibodies, whereas afucosylated PGT121 and A32 did not enhance ADCC activity (>10%) compared to their fucosylated counterparts (Fig. 3b). Since PGT121 neutralizes BG505.T332N and AMC016 efficiently (Supplementary Fig. 2a), the lack of ADCC activity with the afucosylated PGT121 antibody was unexpected and indicates that other factors such as Fc topology and angle of approach are hampering ADCC. Interestingly, HIVIG showed prominent ADCC activity against AMC016, but not against LAI or BG505.T332N infected CEM.NKr-CCR5-luc cells (Fig. 3b and Supplementary Fig. 4b), which might indicate that HIVIG has a preference for binding to AMC016 Env compared to BG505.T332N and LAI. To summarize, afucosylated variants of bNAbs 2G12, N6, PGT151 and PGDM1400 potently eliminate HIV-1 infected cells through NK cell-mediated ADCC.
a ADCC activity of CEM.NKr.CCR5-luc infected cells in the presence of a serial dilution of 2G12 (red; circles) and N6 variants (blue; triangles). Mean + standard deviation of triplicate measurements are plotted, where fucosylated bNAbs are displayed with a closed symbol and afucosylated bNAbs with an open symbol. b Bar graph showing mean ADCC activity + standard deviation of the fucosylated (dark gray) and afucosylated (light gray) Abs at a concentration of 10 μg/ml. Each dot represents the ADCC activity (mean of triplo in Supplementary Fig. 4b) measured against BG505.T332N, LAI or AMC016 infected CEM cells using PBMCs from three different donors. Thus, this figure combines all data plotted in the nine individual graphs in Supplementary Fig. 4b. Percentage ADCC is derived from the raw luciferase data using the following formula: Luciferase count in the presence of effector cells alone minus luciferase count in the presence of antibody and effector cells, divided by luciferase count in the absence of antibody and effector cells, multiplied by 100. Statistical differences between fucosylated and afucosylated bNAbs were determined using an ordinary one way Anova with a Šídák's multiple comparisons test and significant differences are indicated with asterisks. *p < 0.05; **p < 0.01; ***p < 0.001.
Afucosylated anti-HIV-1 bNAbs induce ADCC of reactivated latently infected cells
Expression and density of Env on latently infected cells is likely to be different compared to that on cells during acute infection. To determine ADCC in a latency context, we used a latently infected cell line, ACH-2, and induced viral transcription and Env expression by reactivation with TNFα (Fig. 4a). Reactivation increased the fraction of p24+ ACH-2 cells from ~25% to >85% (Fig. 4a). These reactivated cells were recognized efficiently by the panel of anti-HIV-1 bNAbs, and only weakly by A32 (Supplementary Fig. 5a). NK cell-mediated killing of reactivated ACH-2 cells was measured by the loss of p24+ ACH-2 cells after co-culture with PBMCs in a FACS based ADCC assay (Supplementary Fig. 2b)13. Afucosylated variants of 2G12, N6 and PGT151 effectively induced 10–20% ADCC of p24+ ACH-2 cells, whereas the afucosylated variant of PGT121 and PGDM1400 only showed minor antibody-mediated killing (5–10%; Fig. 4b), which correlates with lack of LAI neutralization and poor binding to reactivated ACH-2 cells of these Abs (Supplementary Figs. 2 and 5). In agreement with potent NK cell-mediated activation, fucosylated 2G12 already induced NK cell-mediated ADCC of ACH-2 cells (Fig. 4b). In parallel to ADCC quantification in the ACH-2 cells, the expression of activation markers was quantified in the effector PBMC cell population after co-incubation. Enhanced CD69 expression, CD107 expression and CD16 shedding was observed in the conditions with high ADCC activity, indicating that PBMC activation correlated with ADCC mediated killing of p24+ ACH-2 cells (Fig. 4b and Supplementary Fig. 5c). Interestingly, afucosylated PGT121 induced maximum shedding of CD16, but this did not translate to strong PBMC activation and ADCC of target cells (Fig. 4b). To summarize, the majority of afucosylated antibodies induce NK cell-mediated ADCC of reactivated ACH-2 cells, which was correlated with expression of activation markers on effector cells.
a Percentage p24+ (black line, circles) and Env+ (red line, squares) ACH-2 cells are plotted after reactivation with various concentrations of TNFα. The adjacent flow cytometry dot plots show Env+ and p24+ cells in unstimulated and TNFα (10 ng/ml) reactivated ACH-2 cells. b Quantification of target cell (ACH-2) killing and effector cell (PBMC) activation using a flow cytometry based ADCC assay for which the gating strategy is described in Supplementary Fig. 5b. Mean + standard deviation of triplicate measurements are plotted for ADCC of ACH-2 cells (top panel), PBMC activation (CD69+CD107+ PBMCs, middle panel) and CD16 shedding (bottom panel) in the presence of 10 µg/ml fucosylated (dark gray) or afucosylated antibody (light gray).
Discussion
The excitement around bNAb therapeutic strategies for PWH has increased substantially after recent human clinical trials showed that bNAb administration has the ability to reduce the replication competent viral reservoir and induce durable CD8+ cell mediated viral suppression16,17,18,57,58. This underlines the potential of bNAbs in HIV-1 curative strategies and hallmarks the beginning of an era in which antibody engineering strategies can be used to tailor the antibody-mediated functions that are contributing to the in vivo efficacy.
In an effort to improve antibody-mediated elimination of HIV-infected cells, we generated a panel of glyco-engineered afucosylated anti-HIV-1 bNAbs and demonstrated that these afucosylated bNAbs induced NK cell-mediated ADCC of HIV-1 infected cells at low antibody concentration in contrast to unmodified bNAbs. Env expression on HIV-1 infected cells is low and often not sufficient for monoclonal IgG1 antibodies to activate effector cells as a result of inefficient FcγR crosslinking59. This antibody opsonization threshold is significantly lower for afucosylated antibodies and therefore NK cell-mediated ADCC can be initiated at lower antigen density or antibody concentration23. This is particularly relevant for the elimination of HIV-1 infected cells at sanctuary sites where the antibody concentration is low due to suboptimal tissue penetration after in vivo administration of bNAbs60.
The angle of approach and stoichiometry of antibodies determines oligomerization of antibodies and FcγR clustering at the immunological synapse13,61,62. We expected stronger antibody effector function with bNAbs binding in a 3:1 stoichiometry (PGT121/N6/2G12) compared to bNAbs binding in a 2:1 (PGT151) or 1:1 (PGDM1400) stoichiometry. Despite the disadvantage in terms of stoichiometry, PGDM1400 and PGT151 were showing high ADCC activity, suggesting that the Fc topology of these antibodies favors proper NK cell activation. ADCC activity against CEM-LAI target cells for afucosylated PGDM1400 was remarkable, since PGDM1400 does not neutralize LAI. Apparently the binding and opsonization of CEM-LAI cells by PGDM1400 is still sufficient to induce ADCC, a phenotype that has been reported previously for a bNAb family member (PGT145)63. In contrast, afucosylated PGT121 demonstrated poor ADCC activity, both in the CEM and ACH-2 ADCC assay, while PGT121 does neutralize AMC016 and BG505.T332N. This is in line with previous data in which strong binding to HIV-1 infected cells was shown for PGT121 but relatively weak Fc effector function potential62. In another study, enhancement of ADCC activity was observed with plant-produced afucosylated PGT121, in contrast to our findings39. The Fc oligomerization of Env-bound PGT121 and resulting topology for FcγR activation seems disadvantageous for strong NK cell activation. This might also explain why Fc-dependent functions were found to be redundant for PGT121 efficacy in SHIV infected macaques64.
Some differences in the magnitude of NK cell-mediated killing by the afucosylated antibodies in our panel are more likely explained by epitope availability and affinity. The A32 inner-domain epitope is a CD4 inducible epitope and is poorly accessible on Env expressed on the HIV-1 infected CEM cells or ACH-2 cells. As a consequence A32 showed poor ADCC activity in our in vitro assay in comparison to other studies where A32 potency is often tested in combination with a CD4 mimetic46,47,65. Thus, it would be of interest to study ADCC activity of the afucosylated A32 Ab in combination with a CD4 mimetic to determine its full potential. Furthermore, future studies with primary CD4+ infected target cells should demonstrate whether ADCC activity of the afucosylated Abs is similar in a more physiological setting in terms of Env conformation. Of note, uninfected CD4+ T-cells that are interacting with shedded gp120 monomers are recognized by the A32 antibody, which could result in bystander killing and is a major disadvantage for the application of A32 in therapeutic strategies66,67.
The efficacy of afucosylated antibodies is also dependent on the engagement and activation of effector cells, such as NK cells and monocytes. The expression of immune checkpoint modulating proteins such as PD-1 and TIGIT on NK cells during chronic HIV-1 infection contributes to an exhausted phenotype with minimal cytotoxic activity54,68,69. ART partially restores the exhaustion profile but the effector cells are not completely recovered and display a dampened cytotoxic profile54. The increased percentage of PD-1+ and TIGIT+ NK cells for PWH on ART in our study also indicates that there is a residual exhaustion profile. The fact that afucosylated N6 bNAb could potently activate these partially exhausted NK cells, indicates that activation through afucosylated antibodies can abrogate these inhibitory signaling events.
Next-generation engineered bNAbs are of considerable interest for therapeutic intervention and cure strategies. In vivo application of glyco-engineered bNAbs offers several advantages compared to other well-known antibody engineering strategies based on amino-acid substitutions. First of all, afucosylated bNAbs specifically enhance FcγRIIIa and do not increase binding to the inhibitory receptor FcγRIIb, which is observed for other protein engineering strategies70. Secondly, afucosylated antibodies occur naturally in the human body, so there is no additional immunogenicity risk and no concern for the development of anti-drug antibodies. Thirdly, the glyco-engineering strategy does not influence FcRn binding and IgG half-life, although bio-distribution might be different71,72. Most importantly, afucosylated anti-CD20 antibodies are FDA approved for treatment of several B-cell malignancies, indicating that these antibodies are well-tolerated and safe and do not lead to aspecific killing or inflammation73. To obtain a homogenous afucosylated bNAb product for in vivo application, instead of the residual 10/20% fucosylation using our decoy substate, afucosylated bNAbs can also be produced in FUT8 Knock-out CHO cells or plant cells34,39. Furthermore, AAV delivery of bNAbs together with a short hairpin RNA targeting the fucosyltransferase allows for continuous in vivo production of afucosylated bNAbs74.
Of note, anti-HIV-1 bNAbs N6 and PGT121 harbor an N-linked glycan in their light chain and heavy chain Fv domain respectively75. The neutralization capacity of N6 and PGT121 afucosylated antibodies was deviating from the fucosylated antibody, suggesting that the reduced fucosylation of the Fv glycan might influence antigen binding and neutralization capacity.
The promising in vivo efficacy of bNAbs in human clinical trials and the role of NK cells in control or elimination of the HIV-1 reservoir justify the identification and generation of antibodies with improved NK cell activation to further enhance bNAb performance. The glyco-engineered afucosylated bNAbs that we describe here have the potential to fulfill this need and improve reservoir reduction in HIV-1 cure strategies by NK cell-mediated killing.
Materials & methods
Antibody production and purification
Monoclonal anti-HIV-1 antibodies were produced as described previously51,76. Briefly, plasmids encoding Ab heavy and light chains were co-transfected in HEK-293F cells (Invitrogen) using PEI-MAX in a 3:1 ratio in OptiMEM. To decrease fucosylation of the N-linked glycan, 0.2 mM of the fucosyltransferase decoy substrate, 2-deoxy-2-fluoro-l-fucose (2FF) (Carbosynth, MD06089), was added to the culture 1 h before transfection33,51. After 5 days, supernatant of transfected cells was harvested and filter sterilized using 0.22 µm Steritop filters (Merck Millipore). Subsequently, antibodies were purified by protein A/G (Pierce) affinity chromatography and buffer exchanged to PBS in a final concentration of 1-2 mg/ml.
Cells
CEM.NKR-CCR5-luc and ACH-2 cell-lines were obtained through the NIH AIDS Reagent program and cultured in IMDM medium (ThermoFisher) supplemented with 10% Fetal Calf Serum (FCS, Gibco) and 100U/ml Penicillin/Streptomycin (P/S, Gibco). TZM-bl cells were obtained through the NIH AIDS reagent program and cultured in DMEM supplemented with 10% FCS 100 U/ml Penicillin and Streptomycin.
PBMCs were isolated from buffy coats (Sanquin blood bank; participants provided their written informed consent to allow use of PBMCs for research purposes) using Ficoll gradient according to manufacturer’s instructions or obtained from left-over PBMC ampules from PWH on ART (>2 year) through the Amsterdam cohort studies. Collection of PBMCs for research purposes was approved by the Amsterdam Cohort Studies (ACS) on HIV infection and AIDS, a collaboration between the Amsterdam Health Service, the Academic Medical Center of the University of Amsterdam, Sanquin Blood Supply Foundation, and the Jan van Goyen Clinic, are part of The Netherlands HIV Monitoring Foundation. All ethical regulations relevant to human research participants were followed. Natural Killer cells were isolated from PBMCs using positive (CD56+ microbeads; Miltenyi) or negative MACS selection (NK cell isolation kit; Miltenyi) according to manufacturer’s instructions.
SDS-Page
Antibody quality and size was validated using a reducing SDS-Page followed by Coomassie blue staining. 4µg of purified antibody was mixed with SDS loading dye (0.125 M Tris-HCl pH 6.8, 20% glycerol, 4% SDS, 0.05% bromephenol blue in milli-Q water) containing 50 mM dithiothreitol (DTT) and incubated 10 min at 95 °C prior to loading on a 10–20% Tris-Glyine gel (Invitrogen). Precision Plus Protein Standard Dual Color (Biorad cat#161-0374) was used as a marker. Antibody heavy and light chains were separated by running gels for 1 h at 200 V at 4 °C in running buffer (25 mM Tris, 192 mM glycine, 0.05% SDS) in a vertical mini-gel electrophoresis system (ThermoFisher). Afterwards, gels were stained using the PageBlue Protein Staining Solution (Thermo Scientific) for 60 min. Destaining of gels was performed using ultrapure water with gentle agitation.
Glycoanalysis
Liquid chromatography – mass spectrometry measurements of immunoglobulin G Fc N-glycosylation were performed as published previously and briefly described in the following77. 5 μg monoclonal antibody (in 1–10 μL) were diluted in 100 μL 100 mM formic acid, incubated for 15 min and dried. Thus denatured antibodies were reconstituted in 20 μL 50 mM ammonium bicarbonate under shaking for 5 min and cleaved by addition of 20 µL of 0.01 µg/µL sequencing grade trypsin (Promega) at 37 °C overnight. Resulting glycopeptides were measured by injecting 200 nL of sample on a trap column, reversed phase separation on a nanoEase M/Z Peptide BEH C18 analytical column (75 μm × 100 mm, particle size 1.7 μm, poresize 130 Å, 1/PK, Waters) at a flow of 600 nL/min and detection by positive-mode electrospray ionisation mass spectrometry (MS)77. The instrumental setup consisted of an Ultimate 3000 RSLCnano system (Dionex/Thermo Fisher Scientific) and an Impact HD quadrupole-time-of-flight(qTOF)-MS (Bruker Daltonics). A linear gradient of solvent A (0.02% TFA in water) and solvent B (95% ACN) was used: 3%B 0 min, 21%B 4.5 min, 50%B 5.5 min, 50%B 8 min, 3%B 9 min, 3%B 11.5 min. Spectra were recorded at a frequency of 1 Hz between m/z 550−1800. The data was processed using LaCyTools version 2.0.1 build 20201216 as previously described, using the following parameters: no alignment; calibration and extraction mass window 0.2 Da and 0.1 Th, respectively; extraction time window 10.0 s.
IgG and FcγR-dimer ELISA
FcγR binding was assessed in ELISA using FcγR-dimer proteins as described previously50. Half-area ELISA microplates or Ni-NTA plates were coated overnight at 4 °C with 2 μg/mL BG505 gp120-D7324 or 2 µg/ml his-tagged BG505 SOSIP.v4.2 respectively48,49. Next day, plates were washes 3 times with 1x Tris-buffered Saline with 0.05% Tween-20 (TBS-T) and subsequently blocked by incubating wells with 100 µl assay buffer (PBS supplemented with 1% BSA, 0.05% Tween-20 and 1 mM EDTA). Then, a 2-fold serial dilution of anti-HIV-1 Abs was added to the wells (starting at 10 µg/ml) and plates were incubated for 90 min at 37 °C. Plates were washed 4x with TBS-T after which biotinylated FcγRIIa-dimer (131H, high affinity polymorphic variant) or FcγRIIIa-dimer (158V, high affinity polymorphic variant) were added to the plates at a concentration of 1 µg/ml. FcγRIIa-dimer and FcγRIIIa-dimer sequences were provided by Bruce Wines, cloned into a pcDNA3.1 expression plasmid and produced as previously described78. To detect binding of biotinylated FcγR-dimers, plates were incubated 60 min at 37 °C with streptavidin-HRP detection antibody (Biolegend; 405210) at a 1:2000 dilution in assay buffer. In parallel to FcγR binding, an anti-IgG ELISA was performed to control for antibody binding strength. Instead of incubating with FcγR-dimers, plates were incubated with a polyclonal goat anti-human IgG HRP antibody (Jackson; 109-036-003; 1:3000) after the incubation of the anti-HIV-1 antibodies. Following incubation, plates were washed 4× with TBS-T. Colorimetric detection was performed by adding develop solution (0.1M NaAc + 0.1M citric acid +1% TMB +0.01% H2O2) and quenching reaction after 5 min with 0.8M H2SO4. Optical density was measured using a SPECTROstar Nano Microplate Reader (BMG LabTech) with a 450 nm filter.
Neutralization assay
The global panel of reference viruses was used to perform neutralization assays as described previously78,79. Briefly, TZM-bl cells were seeded (DMEM, 10% FCS, 100 U/ml Penicillin/Streptomycin) one day prior to neutralization assay to achieve a confluency of 70–80% the next day. Env-pseudotyped HIV-1 viruses of the global panel were produced in HEK-293T cells as described previously49,80. Viruses were pre-incubated for 1 h at RT with a serial dilution of the anti-HIV-1 antibodies, after which the virus+antibody mix was added to the TZM-bl cells in a final volume of 100 μl medium containing 40 μg/ml Dextran and 400 nM Saquinavir. After incubating plates at 37 °C for 72 h, cells were lysed with 1× lysis buffer and incubated on an orbital shaker for 20 min at RT. Then, 25 µl Bright Glo Luciferase (Promega) was added to 5 µl lysed cell suspension to detect luciferase activity with a Glomax plate reader. A dose response non-linear regression fit (GraphPad Prism 9) was used to determine the Ab concentration required to achieve 50% neutralization (IC50) of the virus.
Plate-based NK cell activation
Half-area microplates (Greiner) or Ni-NTA plates (Qiagen) were coated overnight at 4 °C with 2 μg/mL BG505 gp120-D7324 or 5 µg/ml his-tagged BG505 SOSIP.v4.2 respectively48,49. Next day, plates were blocked with 1% BSA in PBS (blocking buffer). After washing plates with Tris-buffered saline (TBS), Env-coated plates were incubated with anti-HIV-1 antibodies diluted in blocking buffer for 1 h at 37 °C. NK cells (stimulated O/N with 10 ng/ml IL-15 after positive/negative isolation) were added onto the ELISA plates at 50,000 cells per well and were incubated for 3 h at 37 °C in the presence of 10 ng/ml IL-15 (Invitrogen) and 5 µg/ml BrefeldynA (Biolegend). Then, NK cells were transferred to a 96 well V-bottom microplate and stained with anti-CD16 BV421 (Clone 3G8; 302038; 1:500; Biolegend), anti-CD56 AF488 (clone HCD56; 1:200; 318311; Biolegend), anti-CD107 APC (Clone H4A3; 1:1000; 328620; Biolegend) and fixable viability dye ef780 (Invitrogen; 65-0865-14; 1:2000). After 30 min incubation at 4 °C, cells were washed 3x with FACS buffer (PBS + 2% FCS), and subsequently fixed with the Fixation/Permeabilization Kit (BD biosciences). After fixation and permeabilization, NK cells were stained intracellularly with anti-IFNγ PE (Clone B27; 1:500; 506506; Biolegend) antibody in Wash/Perm buffer (BD biosciences). After intracellular stain, cells were washed twice in wash buffer and then resuspended in 150 μl FACS buffer for flow cytometry analysis. Samples were measured using a BD FACSymphony A1 Cell Analyzer and analyzed with FlowJo.
Plate-based NK activation PWH
PBMCs were used from left-over ampules from PWH (>2 years on ART) enrolled in the Amsterdam cohort studies (ACS) and PBMCs from healthy blood donors (Sanquin). Due to limited cell numbers, NK cells were not isolated from PBMCs, instead PBMCs were stimulated O/N with 10 ng/ml IL-15 and used directly in the assay. The NK cell activation protocol described above was followed subsequently. To facilitate NK cell characterization in the lymphocyte gate, an anti-CD3 BV510 (clone UCHT-1; 1:200; 300447; BD Biosciences) antibody was included in our flow cytometry panel (Supplementary Fig. 2C). Expression of exhaustion markers on unstimulated PBMCs was determined with flow cytometry using anti-TIGIT APC (clone MBSA43, 1:200, 17-9500-42; ThermoFisher) and anti-PD1 PE (clone EH12.2H7, 1:200; 329905; Biolegend) in addition to anti-CD3/CD16/CD56 stains. This allowed for characterization of TIGIT and PD-1 expression on NK cells.
HEK-cell based NK cell activation
Env expressing HEK-293T cells were obtained by transient transfection of a pCI BG505 gp160-deltaCT plasmid using lipofectamin as described previously78. BG505 expressing HEK cells were co-cultured with CD56+ NK cells in an E:T ratio of 2:1 in the absence or presence of anti-HIV-1 Abs in a final volume of 100 μl IMDM medium. After co-culture for 3 h at 37 °C, cells were stained with anti-CD16 Percp.Cy5.5 (Clone 3G8; 1:500; Biolegend), anti-CD107 APC (Clone H4A3; 1:1000; Biolegend) and fixable viability dye ef780 (Invitrogen; 1:2000). After allowing membrane staining for 30 min at 4 °C, cells were washed 3x with FACS buffer, and then resuspended in FACS buffer for flow cytometry analysis. Samples were read using a BD FACSymphony A1 Cell Analyzer and analyzed with FlowJo.
CEM.NKr.CCR5-luc infection
Plasmids of infectious molecular clones pLAI, pBG505.T332N-LAI55 or pAMC016-LAI56 were transfected in HEK-293T cells as previously described55,56. Virus containing supernatant was collected three days post transfection, filter-sterilized and stored at −80. CEM.NKR.CCR5-luc cells were plated in a 6 or 12-wells plate at a concentration of 1e6/ml and infected with HEK-293T produced virus at a high multiplicity of infection (MOI) in the presence of 5μg/ml DEAE-Dextran. Spinoculation (2 h, 1200 RCF, RT) was performed for BG505.T332N and AMC016 infection experiments. LAI infection did not require spinoculation and was incubated at 37 °C for 2 h. After spinoculation/incubation, cultures were rested at 37 °C for 1 h before washing away virus and resuspending cells in fresh IMDM medium supplemented with 5μg/ml DEAE-Dextran. After two days, 500 µl medium was added to the 12-well infection cultures and at day 4, cells were harvested and washed repeatedly with IMDM culture medium to get rid of viral particles that might interfere with the ADCC assay.
ADCC assay CEM.NKr.CCR5-luc
We used a previously established ADCC assay to measure elimination of HIV-1 infected cells59,66. Briefly, HIV-1 infected CEM cells were plated in a 96-well V-bottom plate at a density of 20.000 cells/well in a volume of 25 µl. 25 µl of antibody, serial dilution in IMDM medium, was subsequently added and incubated at room temperature for 15 min. Overnight rested PBMCs from healthy individuals were added to the wells at an effector target cell ratio of 10:1 in a final volume of 100 µl. Plates were shorty centrifuged (1 min, 300 RCF) to allow efficient contact between effector and target cells, and subsequently incubated for 6 h at 37 °C. After 6 h, plates were centrifuged (5 min, 300 RCF), cells were washed once with PBS, and then resuspended in 50 μl lysis buffer. Plates were incubated on a shaker for 20 min to allow complete cell lysis. Then, 25 µl Bright Glo Luciferase (Promega) was added to 5 µl lysed cell suspension to detect luciferase activity with a Glomax plate reader.
ADCC assay ACH-2
ACH-2 cells were reactivated with 10 ng/ml TNFα in a T-25 flask at a concentration of 1 × 106 cells/ml. After 18 h, unstimulated and reactivated ACH-2 cells were harvested and washed twice to get rid of viral particles. To determine successful reactivation anti-HIV-1 Abs were added to ACH-2 cells (10 µg/ml) and incubated for 1 h at RT. After washing with FACS buffer, antibody binding was detected by incubating with a PE labeled goat anti-human IgG (Southern Biotech) for 30 min at 4 °C. To obtain a fifty-fifty population of p24- and p24+ ACH-2 cells that could serve as target cells in the ADCC assay, unstimulated and reactivated cells were mixed 1:1. These target cells were stained with a CellTrace™ Violet dye (0.25 µM ; ThermoFisher) to allow separation in flow cytometry of target cells from effector cells. ADCC assays were performed in 96-well V-bottom plates (Greiner) in which 20.000 Violet-stained ACH-2 cells were co-incubated with 180:000 PBMCs (E:T = 9:1) in the absence or presence of 10 µg/ml anti-HIV-1 Ab in a final volume of 100 µl. After a short centrifugation step (1 min, 1500 RPM) to stimulate interaction of target and effector cells, plates were incubated for 4 h at 37 °C. Afterwards, cells were washed with 2% FCS in PBS (FACS buffer) and then stained with anti-CD16 PE (Clone 3G8; 1:500; Biolegend), anti-CD107 APC (Clone H4A3; 1:1000; Biolegend), anti-CD69 BV605 (FN50; 1:500; 310937; Biolegend) and fixable viability dye ef780 (Invitrogen; 1:2000). After allowing membrane staining for 1 h at 4 °C, cells were washed 3x with FACS buffer and subsequently fixed with the Fixation/Permeabilization Kit (BD Biosciences). After fixation and permeabilization, cells were stained intracellularly with anti-HIV-1 core antigen FITC (Clone KC57; 1:500; 6604665; BeckmanCoulter) in Wash/Perm buffer (BD Biosciences). After intracellular stain, cells were washed twice in wash buffer and then resuspended in FACS buffer for flow cytometry analysis. Samples were read using a BD FACSymphony A1 Cell Analyzer and analyzed with FlowJo.
Statistics and reproducibility
Statistics were based on at least three independent measurements per test group (n = 3). In all graphs these independent measurements are plotted as well as the standard deviation. Ordinary one-way Anova tests were used to compare between fucosylated and afucosylated groups within the complete panel and were corrected using a Sidak’s multiple comparison test. A Wilcoxon matched-pairs signed rank test was used to compare activation level with PBMCs from the same donor. GraphPad Prism v9.5 was used to perform all statistical analysis.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE81 partner repository with the dataset identifier PXD053363. All source data underlying the graphs in the figures and Supplementary figures is provided in the supplementary data 1 file. Any other data is available from the corresponding author upon request.
References
Ward, A. R., Mota, T. M. & Jones, R. B. Immunological approaches to HIV cure. Semin. Immunol. 51, 101412 (2021).
Deeks, S. G. et al. Research priorities for an HIV cure: International AIDS Society Global Scientific Strategy 2021. Nat. Med. 27, 2085–2098 (2021).
Garrido, C. et al. Interleukin-15-Stimulated Natural Killer Cells Clear HIV-1-Infected Cells following Latency Reversal Ex Vivo. J. Virol. 92, e00235–18 (2018).
Covino, D. A., Desimio, M. G. & Doria, M. Impact of IL-15 and latency reversing agent combinations in the reactivation and NK cell-mediated suppression of the HIV reservoir. Sci. Rep. 12, 18567 (2022).
Papasavvas, E. et al. NK Response Correlates with HIV Decrease in Pegylated IFN-α2a–Treated Antiretroviral Therapy–Suppressed Subjects. J. Immunol. 203, 705–717 (2019).
Marras, F. et al. Control of the HIV-1 DNA Reservoir Is Associated In Vivo and In Vitro with NKp46/NKp30 (CD335 CD337) Inducibility and Interferon Gamma Production by Transcriptionally Unique NK Cells VIRUS-CELL INTERACTIONS crossm. J Virol. 91, 647–664 (2017).
Borducchi, E. N. et al. Antibody and TLR7 agonist delay viral rebound in SHIV-infected monkeys. Nature 563, 360–364 (2018).
Olesen, R. et al. Innate Immune Activity Correlates with CD4 T Cell-Associated HIV-1 DNA Decline during Latency-Reversing Treatment with Panobinostat. J. Virol. 89, 10176–10189 (2015).
Kim, J. T. et al. Latency reversal plus natural killer cells diminish HIV reservoir in vivo. Nat. Commun. 13, 121 (2022).
Sungur, C. M. et al. Human NK cells confer protection against HIV-1 infection in humanized mice. J. Clin. Investig. 132, e162694 (2022).
Bernard, N. F., Kant, S., Kiani, Z., Tremblay, C. & Dupuy, F. P. Natural Killer Cells in Antibody Independent and Antibody Dependent HIV Control. Front. Immunol. 13, 879124 (2022).
Anderko, R. R. & Mailliard, R. B. Mapping the interplay between NK cells and HIV: therapeutic implications. J. Leukoc. Biol. 113, 109–138 (2023).
Bruel, T. et al. Elimination of HIV-1-infected cells by broadly neutralizing antibodies. Nat. Commun. 7, 10844 (2016).
Dupuy, F. P. et al. Antibody-Dependent Cellular Cytotoxicity-Competent Antibodies against HIV-1-Infected Cells in Plasma from HIV-Infected Subjects. mBio 10, e02690–19 (2019).
Mendoza, P. et al. Combination therapy with anti-HIV-1 antibodies maintains viral suppression. Nature 561, 479–484 (2018).
Niessl, J. et al. Combination anti-HIV-1 antibody therapy is associated with increased virus-specific T cell immunity. Nat. Med 26, 222–227 (2020).
Gaebler, C. et al. Prolonged viral suppression with anti-HIV-1 antibody therapy. Nature 606, 368–374 (2022).
Rosás-Umbert, M. et al. Administration of broadly neutralizing anti-HIV-1 antibodies at ART initiation maintains long-term CD8+ T cell immunity. Nat. Commun. 13, 6473 (2022).
Sneller, M. C. et al. Combination anti-HIV antibodies provide sustained virological suppression. Nature 606, 375–381 (2022).
Bournazos, S. et al. Broadly Neutralizing Anti-HIV-1 Antibodies Require Fc Effector Functions for In Vivo Activity. Cell 158, 1243–1253 (2014).
Lu, C.-L. et al. Enhanced clearance of HIV-1–infected cells by broadly neutralizing antibodies against HIV-1 in vivo. Science 352, 1001–1004 (2016).
Duan, S. & Liu, S. Targeting NK Cells for HIV-1 Treatment and Reservoir Clearance. Front. Immunol. 13, 842746 (2022).
Temming, A. R. et al. Functional Attributes of Antibodies, Effector Cells, and Target Cells Affecting NK Cell–Mediated Antibody-Dependent Cellular Cytotoxicity. J. Immunol. 203, 3126–3135 (2019).
Dekkers, G. et al. Decoding the Human Immunoglobulin G-Glycan Repertoire Reveals a Spectrum of Fc-Receptor- and Complement-Mediated-Effector Activities. Front. Immunol. 8, 877 (2017).
Niwa, R. et al. IgG subclass-independent improvement of antibody-dependent cellular cytotoxicity by fucose removal from Asn297-linked oligosaccharides. J. Immunol. Methods 306, 151–160 (2005).
Suzuki, E. et al. A Nonfucosylated Anti-HER2 Antibody Augments Antibody-Dependent Cellular Cytotoxicity in Breast Cancer Patients. Clin. Cancer Res. 13, 1875–1882 (2007).
Li, T. et al. Modulating IgG effector function by Fc glycan engineering. Proc. Natl Acad. Sci. 114, 3485–3490 (2017).
Oosterhoff, J. J., Larsen, M. D., van der Schoot, C. E. & Vidarsson, G. Afucosylated IgG responses in humans – structural clues to the regulation of humoral immunity. Trends Immunol. 43, 800–814 (2022).
Ackerman, M. E. et al. Natural variation in Fc glycosylation of HIV-specific antibodies impacts antiviral activity. J. Clin. Investig. 123, 2183–2192 (2013).
Srpan, K. et al. Shedding of CD16 disassembles the NK cell immune synapse and boosts serial engagement of target cells. J. Cell Biol. 217, 3267–3283 (2018).
Karampatzakis, A. et al. Antibody afucosylation augments CD16-mediated serial killing and IFNγ secretion by human natural killer cells. Front. Immunol. 12, 1–14 (2021).
Felce, J. H. & Dustin, M. L. Natural killers shed attachments to kill again. J. Cell Biol. 217, 2983–2985 (2018).
Dekkers, G. et al. Multi-level glyco-engineering techniques to generate IgG with defined Fc-glycans. Sci. Rep. 6, 36964 (2016).
Sun, T. et al. Functional knockout of FUT8 in Chinese hamster ovary cells using CRISPR/Cas9 to produce a defucosylated antibody. Eng. Life Sci. 15, 660–666 (2015).
Forthal, D. N. et al. Fc-Glycosylation Influences Fcγ Receptor Binding and Cell-Mediated Anti-HIV Activity of Monoclonal Antibody 2G12. J. Immunol. 185, 6876–6882 (2010).
Moldt, B. et al. A Nonfucosylated Variant of the anti-HIV-1 Monoclonal Antibody b12 Has Enhanced Fc RIIIa-Mediated Antiviral Activity In Vitro but Does Not Improve Protection against Mucosal SHIV Challenge in Macaques. J. Virol. 86, 6189–6196 (2012).
Kong, D. et al. A defucosylated bispecific multivalent molecule exhibits broad HIV-1-neutralizing activity and enhanced antibody-dependent cellular cytotoxicity against reactivated HIV-1 latently infected cells. Aids 32, 1749–1761 (2018).
Bardhi, A. et al. Potent In Vivo NK Cell-Mediated Elimination of HIV-1-Infected Cells Mobilized by a gp120-Bispecific and Hexavalent Broadly Neutralizing Fusion Protein. J. Virol. 91, e00937–17 (2017).
Anand, S. P. et al. Enhanced Ability of Plant-Derived PGT121 Glycovariants To Eliminate HIV-1-Infected Cells. J. Virol. 95, e0079621 (2021).
Moldt, B. et al. A Nonfucosylated Variant of the anti-HIV-1 Monoclonal Antibody b12 Has Enhanced FcγRIIIa-Mediated Antiviral Activity In Vitro but Does Not Improve Protection against Mucosal SHIV Challenge in Macaques. J. Virol. 86, 6189–6196 (2012).
Huang, J. et al. Identification of a CD4-Binding-Site Antibody to HIV that Evolved Near-Pan Neutralization Breadth. Immunity 45, 1108–1121 (2016).
Blattner, C. et al. Structural Delineation of a Quaternary, Cleavage-Dependent Epitope at the gp41-gp120 Interface on Intact HIV-1 Env Trimers. Immunity 40, 669–680 (2014).
Trkola, A. et al. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J. Virol. 70, 1100–1108 (1996).
Julien, J.-P. et al. Broadly Neutralizing Antibody PGT121 Allosterically Modulates CD4 Binding via Recognition of the HIV-1 gp120 V3 Base and Multiple Surrounding Glycans. PLoS Pathog. 9, e1003342 (2013).
Sok, D. et al. Recombinant HIV envelope trimer selects for quaternary-dependent antibodies targeting the trimer apex. Proc. Natl Acad. Sci. 111, 17624–17629 (2014).
Ferrari, G. et al. An HIV-1 gp120 Envelope Human Monoclonal Antibody That Recognizes a C1 Conformational Epitope Mediates Potent Antibody-Dependent Cellular Cytotoxicity (ADCC) Activity and Defines a Common ADCC Epitope in Human HIV-1 Serum. J. Virol. 85, 7029–7036 (2011).
Tuyishime, M. et al. Improved killing of HIV-infected cells using three neutralizing and non-neutralizing antibodies. J. Clin. Investig. 130, 5157–5170 (2020).
Sanders, R. W. et al. A Next-Generation Cleaved, Soluble HIV-1 Env Trimer, BG505 SOSIP.664 gp140, Expresses Multiple Epitopes for Broadly Neutralizing but Not Non-Neutralizing Antibodies. PLoS Pathog. 9, e1003618 (2013).
de Taeye, S. W. et al. Immunogenicity of Stabilized HIV-1 Envelope Trimers with Reduced Exposure of Non-neutralizing Epitopes. Cell 163, 1702–1715 (2015).
Wines, B. D. et al. Dimeric FcγR Ectodomains as Probes of the Fc Receptor Function of Anti-Influenza Virus IgG. J. Immunol. 197, 1507–1516 (2016).
Hoepel, W. et al. High titers and low fucosylation of early human anti–SARS-CoV-2 IgG promote inflammation by alveolar macrophages. Sci. Transl. Med. 13, eabf8654 (2021).
Lo, M. et al. Effector-attenuating Substitutions That Maintain Antibody Stability and Reduce Toxicity in Mice. J. Biol. Chem. 292, 3900–3908 (2017).
Jing, Y. et al. Identification of an ADAM17 Cleavage Region in Human CD16 (FcγRIII) and the Engineering of a Non-Cleavable Version of the Receptor in NK Cells. PLoS One 10, e0121788 (2015).
Zhang, X. et al. Analysis of the Characteristics of TIGIT-Expressing CD3−CD56+NK Cells in Controlling Different Stages of HIV-1 Infection. Front. Immunol. 12, 602492 (2021).
Derking, R. et al. Enhancing glycan occupancy of soluble HIV-1 envelope trimers to mimic the native viral spike. Cell Rep. 35, 108933 (2021).
Schorcht, A. et al. The Glycan Hole Area of HIV-1 Envelope Trimers Contributes Prominently to the Induction of Autologous Neutralization. J. Virol. 96, e0155221 (2022).
Gunst, J. D. et al. Early intervention with 3BNC117 and romidepsin at antiretroviral treatment initiation in people with HIV-1: a phase 1b/2a, randomized trial. Nat. Med. 28, 2424–2435 (2022).
Gruell, H. et al. Effect of 3BNC117 and romidepsin on the HIV-1 reservoir in people taking suppressive antiretroviral therapy (ROADMAP): a randomised, open-label, phase 2A trial. Lancet Microbe 3, e203–e214 (2022).
Bruel, T. et al. Lack of ADCC Breadth of Human Nonneutralizing Anti-HIV-1 Antibodies. J. Virol. 91, 433–443 (2017).
Veenhuis, R. T., Clements, J. E. & Gama, L. HIV Eradication Strategies: Implications for the Central Nervous System. Curr. HIV/AIDS Rep. 16, 96–104 (2019).
Murin, C. D. Considerations of Antibody Geometric Constraints on NK Cell Antibody Dependent Cellular Cytotoxicity. Front Immunol. 11, 1635 (2020).
Rossignol, E. D. et al. Mining HIV controllers for broad and functional antibodies to recognize and eliminate HIV-infected cells. Cell Rep. 35, 109167 (2021).
Grunst, M. W. et al. Potent antibody-dependent cellular cytotoxicity of a V2-specific antibody is not sufficient for protection of macaques against SIV challenge. PLoS Pathog. 20, e1011819 (2024).
Parsons, M. S. et al. Fc-dependent functions are redundant to efficacy of anti-HIV antibody PGT121 in macaques. J. Clin. Investig. 129, 182–191 (2018).
Richard, J. et al. Across functional boundaries: Making nonneutralizing antibodies to neutralize HIV-1 and mediate Fc-mediated effector killing of infected cells. mBio 12, e0140521 (2021).
Richard, J. et al. Uninfected Bystander Cells Impact the Measurement of HIV-Specific Antibody-Dependent Cellular Cytotoxicity Responses. mBio 9, e00358–18 (2018).
Lewis, G. K. et al. Knowns and Unknowns of Assaying Antibody-Dependent Cell-Mediated Cytotoxicity Against HIV-1. Front. Immunol. 10, 1025 (2019).
Holder, K. A., Burt, K. & Grant, M. D. TIGIT blockade enhances NK cell activity against autologous HIV‐1‐infected CD4 + T cells. Clin. Transl. Immunol. 10, e1348 (2021).
Porichis, F. et al. Immune Checkpoint Blockade Restores HIV-Specific CD4 T Cell Help for NK Cells. J. Immunol. 201, 971–981 (2018).
Liu, R., Oldham, R., Teal, E., Beers, S. & Cragg, M. Fc-Engineering for Modulated Effector Functions—Improving Antibodies for Cancer Treatment. Antibodies 9, 64 (2020).
Einarsdottir, H. K. et al. Comparison of the Fc glycosylation of fetal and maternal immunoglobulin G. Glycoconj. J. 30, 147–157 (2013).
Sharma, S. K. et al. Influence of Fc Modifications and IgG Subclass on Biodistribution of Humanized Antibodies Targeting L1CAM. J. Nucl. Med. 63, 629–636 (2022).
Klein, C., Jamois, C. & Nielsen, T. Anti-CD20 treatment for B-cell malignancies: current status and future directions. Expert Opin. Biol. Ther. 21, 161–181 (2021).
Termini, J. M., Martinez-Navio, J. M., Gao, G., Fuchs, S. P. & Desrosiers, R. C. Glycoengineering of AAV-delivered monoclonal antibodies yields increased ADCC activity. Mol. Ther. Methods Clin. Dev. 20, 204–217 (2021).
Chuang, G.-Y. et al. Removal of variable domain N-linked glycosylation as a means to improve the homogeneity of HIV-1 broadly neutralizing antibodies. MAbs 12, 1836719 (2020).
Brouwer, P. J. M. et al. Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science 369, 643–650 (2020).
Falck, D., Jansen, B. C., de Haan, N. & Wuhrer, M. High-Throughput Analysis of IgG Fc Glycopeptides by LC-MS. Methods Mol. Biol. 31–47, https://doi.org/10.1007/978-1-4939-6493-2_4 (2017).
Schriek, A. I. et al. Anti-HIV-1 Nanobody-IgG1 Constructs With Improved Neutralization Potency and the Ability to Mediate Fc Effector Functions. Front. Immunol. 13, 893648 (2022).
deCamp, A. et al. Global Panel of HIV-1 Env Reference Strains for Standardized Assessments of Vaccine-Elicited Neutralizing Antibodies. J. Virol. 88, 2489–2507 (2014).
Sanders, R. W. et al. HIV-1 neutralizing antibodies induced by native-like envelope trimers. Science 349, aac4223 (2015).
Perez-Riverol, Y. et al. The PRIDE database resources in 2022: a hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res. 50, D543–D552 (2022).
Acknowledgements
We would like to thank the Amsterdam Cohort Studies (ACS) on HIV infection and AIDS, a collaboration between the Amsterdam Health Service, the Academic Medical Center of the University of Amsterdam, Sanquin Blood Supply Foundation, and the Jan van Goyen Clinic, are part of The Netherlands HIV Monitoring Foundation. We would like to thank the NIH Aids reagent program for sharing materials and cell lines. We would like to thank Jan Nouta and Steinar Gijze for support with the liquid chromatography mass spectrometry measurements and data processing. Furthermore, we would like to thank Judith Burger for the generation of Env-pseudotyped viruses from the global panel. This work was supported by the Aidsfonds under grant number P-53301; amfAR Mathilde Krim fellowship under grant number 110425-73-RKRL; HealthHolland/Aidsfonds under grant number LSHM19101/P-44802; and HealthHolland/AMC under grant number 2019-1167.
Author information
Authors and Affiliations
Contributions
Conceptualization and design of study: S.W.d.T., R.W.S., G.V., M.W., N.K., M.J.v.G. Experimental investigation: S.W.d.T., A.I.S., J.C.U., M.G., J.A.B., D.F. Data analysis: S.W.d.T., A.I.S., D.F., M.J.v.G. Writing original draft: S.W.d.T. Editing and revisions: S.W.d.T., A.I.S., M.G., R.W.S., D.F., G.V., M.W., N.K., M.J.v.G.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks the anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editors: Harry Taylor and Dario Ummarino.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
de Taeye, S.W., Schriek, A.I., Umotoy, J.C. et al. Afucosylated broadly neutralizing antibodies enhance clearance of HIV-1 infected cells through cell-mediated killing. Commun Biol 7, 964 (2024). https://doi.org/10.1038/s42003-024-06659-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s42003-024-06659-8
This article is cited by
-
Pan-caspase inhibitors induce secretion of HIV-1 latency reversal agent lymphotoxin-alpha from cytokine-primed NK cells
Cell Death Discovery (2025)
-
GlycoDash: automated, visually assisted curation of glycoproteomics datasets for large sample numbers
Analytical and Bioanalytical Chemistry (2025)