Abstract
The effects of neurotoxicant cadmium (Cd) exposure on brain development have not been well elucidated. To investigate this, we have herein subjected pregnant mice to low-dose Cd throughout gestation. Using single-cell RNA sequencing (scRNA-seq), we explored the cellular responses in the embryonic brain to Cd exposure, and identified 18 distinct cell subpopulations that exhibited varied responses to Cd. Typically, Cd exposure impeded the development and maturation of cells in the brain, especially progenitor cells such as neural progenitor cells (NPCs) and oligodendrocyte progenitor cells (OPCs). It also caused significant cell subpopulation shifts in almost all the types of cells in the brain. Additionally, Cd exposure reduced the dendritic sophistication of cortical neurons in the offspring. Importantly, these changes led to aberrant Ca2+ activity in the cortex and neural behavior changes in mature offspring. These data contribute to our understanding of the effects and mechanisms of Cd exposure on brain development and highlight the importance of controlling environmental neurotoxicant exposure at the population level.
Similar content being viewed by others
Introduction
Environmental pollutants such as heavy metals have raised great public concern because they are related to the increasing prevalence of multiple diseases, especially neurodevelopmental disorders1,2,3. Research has revealed that exposure to neurotoxicants during brain development is a risk factor for autism in children and neurodegenerative disease in adulthood4,5,6. Cadmium (Cd) is one of the most common environmental pollutants that greatly threatens human health. Cd originating from industrial waste can contaminate water, soil, and air, and then accumulate in meat offal, cereals, root crops, vegetables, seafood, and tobacco7,8,9. Adverse health effects of Cd exposure on kidneys, bones, pancreas, and the brain have been discovered10,11,12. Importantly, Cd has been revealed to be a serious threat to brain development. Extensive observational studies have revealed that prenatal exposure to Cd may lead to chronic brain dysfunctions13,14,15, but the detailed effects and underlying mechanisms are still not fully understood.
It has been proven that Cd can cross the placental barrier16. Therefore, in utero exposure to Cd may occur and should be given special attention because gestation is a critical window of susceptibility to hazardous factors17. Prenatal exposure to Cd has been reported to induce anxiety, hinder gross motor development, impair cognitive function, and cause attention deficit and hyperactivity disorder in children18,19,20. In animal studies, Cd has been revealed to destroy the blood-brain barrier and accumulate in the brain21. The neurogenesis process, axonal development, and the maturation of neurons can be influenced by prenatal Cd exposure as found in zebrafish, mice, and rat models22,23,24. Behavioral changes have also been detected in adults exposed prenatally to Cd25,26. Studies using an embryonic neural stem/progenitor cell (eNPC) model have indicated that Cd exposure inhibits cell proliferation and the maturation of differentiated neurons27,28,29. Although many previous studies have investigated the effects of Cd on brain development, the underlying mechanisms are still poorly understood.
There are different cell types that originate from the neural epithelium during brain development, such as neural progenitor cells (NPCs), oligodendrocyte progenitor cells (OPCs), neurons, and astrocytes. Even the same type of cells could be divided into different subtypes30. This phenomenon is much more complicated in the developing brain31,32,33. These cell subpopulations might respond differently to Cd exposure due to their unique characteristics34. The different responses could be covered in bulk analyses. However, much is still unknown about the specific responses of each cell subpopulation in the developing brain to Cd exposure. A critical goal in the field is to understand how Cd exerts neurodevelopmental toxicity and its underlying mechanisms.
Here, we established a prenatal Cd exposure model using pregnant mice and used a single-cell RNA-seq (scRNA-seq) -based approach to reveal the diverse responses of each cell subpopulation in the embryonic brain to Cd exposure. A series of cell subpopulations of each cell type in the brain were identified. The marker genes were screened to draw the profiles of these cell subpopulations. The DEGs in each cell subpopulation were analyzed to explore the detailed influences and mechanisms of Cd exposure on brain development. The subsequent changes in the electric activity of neural circuits and neural behaviors were also explored. We systematically detected the detailed effects of Cd exposure on each cell subpopulation in the embryonic brain, and the neurotoxic effects of Cd on brain development and brain functions in adulthood.
Results
Cd exposure caused a significant shift of cell populations in the developing brain
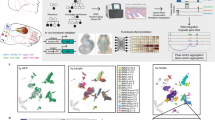
To determine the mechanisms by which Cd exposure influences brain development, we established an animal model by exposing pregnant mice to 3 mg/L cadmium chloride in drinking water throughout gestation. Animal treatment and analyses were carried out as shown in the timeline in Fig. 1a. There was no difference in the body weight of offspring on postnatal day 0 (P0) or P60 (Supplementary Fig. 1a). Additionally, no change in brain weight was detected (Supplementary Fig.1b). The average internal dose of Cd in the fetal brain cortex on embryonic day 15.5 (E15.5), the predominate time point for neuron generation35, was 3.62 ng/g (Fig. 1b). Whole cortex tissue was extracted from 4 fetus from 4 pregnant mice in each group at E15.5 and dissociated into single cells for scRNA-seq. There were 34,540 cells identified in total. These cells were divided into 18 cell clusters according to 3846 genes specifically expressed in these cells (Supplementary Fig. 2), and a significant shift in cell clusters was observed between the control group and Cd-exposed group (Fig. 1c, Supplementary Fig. 3a–c, Supplementary Table S1).
a Schematic of the experimental design and timeline. b The Cd concentration was measured in the embryonic cortex. Data of 4 fetus from 4 pregnant mice in each group are expressed as the mean ± SEM. **p < 0.01, two-tailed Student’s t test. c Combined UMAP plots of cells from control and Cd exposed mice. d UMAP plots of cells by cell population assignment. e Heatmap showing the scaled mean expression of specific marker genes used for cell population determination. f Cell distributions among six determined cell types by percentage in both the control and Cd-exposed groups.
Then, we distinguished these 18 cell clusters using previously identified cell makers in the brain36,37,38. The cell makers of NPCs, OPCs, excitatory neurons, interneurons, astrocytes, and microglia were used to identify the cells (Fig. 1d, Supplementary Table S2). The expression levels of these specific markers and unique genes identified expressed in each cluster were presented in heatmaps (Fig. 1e, Supplementary Fig. 4). The number of each cell population in both the control and Cd-exposed groups were counted. The results revealed that the total number of NPCs and astrocytes increased, while the number of OPCs, excitatory neurons, and interneurons decreased in Cd-exposed mice (Fig. 1f).
Cd exposure delayed the development of the NPC population
The NPC population included 4 clusters distinguished by their specific gene profiles (Fig. 2a, Supplementary Table S3). The numbers of cells in all these 4 clusters were much higher in Cd-exposed mice than in control mice (Fig. 2a). However, the ratio of KI67+ cells did not increase in the telencephalon of E15.5 Cd-exposed mice (Fig. 2b, d, e), which indicated that the cell proliferation rate was not altered. We also found that SOX2+ cells piled up in the telencephalon of Cd-exposed mice, especially in the outer layer (layer 2) (Fig. 2c, f, g). These data indicated that NPC development was impeded after Cd exposure.
a Cell distribution (percentages) of the four NPC clusters in control and Cd-exposed mice. b, c SOX2 and KI67 immunostaining in the E15.5 mouse telencephalon. Scale bar, 50 μm. d–g Statistical results of the number of SOX2+ and KI67+ cells in the telencephalon. Data of 6 fetus from 6 pregnant mice in each group are expressed as the mean ± SEM. **p < 0.01, two-tailed Student’s t test. h Venn analysis of the four NPC cluster marker genes. i Venn analysis of the DEGs in the four NPC clusters in control and Cd-exposed mice. j–m Violin plots showing the expression and distribution of the representative top marker genes in NPC cell clusters. C4, cluster 4; C6, cluster 6; C9, cluster 9; C16, cluster 16.
Analysis of gene profiles revealed that 56, 352, 471, and 358 marker genes were expressed separately in the 4 NPC clusters. However, only 13 marker genes were shared by these 4 clusters (Fig. 2h). There were nearly 100 or more DEGs in each cluster between control and Cd-exposed mice, and only 19 DEGs were shared by these 4 subpopulations (Fig. 2i). These data demonstrated the great heterogeneity of NPCs and their differential responses to Cd exposure.
Representative marker genes and DEGs are listed in Supplementary Table S3. The expression patterns of the typical marker genes of each cluster were presented in violin plots (Fig. 2j–m). These enriched genes were closely related to NPC proliferation, maintenance, neuronal or glial differentiation, and maturation. For example, Dlx1 and Rnd3 in cluster 4 (C4, Fig. 2j); Hmgb2 and Mki67 in C6 (Fig. 2k); Fabp7 and Mt3 in C9 (Fig. 2l); Tpx2 and Pcalf in C16 (Fig. 2m). Some of the DEGs also play a critical role in the fate decision of NPCs and other processes of brain development. For example, Chchd2, Map1b, Dcx, Ncam1, and Hes5 in C4; Mapt, Map2, and Pou2f1 in C6; Ptprz1, H2afz, Slc1a3, and Aldh1l1 in C9; Fau, Gria2, Gng5 and Pou3f2 in C16 (Supplementary Table S3). We then did GO analysis of the DEGs in each cluster. The results revealed that the expression of genes related to neuronal development, neurogenesis, axonal development, and neuronal projection development were significantly influenced (Supplementary Fig. 5a–d).
Cd exposure led to a notable shift in the OPC population
UMAP revealed that Cd exposure caused a significant shift in different OPC clusters (Fig. 3a). The total number and the number of the largest OPC cluster (C0) were remarkably decreased in Cd-exposed mice (Fig. 3b, Supplementary Table S1). Over 640 typical genes were found expressed in the 3 OPC clusters. However, few overlapping genes were found among these 3 clusters (Supplementary Fig. 6a). The results indicated remarkable heterogeneity of the OPC population. The enriched marker genes in each cluster included critical genes regulating OPC functions, such as Ptprz1, Pdgfra, and Vcan. In addition, other genes such as Gap43, Apoe, and Cdkn1a, which play key roles in the cell cycle, proliferation, and apoptosis were also included (Supplementary Table S4).
a Feature plots showing the differential distribution of OPC clusters in control and Cd-exposed mice. b Cell distribution percentages of OPC cell clusters in control and Cd-exposed mice. C0, cluster 0; C13, cluster 13; C15, cluster 15. c Venn analysis of the DEGs in the three OPC clusters in control and Cd-exposed mice. d–f QPCR analysis of the expression of Pdgfα, Vcan, and Ptprz1 in mouse cortical tissue at different developmental stages. Data of 6 fetus from 6 pregnant mice in each group are expressed as the mean ± SEM. Two-tailed Student’s t test. g GO analysis of the DEGs in the largest OPC cluster, C0, revealed that Cd exposure influenced the expression of genes related to multiple functions of brain development. h OPCs were reclustered into 10 subclusters. i Cell distribution percentages of subcluster 0 (S0) to 6 (S6) in control and Cd-exposed mice.
A remarkable number of DEGs, over 900, were found in OPC clusters between control and Cd-exposed mice (Fig. 3c, Supplementary Table S4). However, when we detected the mRNA expression of Pdgfra, Vcan, and Ptprz1 in the whole cortex tissue at E15.5, E17.5, and P0 via bulk real-time PCR analysis, no significant changes were found (Fig. 3d–f), which indicated that the subcluster differences were covered in bulk analysis. Representative DEGs, such as Mapt, Ncam1, Map1b, Gap43, Dcx, and Map2, were key factors involved in cell proliferation, neuronal and oligodendrocyte differentiation, and myelination (Supplementary Table S4). GO analysis of the DEGs in the largest cluster, C0, revealed that cell development, cell projection organization, and nervous system development were significantly influenced by Cd exposure (Fig. 3g). GO analysis of the DEGs in C13 and C15 yielded similar results (Supplementary Fig. 6b, c).
To further explore the heterogeneity of OPCs, we reclustered the OPC population into 10 subclusters (Supplementary Fig. 6d). In particular, the largest OPC cluster, C0, was divided into 7 subclusters (Supplementary Fig. 6d). The marker genes expressed in these subclusters were listed in Supplementary Table S5. The number of cells in subclusters 0, 1, 2, and 6 decreased in Cd-exposed mice, while the number of cells in subclusters 3, 4, and 5 markedly increased (Fig. 3h, i). UMAP showed the distributions of typical genes in each subcluster (Supplementary Fig. 6e). Taken together, these findings demonstrated the significant heterogeneity of the OPC population and the diverse responses of OPCs to Cd exposure.
Cd exposure disturbed the development of neuronal subpopulations
The effects of Cd exposure on progenitor cells would finally influence the generation of neurons. Thus, we analyzed the detailed changes in excitatory neuron and interneuron populations. UMAP revealed that Cd exposure caused extensive changes in the excitatory neuron population (Fig. 4a). The total number of excitatory neurons was slightly decreased in Cd-exposed mice. However, the number of excitatory neurons in C1 and C7 increased, while the number of excitatory neurons in C3 and C8 decreased in response to Cd exposure (Fig. 4b). Two clusters of interneurons were identified, and the number of cells in both of these clusters was decreased in Cd-treated mice (Fig. 4c).
a Feature plots revealing the differential distribution of excitatory neurons in control and Cd-exposed mice. b Cell distribution percentages of excitatory neuron clusters in control and Cd-exposed mice. C1, cluster 1; C3, cluster 3; C7, cluster 7; C8, cluster 8. c Cell distribution percentages of interneuron clusters in control and Cd-exposed mice. C2, cluster2; C5, cluster 5. d Venn analysis of excitatory neuron cluster marker genes and DEGs in the four excitatory neuron clusters in control and Cd-exposed groups. e Venn analysis of interneuron cluster marker genes and DEGs in control and Cd-exposed mice. f–h Violin plots showing the expression and distribution of the representative top marker genes in C1, C3, and C8 excitatory neuron clusters. i The mRNA expression of Dcx, Neurod2, Neurod6, and Tubb3 in mouse cortical tissue at different developmental stages. Data of 6 fetus from 6 pregnant mice in each group are expressed as the mean ± SEM. *p < 0.05, two-tailed Student’s t test.
Analysis of the identified genes and DEGs in the excitatory neuron clusters revealed remarkable heterogeneity of these cells and their responses to Cd exposure. A total of 719 genes and 768 DEGs were found in the 4 excitatory neuron clusters (Fig. 4d, Supplementary Table S6). The numbers of identified genes and DEGs in interneurons were much lower than those in excitatory neurons. However, interneurons also exhibited heterogeneity (Fig. 4e, Supplementary Table S6). The typical genes which played a critical role in neuronal development were presented in violin plots to show their expression patterns. Such as Neurod1, Eomes, Rnd2, and Neurog2 in C1 (Fig. 4f); Sox5 and Fezf2 in C3 (Fig. 4g); Syt4 and Satb2 in C8 (Fig. 4h). GO analysis of the DEGs in each excitatory neuron cluster demonstrated that neuronal differentiation, projection, and synapse development were significantly influenced by Cd exposure (Supplementary Fig. 7a–d). However, bulk real-time PCR analysis of previous established typical neuronal markers in the whole cortex tissue revealed that the expression of Tubb3 was slightly inhibited only at E17.5 (Fig. 4i), no difference in the expression of Dcx, Neurod2, and Neurod6 was detected at E15.5, E17.5 or P0 (Fig. 4i). These results suggested again that the Cd-induced changes were submerged in bulk analysis.
Excitatory neurons were further divided into 12 subclusters according to their unique gene profiles (Supplementary Fig. 8a, Supplementary Table S7). Among these subclusters, 2, 3, 5, and 6 were the most significantly changed groups, the number of which also achieved a dominant position (Supplementary Fig. 8b). The violin plots revealed the expression patterns of some the key identified genes in these subclusters, such as Vcan, Satb2, Ptprz1, Neurod1, Neurog2, and Gadd45g (Supplementary Fig. 8c–f). Other identified genes of each subcluster were listed in Supplementary Table S7. All these data demonstrated that the development of different neuronal subpopulations in the embryonic brain was affected heterogeneously by Cd exposure.
Pseudotime analysis verified that Cd exposure retarded brain development
To further explore the effects of Cd exposure on the chronological development and maturation of cells in the brain, we did a pseudotime map analysis. A pseudotime map was used to determine the developmental trajectories of cells by identifying cells with similar marker genes. The cells were defined into 7 stages according to the developmental stage (Fig. 5a). The distribution of all 18 cell clusters was located in the pseudotime map to identify their developmental stages (Fig. 5b). We found a distinctive shift of cell clusters on the pseudotime map caused by Cd exposure (Fig. 5c). As for the NPC populations, we found that the cells were hindered in the early developmental stage by Cd exposure in all the clusters (Fig. 5d). For OPC populations, the decreased cell number in Cd-exposed mice all happened in the later stages of development. However, in the early stages, especially in stage 1, the cell plots were much denser in the Cd-exposed group (Fig. 5e). For excitatory neuron and interneuron populations, although the influences of Cd exposure seemed muddledness due to the changes in cell number in the pseudotime map, an overall retardance of neuronal development was observed (Fig. 5f, g). Similar results have been found in astrocytes (Fig. 5h).
a Seven stages were clarified in the pseudotime map. b Distribution of different cell clusters in the pseudotime map. C10, C11, and C14 were not included due to their different origins and unclear definitions. c Cell distribution in the pseudotime map for control and Cd-exposed mice. d NPC cell cluster distribution in the pseudotime map. e OPC cell cluster distribution in the pseudotime map. f Excitatory neuron cluster distribution in the pseudotime map. g Interneuron cluster distribution in the pseudotime map. h Astrocyte cluster distribution in the pseudotime map.
Embryonic Cd exposure impaired the morphology and population Ca2+ transients of cortical neurons in adulthood
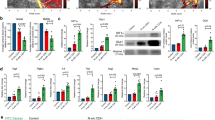
To evaluate the influence of embryonic Cd exposure on offspring mice in adulthood, we first examined the morphological changes in neurons when the offspring mice grew up to 2-month-old age. The total length of neuronal processes and the branches were not influenced by Cd exposure (Fig. 6a–c, e–g). However, the density of spines in the neuronal processes was significantly decreased in Cd-exposed mice (Fig. 6a, d, e, h). The expression of the spine development-related genes Syn1, Syn2 and Psd95 was detected at E15.5, E17.5, and P0. The data revealed that Syn2 and Psd95 expression was inhibited by Cd exposure at P0 (Fig. 6i–k).
a–d Morphological changes in neuronal processes and spine density in excitatory neurons. a Scale bar, 50 μm for the up panel and 10 μm for the down panel. b Data of 4 offspring from 4 pregnant mice in each group are expressed as the mean ± SEM. The circles in the graph represent individual cells quantified. n = 43 for the control group, n = 51 for the Cd-exposed group; (c) n = 44 for the control group, n = 53 for the Cd-exposed group; (d) n = 38 for the control group, n = 32 for the Cd-exposed group. b, d **p < 0.01, two-tailed Student’s t test. c Two-way analysis of variance (ANOVA) followed by Bonferroni’s post hoc test. e–h Morphological changes in interneurons. e Scale bar, 50 μm for the up panel and 10 μm for the down panel; (f) n = 81 for the control group, n = 62 for the Cd-exposed group; (g) n = 85 for the control group, n = 69 for the Cd-exposed group; (h) n = 51 for the control group, n = 53 for the Cd-exposed group. f, h **p < 0.01, two-tailed Student’s t test. (g) Two-way analysis of variance (ANOVA) followed by Bonferroni’s post hoc test. i–k The expression of genes related to spine development. Data of 6 fetus from 6 pregnant mice in each group are expressed as the mean ± SEM. *p < 0.05, **p < 0.01, two-tailed Student’s t test.
To assess the influence of Cd-induced morphological impairment on neural circuit functions, we monitored the Ca2+ activity of cortical neurons in free-behaving adult mice exposed to Cd in utero (Fig. 7a). A genetically encoded Ca2+ indicator GCaMP6s was expressed in the auditory cortex (AuC) of the cerebral cortex (Fig. 7b). NeuN staining of serial sections confirmed the expression of GCaMP6s in AuC neurons (Fig. 7c). Fiber photometry was used to record population Ca2+ activity. From the changes in Ca2+ transients, we found that the latency of neurons to respond to sound stimuli did not change (Fig. 7d–j). However, embryonic Cd exposure prolonged the half-width of the Ca2+ signal in adult cortical neurons (Fig. 7d–i, k) at 8944 Hz stimuli. Importantly, the amplitude and values of⊿F/F, were remarkably reduced in Cd-exposed mice at 4913 Hz and 16,284 Hz stimuli (Fig. 7d–i, l). Taken together, these data revealed that embryonic Cd exposure disturbed neural circuit functions in adults.
a Schematic of the experimental protocol. b Location of the AAV-transfected region in the cortex. c Immunostaining images of GCaMP6s and NeuN. Top, average (purple) trace of neural Ca2+ transients in the auditory cortices of control mice at the onset of 4913 Hz (d), 8944 Hz (e), and 16284 Hz (f) pure tones (black bar). The average trace was from 28 trials from four mice. The shaded area represented SEM. Bottom, color-coded intensities of neural Ca2+ transients in the cortex from different trials. Data of 4 offspring from 4 pregnant mice in each group are expressed as the mean ± SEM. The circles in the graph represent individual trials quantified. g–i Average trace of neural Ca2+ transients and color-coded intensities in the cortex in Cd-exposed mice. Data from 27 trials from four mice. j The latencies of Ca2+ transients in response to 4913 Hz, 8944 Hz, and 16284 Hz pure tones. Two-sided Wilcoxon rank-sum test. k The half-widths of Ca2+ transients. Two-sided Wilcoxon rank-sum test. 8944 Hz, *p < 0.05. I Amplitudes of the half-widths of Ca2+ transients. Two-sided Wilcoxon rank-sum test. 4913 Hz, **p < 0.01; 16284 Hz, *p < 0.05.
Embryonic Cd exposure impaired the neurobehaviors of adult mice
To further evaluate the influence of embryonic Cd exposure on offspring mice in adulthood, we carried out a series of behavioral tests. The open field test revealed that Cd-exposed mice exhibited a lack of exploratory activities. Both the time spent and total distance traveled in the center area in the open field test were decreased in the Cd-exposed mice (Fig. 8a, b). Cd-exposed mice had much sparser movement traces. Especially, they lacked center area traces compared with control mice (Fig. 8c). Next, we did a tail suspension test and an elevated plus maze, which were used to examine depression-like behavior of mice. We found that Cd-exposed mice had a much longer immobility time than control mice in the tail suspension test (Fig. 8d), and spent a much shorter time in the open arms in the elevated plus maze (Fig. 8e). These results suggested that Cd-exposed mice exhibited much higher depression-like behavior.
a–c Performance of mice in the open field test. a, b Cd-exposed mice traveled shorter distances in the center area and spent less time in the center area. Data of 9 offspring from 6 pregnant mice in each group are expressed as the mean ± SEM. *p < 0.05, **p < 0.01, two-tailed Student’s t test. c Movement traces of the mice showing that Cd exposure decreased the movement in the center area. d Mice in the Cd exposure group exhibited a much longer immobility time than control mice in the tail suspension test. Data of 10 offspring from 6 pregnant mice in each group are expressed as the mean ± SEM. **p < 0.01, two-tailed Student’s t test. e Cd-exposed mice spent less time in the open arms in the elevated plus maze. Data of 9 offspring from 6 pregnant mice in each group are expressed as the mean ± SEM. *p < 0.05, two-tailed Student’s t test. f, g Results of the passive avoidance test revealed the impairment of learning and memory functions in mice after Cd exposure. Data of 10 offspring from 6 pregnant mice in each group are expressed as the mean ± SEM. *p < 0.05, **p < 0.01, two-tailed Student’s t test. h–j Results of the Morris water maze test revealed the impairment of learning and memory in Cd-exposed mice. h Escape latencies on all 5 training days. *p < 0.05, **p < 0.01, two-way repeated measures analysis of variance (ANOVA) followed by Bonferroni’s post hoc test. i Time spent in the target quadrant during the probe trial. *p < 0.05, two-tailed Student’s t test. j Representative searching trail of mice in the probe trial. Data of 10 offspring from 6 pregnant mice in each group are expressed as the mean ± SEM.
Cognitive functions were first evaluated using a passive avoidance test. Cd-exposed mice had a much shorter latency in the retention test (Fig. 8f), and much higher numbers to enter the dark area (Fig. 8g). In the Morris water maze, Cd-exposed mice took much longer time to find the platform beginning on day 4 of the training process compared with control mice (Fig. 8h). In the probe trial, Cd-exposed mice spent much less time in the target quadrant (Fig. 8i, j). There was no difference in the average swimming speed between the control group and the Cd-exposed group (Supplementary Fig. 9), suggesting that 3 mg/L Cd exposure did not directly affect the exercise ability of mice. The results demonstrated that embryonic Cd exposure impaired the learning and memory abilities of the mice. Together, these data proved that the influences of Cd on cell development during gestation ultimately caused behavioral changes in adult mice.
Discussion
The effects and mechanisms of Cd exposure on brain development have not been fully elucidated. In this study, we revealed the diverse responses of brain cells in response to Cd exposure during embryonic development. We found that Cd exposure delayed the differentiation of embryonic NPCs and OPCs, and caused significant subpopulation shifts in these progenitor cells. Additionally, Cd exposure disturbed the generation and subpopulation distribution of excitatory neurons and interneurons, and hindered the maturation of these neurons. Furthermore, we sorted out the gene markers of each subpopulation and explored DEGs in specific cell subpopulations after Cd exposure. Finally, we uncovered that the influences of embryonic Cd exposure on brain development led to abnormal electrical activities in cortical neurons and impaired neural behaviors in adult offspring mice. These data contribute to the understanding of the effects and mechanisms of the neurodevelopmental toxicity of Cd.
Confirming the internal exposure doses of Cd is crucial for comparing its effects to human exposure outcomes. The Environmental Protection Agency in the United States39, the European Union40, and China41 have all set the maximum contaminant level for Cd in drinking water as 5 μg/L. A previous study has demonstrated that exposure of pregnant mice to 150 mg/L Cd in drinking water is equivalent to a blood Cd concentration of 4.45 μg/L on E1825, which is lower than the threshold of Cd (5 μg/L) for medical surveillance in Occupational Safety & Health Administration, USA42. The peak blood Cd level in mice resulting from 3 mg/L Cd exposure in drinking water has been confirmed closing to the blood Cd concentration in specific populations, which ranges from 0.24 to 1.94 μg/L43,44,45,46. The average internal exposure dose of Cd in the fetal brain in our experiments was 3.62 ng/g, which is also lower than the Cd content previously detected in human placental tissues47,48. Thus, the dose used in our experiments has strong environmental implications. These findings also indicate that the neurodevelopmental toxicity of Cd, even at low doses, should be considered.
Developmental brain disorders and neural behavior changes have been explored in children who exposed to Cd during gestation19,49. Similar evidence has been found in animal-based studies25,50. According to studies in humans and experimental animals, the underlying mechanisms are that Cd exposure impairs the functions of the blood-brain barrier21, induces neuroinflammation51, disturbs the development of monoaminergic, GABAergic, and glutamatergic systems22, hinders myelin and axonal development24, and influences spontaneous electrical activity52. However, these findings cannot fully explain the effects and mechanisms of Cd-induced developmental neurotoxicity. The problem could be much more serious at the cellular and molecular levels than previously reported. One major reason is that there are diverse cell subpopulations in the brain, and Cd-induced changes can be covered in bulk analysis because these cell subpopulations exhibit different reactions to Cd exposure.
Here, our data demonstrated the diverse responses of cells in the embryonic brain to Cd exposure. Even within the same types of cells, we discovered varying responses at the subpopulation level. We also revealed that each cell subpopulation had its unique DEGs after Cd exposure. This finding is important because these DEGs could be submerged in bulk analysis. In this study, when we assessed the impact of Cd on brain development using bulk real-time PCR and immunostaining analysis with commonly used markers, most of the changes at subpopulation level were covered up. Exploring the DEGs in each cell subpopulation via scRNA-seq analysis allowed us better understand the effects of Cd exposure on brain development and the underlying mechanisms involved. We also explored new sensitive genes that could be used for evaluating the developmental neurotoxicity of Cd in the brain.
NPCs give rise to OPCs, neurons, and astrocytes at different time points during brain development. Additionally, OPCs differentiate into oligodendrocytes and can also transdifferentiate into other glial cells53. Based on the changes in the number of each subpopulation, we concluded that Cd exposure hindered the differentiation of progenitor cells, including NPCs and OPCs, and impeded the maturation of neurons during brain development. This conclusion was further supported by pseudotime analysis results, which revealed that most of the cells in the brain were arrested in an immature state in Cd-exposed mice. Analysis of the functions of the feature genes of the predominately altered NPC and OPC subpopulations and the DEGs also supported this conclusion. These detailed results revealed that one of the key mechanisms by which Cd influences brain development to retard the normal development and maturation of cells in the brain. This finding is different from previous in vitro observations that Cd exposure inhibits the proliferation of NPCs27,34. This could be due to the differences between the in vivo and in vitro models used and the actual internal exposure doses of Cd. However, previous research using an in vitro model also suggested that neuronal differentiation and maturation was inhibited by Cd exposure34,54. The inhibitory effects of Cd exposure on neurogenesis have also been demonstrated in the embryonic and adult brain in vivo23,55. These findings reinforce our conclusion that Cd hindered the maturation of brain cells.
Another critical mechanism of Cd-induced neurodevelopmental toxicity was changes in cell subpopulation shifts. In particular, a significant shift was found in the NPC and OPC subpopulations after Cd exposure. Additionally, a large number of DEGs had been identified in these subpopulations. A series of meaningful changes in brain development were revealed by GO analysis of these DEGs. Most of these changes could hardly be revealed in the bulk analysis28,56. The subpopulation shifts in progenitor cells will ultimately influence their fate decision and cause significant alterations in terminal differentiation. Thus, changes in NPCs and OPCs might be one of the key mechanisms by which Cd exposure retarded and disturbed neuronal and glial differentiation in the brain. GO analysis of the DEGs in these subpopulations confirmed this conclusion, which was also further verified by the subsequent analysis of neurons.
Previous studies have revealed that Cd exposure causes oxidative stress and disturbs autophagy in cultured neurons and cortical tissues57,58. Other mechanisms, such as calcium ion (Ca2+) transfer and glycolysis promotion, have also been explored59,60. The variations were much more complicated among the subpopulations we identified. The analysis of neurons revealed a remarkable disturbance in the subpopulation distribution of excitatory neurons and interneurons. The molecular mechanisms were largely explored by DEG analysis using subpopulation data. The results suggested that Cd might also directly influence neuronal development. However, because the number of cells increased in some neuronal subpopulations while decreased in others, the typical DEGs changed in these subpopulations could be covered by this contradictory variation in bulk analysis. Thus, although no alteration was found in the bulk analysis, significant changes were detected at the subpopulation level. The changes in excitatory neuron and interneuron subpopulations provide a reasonable explanation for how Cd exposure impairs neuronal circuit formation. Subpopulation shifts in excitatory neurons and interneurons finally disturbed neuronal circuit formation. The impairment of neuronal spine development also contributed to the disturbance of neuronal circuits. Here, the prolonged half-width of Ca2+ transients following exposure to 8944 Hz stimuli indicated abnormal neuronal excitability in Cd-exposed mice. The reduced amplitudes at 4913 Hz and 16284 Hz stimuli revealed that Cd exposure decreased the sensitivity of AuC neurons. These data explored an abnormal electrical activity of cortical neurons in free-moving mice when they grew up, which indicated impaired neuronal circuit formation. The results demonstrated that Cd-induced impairments of embryonic brain development could not be fully reversed after Cd was removed. This conclusion was further confirmed by the abnormal neural behaviors found in Cd-exposed mice. Both cognitive and emotional functions were impaired in Cd-exposed mice. These results suggested that the hazardous effects of Cd exposure on neuronal circuit formation might be universal and affect multiple brain regions. Importantly, the data from free-moving mice provided direct evidence that Cd impaired neural behaviors by disturbing neuronal circuit functions. Cd-induced changes in neural behavior in humans and experimental animals have been wildly reported in previous studies61,62,63. However, the underlying mechanisms have not been fully explored. Here, we uncovered the detailed changes at the subpopulation level using single-cell technologies. These findings contributed new insight into the mechanisms by which Cd exposure affects brain development. Additionally, these findings contributed to the understanding of how embryonic neurotoxicant exposure causes neurodevelopmental disorders and neurodegenerative disease in adulthood. This study also indicated that more single-cell-based technologies should be applied to fully explore the detailed neurotoxic effects of Cd and the underlying mechanisms, especially, to explore the sensitive cell subpopulations and sensitive genes responsible for the neurodevelopmental toxicity of neurotoxicants such as Cd.
In summary, we revealed the diverse responses of brain cells to Cd exposure based on single-cell technologies. Two major effects and mechanisms by which Cd exposure affects brain development were uncovered here (Fig. 9). One is that Cd influenced embryonic brain development by retarding the normal development and maturation of cells in the brain. Another was that Cd exposure caused cell subpopulation shifts during embryonic brain development. These changes ultimately disturbed the neural circuit functions and neural behaviors of the mice when they grew up. Our data also provided direct evidence in free-moving mice that Cd exposure impaired neural behaviors by disturbing neuronal circuit functions. Our research provided new insights into the neurotoxic effect of Cd in brain development. Additionally, our findings also indicated that the influences of Cd exposure on brain development could be much greater and sophisticated than previously reported.
Methods
Animals and treatment
Eight-week-old C57BL/6J mice were used in the experiments. The mice were maintained in 23 ± 1 °C, 55 ± 5% humidity, and 12-h light/dark cycle conditions. All mice were acclimatized for 1 week before mating. Then, female mice were mated with males overnight, and the next morning the appearance of a vaginal plug was identified as E0.5. The pregnant mice were randomly assigned to either the control group (drinking water) or the Cd exposure group (drinking water containing 3 mg/L Cd, Sigma-Aldrich, Inc., MO, USA). The duration of Cd exposure was from E0.5 to E21. The dose of Cd applied in our study was based on previous reports43,44,55. The detailed timeline of animal treatment and detection was presented in Fig. 1a. Both male and female fetus or offspring were used in the experiments. All animal experimental protocols were approved by the Animal Care and Use Committee of Army Military Medical University (ethical approval number: AMUWEC20218102).
Inductively coupled plasma-mass spectrometry
The cortical tissues of 8 fetus from 8 pregnant mice in each group were collected at E15.5 and digested in 2 mL of concentrated nitric acid with a graphite digestion instrument at 120 °C for 30 min. When the solution became clear and transparent, the sample was cooled, and ultrapure water was added to 10 mL. The concentration of Cd in the cortex was measured using an inductively coupled plasma mass spectrometer (ICP-MS, NexION2000 ICP-MS, PerkinElmer, USA). The operating parameters of the ICP-MS were as follows: radiofrequency power, 1.6 kW; cooling gas flow rate, 18 L/min; auxiliary gas flow rate, 1.0 L/min; nebulizer gas flow rate, 1.02 L/min; oxygenating device flow, 0.1 L/min; peristaltic pump speed, 35 rpm; and cooling temperature, −5 °C. The samples were analyzed in triplicate. The concentration of Cd in cortical tissues was expressed as ng per g wet weight.
Single-cell RNA-seq
Cortical tissues of 4 E15.5 fetus from 4 pregnant mice in each group were isolated and digested with accutase and pancreatin (Gibco, USA). The 4 samples from the control group and the 4 samples from the Cd-exposed group were separately mixed for subsequent analysis. Cells were collected and resuspended in PBS containing 0.04% bovine serum albumin (BSA) and then filtered through a 40 μm cell strainer. Next, the cells (cell viability > 90%) were loaded into the 10X Genomics Chromium Single Cell Platform (10X Genomics, USA) at a concentration of 1000 cells/μL. Library quantification and quality control were performed with a Qubit4.0 fluorometer (Thermo Fisher Scientific, USA) and Agilent 2100 bioanalyzer (Agilent Technologies, USA). The final library pool was sequenced on an Illumina Nova6000 instrument by Shanghai Personalbio (Shanghai, China) using 150-base-pair paired-end reads.
Cell Ranger (version 6.0) was used to map reads to the GRCm38 mouse genome and acquire gene counts to generate expression matrix files. Dimension-reduction and clustering were performed by the R package Seurat (V4.1.1). Then, the objects were normalized and scaled using the NormalizeData and ScaleData functions. The cells were separated into different clusters with FindClusters based on the top 20 principal components with a resolution of 0.8. The cell clusters were visualized using uniform manifold approximation and projection (UMAP).
The Wilcoxon algorithm was used to analyze the differential expression of genes in cells within a cluster and among other clusters to identify marker genes. Differentially expressed genes (DEGs) were analyzed using the Seurat FindMarkers function based on the Wilcoxon likelihood-ratio test. Gens with an average logFC>1 or <−1 and adjusted p < 0.05 were considered DEGs. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis were performed with the “clusterProfiler” package. The cell type annotation of each cluster was done based on the expression from DEG and canonical markers in the literature with Featureplot and violin plots. Pseudotime trajectory analysis was performed with Monocle2. For the construction of the trajectory, Seurat v3.1.2 FindVairableFeatures was used.
RNA isolation and real-time PCR analysis
Cortical tissues of 6 fetus from 6 pregnant mice in each group were used in the experiments. Briefly, total RNA was isolated from the cortical tissues of fetus and offspring using TRIzol reagent (TaKaRa, Otsu, Japan). cDNA was reverse-transcribed using a reverse-transcription PCR kit (TaKaRa, Tokyo, Japan). The expression of the selected target genes was measured using a Bio-Rad IQ5 Detection System (Bio-Rad, Hercules, CA, USA) with specific primers (Supplementary Table S8). Fold changes were calculated using the 2−ΔΔCT method64.
Immunohistological staining and quantification
Immunohistological staining was performed as we described previously65. Briefly, the whole brain was fixed in 4% paraformaldehyde for 24 h and then dehydrated with 30% sucrose. The brain tissues were cut into eight-μm-thick sagittal sections. The sections were then blocked in 3% BSA containing 0.25% Triton X-100 for 1 h, and then incubated with primary antibodies overnight at 4 °C (anti-KI67, ab15580, rabbit, 1:100, Abcam, UK; anti-SOX2, AB5603, rabbit, 1:200, Millipore, USA). Alexa Fluor-conjugated secondary antibodies were used for cell visualization (donkey anti-rabbit,1:400, Invitrogen, USA). All images were captured using a confocal scanning laser microscope (Leica, SP8, Germany). Six fetus from 6 pregnant mice in each group were used for statistical analysis. Cell quantification was manually performed in a double-blinded manner. One-fifth of the serial cortex sections were collected for quantification. Cell counting and thickness measurements were performed with ImageJ software.
Golgi staining
Dendritic development in the cortex of the offspring was assessed by Golgi staining using a Rapid Golgi Stain Kit (FD NeuroTechnologies, MD). Briefly, whole brain tissue was immediately fixed in fixative for 48 h. Three-mm-thick brain tissue sections were cut and soaked in Golgi-Cox staining solution for 14 days in a cool and ventilated place away from light. The brain tissue was then immersed in 80% glacial acetic acid overnight, and placed in 30% sucrose. After dehydration, the brain tissue was cut into 100-μm-thick sections, pasted on a gelatine slide, and dried in the dark. The dried sections were treated with concentrated ammonia water for 15 min and acid hardening fixative solution for another 15 min. Multilayer images were obtained by a digital slice scanner (3DHISTECH, Hungary). Dendritic morphology and dendritic spines were analyzed using ImageJ software with the NeuronJ and Sholl plugins. Four offspring from 4 pregnant mice in each group were used for quantification.
Virus injection and optical-fiber-based Ca2+ recording in freely behaving mice
Four offspring from 4 pregnant mice in each group were used in the experiments. Ca2+ recording in freely behaving mice was carried out as previously reported66. Briefly, mice were anaesthetized with 1–2% isoflurane by inhalation. A 0.5 mm × 0.5 mm craniotomy was made at −3.1 mm anteroposterior (AP) and 3.8 mm mediolateral (ML) from bregma. A total of 80 nL AAV-hSyn-GCaMP6s (AAV2/9, titer: 4.0 × 1012 viral particles/mL) was injected into each site, starting at a depth of −0.25 mm, at a 20-degree dorsoventral (DV, from the dura) slope. The mice were allowed to recover for 4 weeks before Ca2+ recordings.
Optical-fiber-based Ca2+ signal recordings were performed using a custom-built setup (FOM-01M FiberOptoMeter, Suzhou Institute of Biomedical Engineering and Technology, China). An optical fiber (200 μm in diameter, NA 0.39, FT200UMT, Thorlabs, USA) was slowly inserted into the auditory cortex (AuC). The fiber tip was typically advanced to a depth of 1.25 mm at a 20-degree DV slope from the dura and then fixed to the skull with dental cement (Tetric EvoFlow; Ivoclar Vivadent Corporate). Ca2+ signals were recorded at 2000 Hz with customized acquisition software written in LabView (National Instrument, USA). To record Ca2+ signal in the AuC with the optical fiber, an electrostatic speaker placed ~5 cm away from the ears of the mice was used to deliver a pure tone (4913 kHz, 8944 kHz, and 16284 kHz, 70 dB sound pressure level, 1-second duration). The Ca2+ signals were calculated as relative fluorescence changes as follows:⊿F/F = (F − Fbaseline)/Fbaseline, where Fbaseline is the average of the fluorescence intensity during current recording. The brain sections were prepared for histology after the end of the fiber recording.
Behavior tests
For each test, 9 to 10 offspring from 6 pregnant mice in each group were used. To minimize the number of animals and avoid interference from different tests, each group of mice was subjected to only 2 or 3 tests and allowed to rest for 1 week between tests.
Tail suspension test
The tail suspension test was carried out to assess depression-like behavior in the mice67,68. Briefly, the mice were suspended for 6 min by tape positioned 2 cm from the end of the tail so that the heads of the mice were ~20 cm above the floor. The total immobility time in the last 4 min was recorded. When the mouse stopped moving its limbs and body, it was considered to be in an immobile state.
Open field test
The open field test was performed using a Multi Conditioning System (TSE Systems, Germany) as described in a previous study69. Briefly, the mice were individually placed in the center of a 30 × 30 × 30-cm black box in a completely airtight case and the distance traveled and time spent in the center area were recorded during a 5-min trial to evaluate anxiety-like behavior.
Elevated plus maze test
The elevated plus maze test was used to assess anxiety behavior70. The elevated plus maze consisted of two open arms (30 × 5 × 1 cm) and two closed arms (30 × 5 × 15 cm) (RWD Life Science Co., Ltd, Shenzhen, China) raising 50 cm above floor level. The mice were placed in the center of the maze facing a closed arm and away from the experimenter. The total time spent in the open arms was recorded over 5-min trial with a SMART3.0 behavioral video tracking system (Panlab, Spain).
Passive avoidance test
The passive avoidance test was used to evaluate aversive memory processing with a Multi Conditioning System 2.0 (TSE Systems, Germany)69. The main apparatus consisted of a lighted compartment (15 × 30× 26 cm) and a dark compartment (15 × 30 × 26 cm), divided by an electronically controlled door. The floor was made up of 3-mm diameter stainless steel rods spaced 1 cm apart. The avoidance task comprised a training phase and a testing phase. During the training phase, the mice were put into the lighted compartment, and the door was opened. Once the mouse entered the dark compartment, the door was closed, and a weak electrical stimulus (0.5 mA, 2 s) was delivered through the stainless-steel floor. The latency to the first entry into the dark compartment was recorded as the acquisition time. The mice were given a maximum of 300 s to enter the dark compartment. The testing phase was performed after 24 h. The mice were placed in the lighted compartment, and the latency to the first entry into the dark compartment was measured as the retention time. The number of entries into the dark compartment was also recorded, and the cut-off time was 300-s.
Morris water maze test
The Morris water maze test was performed to assess learning and memory ability as we previously described71. Briefly, the apparatus consisted of a pool with a diameter of 120 cm and a depth of 50 cm, and a platform with a diameter of 9 cm and a height of 30 cm (RWD Life Science Co., Ltd, Shenzhen, China). The mice received 4 training sessions, in which they were allowed to search the pool for the hidden platform, every day for 5 consecutive days. The mice were guided to the platform and allowed to stay there for 15 s if they failed to find the platform within 60 s. On the 6th day, the mice were allowed to swim freely for 60 s in the pool without the platform. The average swimming speed, latency to reach the platform, and time spent in the target quadrant were recorded using a SMART3.0 behavioral video tracking system (Panlab, Spain).
Statistics and reproducibility
The data are expressed as the mean ± standard error of the mean (SEM). The data were analyzed using GraphPad Prism 8 and IBM SPSS version 22.0. Two-tailed unpaired student’s t test and two-way analysis of variance (ANOVA) followed by Bonferroni’s post hoc test were performed to analyze the differences between the control group and the Cd exposure group when the data were normally distributed and had homogeneous variances; otherwise, a two-sided Wilcoxon rank-sum test was performed. Statistical significance was set at p < 0.05.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Single-cell RNA-sequence raw data are available in the Sequence Read Archive (SRA) under the accession number PRJNA1131648. The source data behind the graphs in the paper can be found in Supplementary Data 1.
Code availability
Cell Ranger (version 6.0), R package Seurat (V4.1.1), and Seurat v3.1.2 are used for data analysis of the single-cell RNA-seq. ImageJ software with the NeuronJ and Sholl plugins are used for evaluation of the dendritic morphology and spines. LabView (National Instrument, USA) are used to record Ca2+ signals. The Multi Conditioning System 2.0 (TSE Systems, Germany) and SMART3.0 behavioral video tracking system (Panlab, Spain) are used for data collection and analysis in behavior tests. GraphPad Prism 8 and IBM SPSS version 22.0 are used for statistical analysis. The detailed information is descripted in the Methods section of the paper.
References
Sarigiannis, D. A. et al. Neurodevelopmental exposome: the effect of in utero co-exposure to heavy metals and phthalates on child neurodevelopment. Environ. Res. 197, 110949 (2021).
Alampi, J. D. et al. Association between gestational exposure to toxicants and autistic behaviors using Bayesian quantile regression. Am. J. Epidemiol. 190, 1803–1813 (2021).
Nyanza, E. C. et al. Effects of prenatal exposure and co-exposure to metallic or metalloid elements on early infant neurodevelopmental outcomes in areas with small-scale gold mining activities in Northern Tanzania. Environ. Int. 149, 106104 (2021).
Leppert, B. et al. Association of maternal neurodevelopmental risk alleles with early-life exposures. Jama. Psychiatry. 76, 834–842 (2019).
Pu, Y. et al. Maternal glyphosate exposure causes autism-like behaviors in offspring through increased expression of soluble epoxide hydrolase. Proc. Natl Acad. Sci. USA 117, 11753–11759 (2020).
Reuben, A. et al. Association of childhood lead exposure with MRI measurements of structural brain integrity in midlife. JAMA 324, 1970–1979, (2020).
Wang, Y. et al. Source apportionment of soil heavy metals: a new quantitative framework coupling receptor model and stable isotopic ratios. Environ. Pollut. 314, 120291 (2022).
Martinkova, E. et al. Cadmium isotope fractionation of materials derived from various industrial processes. J. Hazard. Mater. 302, 114–119 (2016).
Wang, R. et al. Cadmium in food: Source, distribution and removal. Food Chem. 405, 134666 (2023).
Zhang, Y. et al. Health risk assessment of cadmium exposure by integration of an in silico physiologically based toxicokinetic model and in vitro tests. J. Hazard. Mater. 443, 130191 (2023).
Luo, H. et al. Cadmium exposure induces osteoporosis through cellular senescence, associated with activation of NF-kappaB pathway and mitochondrial dysfunction. Environ. Pollut. 290, 118043 (2021).
Hong, H. et al. Cadmium exposure impairs pancreatic beta-cell function and exaggerates diabetes by disrupting lipid metabolism. Environ. Int. 149, 106406 (2021).
Gari, M. et al. Prenatal exposure to neurotoxic metals and micronutrients and neurodevelopmental outcomes in early school age children from Poland. Environ. Res. 204, 112049 (2022).
Skogheim, T. S. et al. Metal and essential element concentrations during pregnancy and associations with autism spectrum disorder and attention-deficit/hyperactivity disorder in children. Environ. Int. 152, 106468 (2021).
Liu, J. et al. Interaction of prenatal bisphenols, maternal nutrients, and toxic metal exposures on neurodevelopment of 2-year-olds in the APrON cohort. Environ. Int. 155, 106601 (2021).
Everson, T. M. et al. Placental expression of imprinted genes, overall and in sex-specific patterns, associated with placental cadmium concentrations and birth size. Environ. Health Perspect. 127, 57005 (2019).
Raja, G. L., Subhashree, K. D. & Kantayya, K. E. In utero exposure to endocrine disruptors and developmental neurotoxicity: Implications for behavioural and neurological disorders in adult life. Environ. Res. 203, 111829 (2022).
Karakis, I. et al. Maternal metal concentration during gestation and pediatric morbidity in children: an exploratory analysis. Environ. Health Prev. Med. 26, 40 (2021).
Ma, C. et al. Association of prenatal exposure to cadmium with neurodevelopment in children at 2 years of age: The Japan Environment and Children’s Study. Environ. Int. 156, 106762 (2021).
Liu, C. et al. Association of both prenatal and early childhood multiple metals exposure with neurodevelopment in infant: a prospective cohort study. Environ. Res. 205, 112450 (2022).
Zhang, T. et al. Cadmium-induced dysfunction of the blood-brain barrier depends on ROS-mediated inhibition of PTPase activity in zebrafish. J. Hazard. Mater. 412, 125198 (2021).
Antonio, M. T., Peinado, V., Gonzalez, J. C. & Leret, M. L. Effects of maternal cadmium administration on development of monoaminergic, GABAergic and glutamatergic systems. Environ. Toxicol. Pharm. 29, 87–90 (2010).
Chow, E. S., Hui, M. N., Lin, C. C. & Cheng, S. H. Cadmium inhibits neurogenesis in zebrafish embryonic brain development. Aquat. Toxicol. 87, 157–169 (2008).
Rai, N. K. et al. Exposure to As, Cd, and Pb-mixture impairs myelin and axon development in rat brain, optic nerve and retina. Toxicol. Appl. Pharm. 273, 242–258 (2013).
Liu, W. B. et al. Environmental cadmium exposure during gestation impairs fetal brain and cognitive function of adult offspring via reducing placenta-derived E2 level. Chemosphere 307, 135668 (2022).
Minetti, A. & Reale, C. A. Sensorimotor developmental delays and lower anxiety in rats prenatally exposed to cadmium. J. Appl Toxicol. 26, 35–41 (2006).
Luo, H. et al. Cadmium inhibits neural stem/progenitor cells proliferation via MitoROS-dependent AKT/GSK-3beta/beta-catenin signaling pathway. J. Appl. Toxicol. 41, 1998–2010 (2021).
Deng, P. et al. Transcriptomic insight into cadmium-induced neurotoxicity in embryonic neural stem/progenitor cells. Toxicol. In Vitro 62, 104686 (2020).
Song, B. et al. Single-cell transcriptomic analysis reveals the adverse effects of cadmium on the trajectory of neuronal maturation. Cell Biol. Toxicol. https://doi.org/10.1007/s10565-022-09775-5 (2022).
Zeisel, A. et al. Molecular architecture of the mouse nervous system. Cell 174, 999–1014.e1022 (2018).
Zeng, B. et al. The single-cell and spatial transcriptional landscape of human gastrulation and early brain development. Cell Stem Cell 30, 851–866.e857 (2023).
Gong, Y. et al. Ex utero monkey embryogenesis from blastocyst to early organogenesis. Cell 186, 2092–2110.e2023 (2023).
Herring, C. A. et al. Human prefrontal cortex gene regulatory dynamics from gestation to adulthood at single-cell resolution. Cell 185, 4428–4447.e4428 (2022).
Song, B. et al. Single-cell RNA sequencing of mouse neural stem cell differentiation reveals adverse effects of cadmium on neurogenesis. Food Chem. Toxicol. 148, 111936 (2021).
Noctor, S. C., Flint, A. C., Weissman, T. A., Dammerman, R. S. & Kriegstein, A. R. Neurons derived from radial glial cells establish radial units in neocortex. Nature 409, 714–720 (2001).
Zhong, S. et al. A single-cell RNA-seq survey of the developmental landscape of the human prefrontal cortex. Nature 555, 524–528 (2018).
Polioudakis, D. et al. A single-cell transcriptomic atlas of human neocortical development during mid-gestation. Neuron 103, 785–801.e788 (2019).
Fan, X. et al. Single-cell transcriptome analysis reveals cell lineage specification in temporal-spatial patterns in human cortical development. Sci. Adv. 6, eaaz2978 (2020).
EPA, USEPA. Edition of the drinking water standards and health advisories. Washington, DC: U.S. Environmental Protection Agency, Office of Water. EPA822-F-18-001. https://www.epa.gov/system/files/documents/2022-01/dwtable2018.pdf (2018).
Final review of scientific information on cadmium, version of December 2010. United Nations Environment Programme, https://www.unep.org/resources/report/final-review-scientific-information-cadmium (2010).
Standards for drinking water quality. National Disease Control and Prevention Administration, the People’s Republic of China. GB 5749-2022. https://www.ndcpa.gov.cn/jbkzzx/c100201/common/content/content_1665979083259711488.html.
Faroon, O. et al. in Toxicological Profile for Cadmium (Agency for Toxic Substances and Disease Registry, 2012).
Wang, H., Zhang, L., Abel, G. M., Storm, D. R. & Xia, Z. Cadmium exposure impairs cognition and olfactory memory in male C57BL/6 mice. Toxicol. Sci. 161, 87–102 (2018).
Wang, H., Abel, G. M., Storm, D. R. & Xia, Z. Adolescent cadmium exposure impairs cognition and hippocampal neurogenesis in C57BL/6 mice. Environ. Toxicol. 37, 335–348 (2022).
Sakellari, A. et al. Predictors of cadmium and lead concentrations in the blood of residents from the metropolitan area of Athens (Greece). Sci. Total Environ. 568, 263–270 (2016).
Zhang, L. L. et al. Baseline blood levels of manganese, lead, cadmium, copper, and zinc in residents of Beijing suburb. Environ. Res 140, 10–17 (2015).
Esteban-Vasallo, M. D., Aragones, N., Pollan, M., Lopez-Abente, G. & Perez-Gomez, B. Mercury, cadmium, and lead levels in human placenta: a systematic review. Environ. Health Perspect. 120, 1369–1377 (2012).
Xu, L. et al. Differential proteomic expression of human placenta and fetal development following e-waste lead and cadmium exposure in utero. Sci. Total Environ. 550, 1163–1170 (2016).
Tung, P. W. et al. Association between placental toxic metal exposure and NICU Network Neurobehavioral Scales (NNNS) profiles in the Rhode Island Child Health Study (RICHS). Environ. Res 204, 111939 (2022).
Ouyang, L. et al. Cognitive outcomes caused by low-level lead, cadmium, and mercury mixture exposure at distinct phases of brain development. Food Chem. Toxicol. 175, 113707 (2023).
Huang, Y. et al. Exposure to cadmium induces neuroinflammation and impairs ciliogenesis in hESC-derived 3D cerebral organoids. Sci. Total Environ. 797, 149043 (2021).
Kocovic, D. M. et al. Cadmium versus lanthanum effects on spontaneous electrical activity and expression of connexin isoforms Cx26, Cx36, and Cx45 in the Human Fetal Cortex. Cereb. Cortex 30, 1244–1259 (2020).
Weng, Q. et al. Single-cell transcriptomics uncovers glial progenitor diversity and cell fate determinants during development and gliomagenesis. Cell Stem Cell 24, 707–723.e708 (2019).
Tasneem, S., Farrell, K., Lee, M. Y. & Kothapalli, C. R. Sensitivity of neural stem cell survival, differentiation and neurite outgrowth within 3D hydrogels to environmental heavy metals. Toxicol. Lett. 242, 9–22 (2016).
Wang, H., Abel, G. M., Storm, D. R. & Xia, Z. Cadmium exposure impairs adult hippocampal neurogenesis. Toxicol. Sci. 171, 501–514 (2019).
Hudson, K. M. et al. Transcriptomic, proteomic, and metabolomic analyses identify candidate pathways linking maternal cadmium exposure to altered neurodevelopment and behavior. Sci. Rep. 11, 16302 (2021).
Li, M. et al. Melatonin antagonizes cadmium-induced neurotoxicity by activating the transcription factor EB-dependent autophagy-lysosome machinery in mouse neuroblastoma cells. J. Pineal Res. 61, 353–369 (2016).
Al Olayan, E. M., Aloufi, A. S., AlAmri, O. D., El-Habit, O. H. & Abdel Moneim, A. E. Protocatechuic acid mitigates cadmium-induced neurotoxicity in rats: role of oxidative stress, inflammation and apoptosis. Sci. Total Environ. 723, 137969 (2020).
Bovio, F., Melchioretto, P., Forcella, M., Fusi, P. & Urani, C. Cadmium promotes glycolysis upregulation and glutamine dependency in human neuronal cells. Neurochem. Int. 149, 105144 (2021).
Wang, T. et al. Ca(2+) transfer via the ER-mitochondria tethering complex in neuronal cells contribute to cadmium-induced autophagy. Cell Biol. Toxicol. 38, 469–485 (2022).
Nagymajtenyi, L., Schulz, H. & Desi, I. Behavioural and functional neurotoxicological changes caused by cadmium in a three-generational study in rats. Hum. Exp. Toxicol. 16, 691–699 (1997).
Hill, D. S. et al. Autism-like behavior and epigenetic changes associated with autism as consequences of in utero exposure to environmental pollutants in a mouse model. Behav. Neurol. 2015, 426263 (2015).
Liu, Z., Cai, L., Liu, Y., Chen, W. & Wang, Q. Association between prenatal cadmium exposure and cognitive development of offspring: a systematic review. Environ. Pollut. 254, 113081 (2019).
Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408 (2001).
He, Z. et al. SOX2 modulated astrocytic process plasticity is involved in arsenic-induced metabolic disorders. J. Hazard. Mater. 435, 128942 (2022).
Zhang, Q. et al. Locomotion-related population cortical Ca(2+) transients in freely behaving mice. Front. Neural Circuits 11, 24 (2017).
Poleszak, E. et al. Antidepressant-like activity of typical antidepressant drugs in the forced swim test and tail suspension test in mice is augmented by DMPX, an adenosine A(2A) receptor antagonist. Neurotox. Res 35, 344–352 (2019).
Chen, C. et al. Astrocyte-specific deletion of Sox2 promotes functional recovery after traumatic brain injury. Cereb. Cortex 29, 54–69 (2019).
Zhou, C. et al. Histone hypoacetylation contributes to neurotoxicity induced by chronic nickel exposure in vivo and in vitro. Sci. Total Environ. 783, 147014 (2021).
Takagi, T., Nishizaki, Y., Matsui, F., Wakamatsu, N. & Higashi, Y. De novo inbred heterozygous Zeb2/Sip1 mutant mice uniquely generated by germ-line conditional knockout exhibit craniofacial, callosal and behavioral defects associated with Mowat-Wilson syndrome. Hum. Mol. Genet. 24, 6390–6402 (2015).
Chen, C. et al. Thyroid hormone promotes neuronal differentiation of embryonic neural stem cells by inhibiting STAT3 signaling through TRalpha1. Stem Cells Dev. 21, 2667–2681 (2012).
Acknowledgements
This work was supported by grants from the Chongqing Talents: Excellent Scientist Project (Grant No. cstc2024ycjh-bgzxm0069), and the National Natural Science Foundation of China (Grant No. U2130107 and 31870842).
Author information
Authors and Affiliations
Contributions
Conceived and designed experiments: C.C. and Z.Z. Performed the experiments: Q.M., Z.Y., C.Y., M.L., M.G., P.D., M.H., Y.L., K.Z., H.P., and M.Q. Analyzed the data: C.C., Q.M., and Z.Y. Wrote the manuscript: C.C. Revised the article: Z.Y. and Z.Z. All authors approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks the anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editors: Christoph Anacker and Benjamin Bessieres. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Ma, Q., Yang, Z., Yang, C. et al. A single-cell transcriptomic landscape of cadmium-hindered brain development in mice. Commun Biol 7, 997 (2024). https://doi.org/10.1038/s42003-024-06685-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s42003-024-06685-6