Abstract
Colorectal cancer (CRC) ranks as the second most lethal cancer worldwide because of its high rate of metastasis, and approximately 20% of CRC patients have metastases at initial diagnosis. Metabolic reprogramming, a hallmark of cancer cells, has been implicated in the process of metastasis. We previously demonstrated that fucosyltransferase 2 (FUT2) promotes the malignancy of CRC cells, however, the underlying mechanisms remain unclear. Here, bioinformatic analysis revealed that FUT2 is associated with the malignant phenotype and fatty acid metabolism in CRC. FUT2 knockdown decreased glucose uptake and de novo fatty acid synthesis, which in turn inhibited the proliferation and metastasis of CRC cells. Mechanistically, FUT2 promotes YAP1 nuclear translocation and stabilizes mSREBP-1 by fucosylation, thus promoting de novo fatty acid synthesis in CRC cells. In summary, this study demonstrates that FUT2 promotes the proliferation and metastasis of CRC cells by reprogramming fatty acid metabolism via YAP/TAZ signaling and SREBP-1, indicating that FUT2 might be a potential target for developing therapeutic strategies against CRC.

Similar content being viewed by others
Introduction
Colorectal cancer (CRC) is one of the most common and second deadly cancers globally, and its incidence and mortality are increasing rapidly1,2. The high mortality of CRC is due to tumor metastasis, which accounts for 90% of CRC deaths and less than 20% of the 5-year survival rates3. CRC can spread through the body and form distant metastases in the lymph nodes, lungs, liver, peritoneum and abdominal cavity4. Even with considerable progress in the detection and management of CRC, the survival rates for patients are still significantly low, primarily due to tumor metastasis or recurrence5,6. Therefore, it is urgently necessary to clarify the underlying mechanisms of CRC metastasis and develop new therapies to improve the outcomes of CRC patients. Metabolic reprogramming is commonly observed in cancers, and recognized as a hallmark of cancer7. Among these dysregulated metabolic pathway, abnormal lipid metabolism plays a major role in tumorigenesis and metastasis8. Lipid metabolic reprogramming provides cancer cells with components for the synthesis of cellular membranes, generation of energy, and signal transduction9,10. Research has showed that dysregulated lipid metabolism plays a vital role in facilitating CRC invasion and metastasis11,12. However, the mechanisms underlying the reprogramming of lipid metabolism in CRC remain poorly understood.
Glycosylation, a ubiquitous and essential post-translational modification, plays crucial roles in cell growth, cell adhesion, immune tolerance, and metabolic reprogramming etc13,14. Accumulating evidences show that aberrant glycosylation, a common feature of cancer cells, plays important roles in metabolic reprogramming in cancer15,16. Researchers have shown that fucosyltransferases are abnormally regulated in CRC and linked to multiple aspects of cancer pathology17. Our prior research revealed that fucosyltransferase 2 (FUT2), which is responsible for the addition of fucose to the α-1, 2 linkage at glycan termini, was upregulated in CRC, and promoted the progression of CRC18. However, the role of FUT2 in CRC metastasis is unclear.
The Hippo signaling pathway, an evolutionarily conserved pathway, regulates multiple biological processes and plays an important role in the tumorigenesis and metastasis of cancer, including metabolic reprogramming19. As a key regulator of the Hippo pathway, YAP is predominantly regulated by phosphorylation20. When Hippo signaling is activated, YAP is inactivated by phosphorylation and constrained in the cytoplasm. In contrast, when Hippo is inactive, YAP is activated by dephosphorylation and shuttled into the nucleus21. Recently, studies have indicated that YAP plays important role in metabolic regulation, such as lipogenesis, fatty acid degradation, glycolysis and glutamate decomposition, and has received increasing attention of the metabolism reprogramming in cancer cells22. The effect of YAP nuclear-cytoplasmic transport can be regulated by phosphorylation, methylation, ubiquitination, and glycosylation. The OGT-mediated glycosylation of YAP induces YAP dephosphorylation and activation to promote tumor growth and metastasis23. The polypeptide N-acetylgalactosaminyltransferase 12 (GALNT12) promotes YAP1 nuclear translocation, which activates key downstream genes, therefore promoting fibrosarcoma growth24.
In this study, we confirmed that upregulated FUT2 was associated with lipid metabolism in CRC. FUT2 promoted the transcription of glucose transporter type 1 (GLUT1) and fatty acid synthases through YAP/TAZ signaling, through which YAP1 was dephosphorylated and activated by fucosylation in CRC cells. Additionally, the fucosylation of mSREBP-1 by FUT2 enhances its stability, further promoting de novo fatty acid synthesis, thereby promoting CRC metastasis. This study provides new insight into the mechanism of lipid metabolism in CRC and potential targets for future therapies.
Results
FUT2 is upregulated in CRC and correlated with clinical outcomes
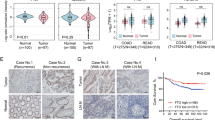
We previously reported that FUT2 is upregulated in clinical CRC tissue, compared with matched normal tissues from the same patients18. Here, the data from The Cancer Genome Atlas (TCGA) and Genote-Tissue Expression (GTEx) databases revealed that FUT2 gene expression is elevated in CRC tumors compared with normal colorectal tissues (Fig. 1A). Immunohistochemistry results from the Human Protein atlas (HPA) database also revealed that FUT2 was significantly elevated in CRC tissues compared with normal tissues (Fig. 1B). Additionally, receiver operating characteristic (ROC) curve analysis revealed that FUT2 had an area under the curve (AUC) value of 0.78 (sensitivity 98%, specificity 55%) (Fig. 1C), indicating that FUT2 could aid in the diagnosis of CRC. On the basis of the expression levels of FUT2, the correlation between FUT2 and CRC clinical parameters was investigated via the chi-square test. The results revealed that the expression of FUT2 was associated with race, M stage, N stage, and pathologic stage (Supplementary Table 1). The stromal and ESTIMATE scores were higher in the high-FUT2 expression group than in the low-FUT2 expression group, while no notable variance was observed in the immune scores (Fig. 1D). To further investigate the role of FUT2 in the tumor microenvironment, we analyzed the single-cell RNA (scRNA) sequence dataset GSE146771 from the Tumor Immune Single-cell Hub (TISCH) database. TSNE plots displayed thirteen distinct cell clusters (Fig. 1E), in which FUT2 was predominantly localized in malignant tumor cells (Fig. 1F, G). Notably, elevated fatty acid metabolism was also detected in malignant tumor cells (Fig. 1H). Expanding our investigation to TCGA-CRC patients, we found that the lipid metabolism pathway significantly impacts CRC prognosis in high-risk patients (Supplementary Fig. 1A–D). Furthermore, gene set enrichment analysis (GSEA) demonstrated that FUT2 was positively correlated with fatty acid metabolism in CRC patients (P = 0.00052, NES = 1.78) (Fig. 1I). These findings suggest that FUT2 is associated with fatty acid metabolism in CRC.
A The mRNA levels of FUT2 in normal (n = 359) and CRC (n = 616) specimens were quantified. Relevant data processing details of the specimens were provided in the methods section. B Representative immunohistochemistry (IHC) images from the Human Protein Atlas (HPA) illustrating the expression of FUT2 in CRC and normal tissues. Scale bar represents 200 μm. C Receiver operating characteristic (ROC) curve analysis of FUT2 mRNA expression in CRC (n = 975), showing an AUC of 0.782, with a sensitivity of 98% and specificity of 55%. Relevant data processing details were provided in the methods section. D Tumor microenvironment (TME) score categorized by the expression levels of FUT2 in the TCGA-CRC dataset, indicating its correlation with the stromal score. Relevant data processing details were provided in the methods section. E TSNE plot illustrating cell type clustering in the GSE146771 dataset, highlighting the distribution of various cell populations. Relevant data processing details were provided in the methods section. F TSNE plot depicting the distribution of FUT2 expression across different cell subsets within the GSE146771 dataset, indicating its correlation with malignancy. Relevant data processing details were provided in the methods section. G Violin plot representing FUT2 expression at the single-cell level within the GSE146771 dataset, showing variability among different cell types. Relevant data processing details were provided in the methods section. H TSNE plot of the gene set associated with fatty acid metabolism across cell subsets within the GSE146771 dataset, providing insights into metabolic reprogramming. Relevant data processing details were provided in the methods section. I GSEA comparing high and low FUT2 expression groups in the TCGA-CRC dataset, revealing a significant enrichment of fatty acid metabolism pathways (P = 0.00052, NES = 1.78). Relevant data processing details were provided in the methods section. Statistical significance was determined using Wilcoxon rank test, with *p < 0.05, ****p < 0.0001, ns: not significant.
FUT2 enhances fatty acid synthesis in CRC cells
To investigate the biological role of FUT2 in lipid metastasis in CRC cells, specific shRNAs targeting FUT2 were introduced into SW480 and DLD-1 cells to silence FUT2 expression in CRC cells, and the effects were assessed by Western blot. Compared with that in NC cells, the expression of FUT2 in sh-FUT2 cells was significantly lower (Fig. 2A). To ensure that the fucosylation was catalyzed by FUT2, not FUT1, which also catalyze the addition of alpha 1, 2-linked fucose to glycans, the expression of FUT1 was investigated. As shown in Fig. 2B, the expression of FUT1 was not affected by FUT2 knockdown. FUT2 knockdown significantly decreased the intracellular levels of triglyceride (TG), total cholesterol (T-CHO) and free fatty acid (FFA) in SW480 and DLD-1 cells (Fig. 2C). The lipid content, as detected by the fluorescent lipophilic stain Nile Red, revealed that FUT2 knockdown significantly reduced the neutral lipids in SW480 and DLD-1 cells (Fig. 2D). Additionally, FUT2 knockdown markedly reduced the levels of TG and T-CHO in subcutaneous tumor models (Fig. 2E). To investigate whether the FUT2 induced increase in lipid accumulation was due to its regulation of the de novo fatty acid synthesis pathway, the key enzymes involved in de novo fatty acid synthesis were examined. As shown in Fig. 2F, knockdown of FUT2 significantly decreased the expression of acetyl-CoA carboxylase α (ACC 1), ATP citrate lyase (ACLY), carnitine palmitoyltransferase 1 A (CPT1A), and fatty acid synthetase (FASN), whereas the levels of adipose triglyceride lipase (ATGL) were markedly increased in both SW480 and DLD-1 cells. These results indicate that FUT2 enhances de novo fatty acid synthesis in CRC cells.
A SW480 and DLD-1 cells were stably transfected with FUT2-shRNA, and then western blot analysis of FUT2 (A)/FUT1 B expression was determined. The relative density of protein bands was quantified, normalized to β-actin/GAPDH, and fold changes were shown in histograms from three independent experiments. C SW480 and DLD-1 cells were transfected with FUT2-siRNA, and the levels of TG, T-CHO, and FFA were detected. Data shown are mean ± SD; n = 3 samples. D SW480 and DLD-1 cells were transfected with FUT2-siRNA, and then stained with Nile red to evaluate intracellular neutral lipid accumulation. Red fluorescence signals were captured and visualized through a fluorescent microscope using constant fluorescence parameters explained in the methods section: scale bar, 50 μm. E BALB/c nude mice were subcutaneously implanted with the FUT2-knockdown (sh-FUT2) or control (NC) SW480 cells. After 30 days of transplantation, the levels of TG and T-CHO in tumor specimens (n = 6 mice/group) were detected. F SW480 and DLD-1 cells were transfected with FUT2-shRNA, and then western blot analysis of key lipid metabolism enzymes, including ACLY, ACC1, FASN, CPT1A, and ATGL expressions were determined. The relative density of protein bands was quantified, normalized to β-actin or GAPDH, and fold changes were shown in histograms from three independent experiments. Statistical significance was determined using one-way ANOVA using the Bonferroni post-hoc test, with *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, ns: not significant.
FUT2 promotes CRC cell growth and metastasis by enhancing fatty acid synthesis
The reprogramming of fatty acid metabolism can accelerate the development and progression of CRC25. To further explore whether FUT2-mediated fatty acid synthesis promotes CRC cell growth and metastasis, palmitic acid, the most common long-chain fatty acid, was used. The results showed that palmitic acid treatment markedly promoted cell proliferation and colony formation, which were significantly attenuated by FUT2 knockdown (Fig. 3A, B). As shown in Fig. 3C, FUT2 knockdown markedly suppressed palmitic acid-induced migration and invasion of CRC. Overexpression of FUT2 significantly increased the migration and invasion of CRC cells (Fig. 3D and Supplementary Fig. 2). Meanwhile, treated with orlistat, a fatty acid synthase inhibitor, the migration and invasion of CRC cells were significantly reduced with or without FUT2 overexpression. These results suggest that FUT2 promotes CRC cell growth and metastasis by enhancing fatty acid synthesis.
SW480 and DLD-1 cells were transfected with FUT2-shRNA, and then CCK8 assay (A) and colony formation assay (B) were performed to assess the proliferative capacity of cells with or without 50 μM palmitic acid. Data shown are mean ± SD; n = 3 samples. SW480 and DLD-1 cells transfected with FUT2-shRNA or FUT2/Lv105. After transfection, cells were treated with 50 μM palmitic acid (C) or 50 μM orlistat (D) for 24 h and then transwell assay was performed. The number of migrating/invading cells was quantified using Image J software. Scale bar represents 50 μm. Data shown are mean ± SD; n = 3 samples. E SW480 cells were transiently transfected with FUT2-siRNA, and then stained with phalloidin to evaluate intracellular cytoskeleton organization. Green fluorescence signals were captured and visualized through a fluorescent microscope using constant fluorescence parameters explained in the methods section: scale bar, 50 μm. F SW480 cells were transiently transfected with FUT2/Lv105. After transfection, the cells were treated with orlistat (50 μM) for 24 h, and then stained with phalloidin to evaluate intracellular cytoskeleton organization. Green fluorescence signals were captured and visualized through a fluorescent microscope using constant fluorescence parameters explained in the methods section: scale bar, 50 μm. Statistical significance was determined using one-way ANOVA using the Bonferroni post-hoc test, with *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, ns: not significant.
Elevated levels of membrane lipid rafts, which affect signaling and protein localization at the cell membrane, in turn affect the dynamics of the cytoskeleton, which plays a crucial role in tumor progression, including migration, invasion, and colonization at new sites26,27. Phalloidin staining revealed that FUT2 knockdown significantly reduced cytoskeleton formation (Fig. 3E), and FUT2 overexpression increased cytoskeleton formation (Fig. 3F). The FUT2-mediated increase in cytoskeleton formation was significantly reduced by orlistat treatment (Fig. 3F). The results showed that blocking fatty acid synthesis significantly inhibited cytoskeleton formation, confirming the role of FUT2 in the regulation of lipid metabolism in CRC cells.
Dysregulated fatty acid metabolism can cause cellular stress, such as oxidative stress, which in turn affects cytoskeletal stability and cell viability28. The results revealed that FUT2 knockdown significantly increased the levels of ROS and MDA (Fig. 4A, B). Moreover, FUT2 knockdown markedly decreased the level of cellular glutathione (GSH), an essential antioxidant that protects cells against various forms of prooxidants and free radicals (Fig. 4C). Increased lipid peroxidation is lethal to cells, whereas ferroptosis, a form of non-apoptotic cell death, is triggered by excessive lipid peroxidation-induced damage and iron accumulation29. To further investigate the role of FUT2 in CRC, the effect of FUT2 on ferroptosis was detected. FUT2 knockdown significantly decreased the expression of FTH and GPX4 and increased the expression of TFR1 (Fig. 4D). We further evaluated the role of FUT2 in ferroptosis by assessing RSL3-induced ferroptosis with or without ferrostatin-1 (Fer-1), an inhibitor of ferroptosis. FUT2 knockdown significantly increased the MDA production of RSL-3 treated SW480 cells (Fig. 4E), whereas FUT2 overexpression markedly decreased the MDA production of RSL-3 treated SW480 cells (Fig. 4F). Moreover, after Fer-1 treated, the MDA production of SW480 cells was not affected by FUT2 knockdown or overexpression. Induction of cellular ferroptosis has already been shown to overcome cancer resistance to cisplatin30. We further investigated the impact of FUT2 on CRC cells following treatment with cisplatin by viability assays. Notably, FUT2 knockdown enhanced the sensitivity of CRC cells to cisplatin (Fig. 4G).
A SW480 and DLD-1 cells were transfected with FUT2-shRNA, and then the ROS level was assessed by flow cytometry. The bar graph showing ROS level was expressed as a percentage of the control. Data shown are mean ± SD; n = 3 samples. B, C SW480 cells were transfected with FUT2-shRNA, and then the levels of MDA, GSH and glutathione disulfide (GSSG) levels were assessed. Data shown are mean ± SD; n = 3 samples. D SW480 cells were transfected with FUT2-shRNA, and then western blot analysis of key ferroptosis markers, including GPX4, FTH, and TFR1 expressions were determined. The relative density of protein bands was quantified, normalized to α-tubulin, and fold changes were shown in histograms from three independent experiments. SW480 cells were transiently transfected with FUT2-shRNA (E) or FUT2/Lv105 (F). After transfection, the cells pre-treated with RSL3 (2 μM) were treated with ferrostatin (Fer-1) (1 μM) for 24 h, and then the level of MDA was determined. Data shown are mean ± SD; n = 3 samples. G Cell viability assay in SW480 cells treated with indicated concentrations of cisplatin for 24 h, showing that FUT2 expression modulates cisplatin sensitivity. Data shown are mean ± SD; n = 3 samples. Statistical significance was determined using one-way ANOVA using the Bonferroni post-hoc test, with *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, ns: not significant.
FUT2 promotes the migration and invasion of CRC cells via YAP/TAZ signaling
To investigate the underlying mechanism of FUT2 in CRC, GO and KEGG enrichment analyses were conducted on genes differentially expressed in relation to FUT2 in the TCGA-CRC dataset. GO analysis revealed that FUT2 was associated with cell adhesion, regulation of the actin cytoskeleton, and fatty acid synthesis (Fig. 5A), and KEGG analysis revealed that FUT2 was significantly linked to the Hippo pathway (Fig. 5B). To confirm this, key factors of the Hippo pathway, including CTGF and CYR61, which are critical in the metastatic process of tumor cells, were tested. FUT2 knockdown had no effect on the expression of YAP1 or LATS1 in SW480 cells, whereas the expression of TAZ, CTGF and CYR61 was significantly reduced, and the phosphorylation of YAP1 (Ser127) was significantly increased (Fig. 5C). Additionally, the effects of FUT2 on YAP1 distribution were analyzed via a cell fractionation assay. Compared with NC, FUT2 knockdown significantly decreased the nuclear fraction of YAP1 and increased the cytoplasmic fraction of YAP1 in SW480 cells (Fig. 5D), indicating that FUT2 promotes YAP1 translocation into the nucleus. The level of TAZ in the nucleus was significantly decreased in FUT2 knockdown SW480 cells, compared with that in NC cells (Fig. 5D). The expression of CTGF, a downstream factor of the YAP/TAZ signaling, was significantly reduced by FUT2 knockdown or YAP1/TAZ knockdown (Fig. 5E, F). Verteporfin, an inhibitor of YAP1, was used to investigate whether FUT2 promotes CRC cell metastasis through YAP/TAZ signaling. As shown in Fig. 5G, the migration and invasion of CRC cells were significantly reduced in simultaneously knockdown of FUT2 and verteporfin treatment CRC cells, compared with that in knockdown FUT2 or verteporfin treatment CRC cells. Overexpression of FUT2 increased the migration and invasion of CRC cells, which was significantly suppressed by verteporfin treatment (Fig. 5H). These findings suggest that FUT2 promotes the migration and invasion of CRC cells through the YAP/TAZ signaling, at least partially.
A total of 2910 genes regulated by FUT2 were generated from the TCGA-CRC dataset, and then GO (A) and KEGG (B) pathway enrichment analyses of 2910 genes were performed. Relevant data processing details are provided in the Methods section. C SW480 cells were transfected with FUT2-shRNA, and then western blot analysis of CYR61, CTGF, TAZ, LATS1, p-YAP1, and YAP1 expressions were determined. The relative density of protein bands was quantified, normalized to GAPDH, and fold changes were shown in histograms from three independent experiments. D SW480 cells were transfected with FUT2-shRNA, and then western blot analysis of the intracellular distribution of YAP1 and TAZ expressions were determined. The relative density of protein bands was quantified, normalized to α-tubulin or PCNA, and fold changes were shown in histograms from three independent experiments. SW480 cells were transfected with FUT2-shRNA, YAP1-siRNA (E) or TAZ-siRNA (F), and then western blot analysis of CTGF expressions were determined. The relative density of protein bands was quantified, normalized to GAPDH, and fold changes were shown in histograms from three independent experiments. SW480 and DLD-1 cells were transfected with FUT2-shRNA (G) or FUT2/Lv105 (H). After transfection, the cells were treated with Verteporfin (500 nM) for 24 h, and then transwell assay was performed. The number of migrating/invading cells was quantified via Image J software. Scale bar represents 50 μm. Data shown are mean ± SD; n = 3 samples. Scale bar represents 50 μm. Statistical significance was determined using one-way ANOVA using the Bonferroni post-hoc test, with *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, ns: not significant.
YAP1 nuclear transport is regulated by fucosylation
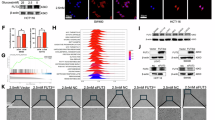
To investigate whether YAP1 was fucosylated by FUT2, co-immunoprecipitation and lectin blotting analysis were performed. Co-immunoprecipitation experiment showed that FUT2 was interacts directly with YAP1, but not with TAZ (Fig. 6A). The UEA-1 pull-down assays showed that the fucosylation of YAP1 was markedly reduced by FUT2 knockdown (Fig. 6B), and the glycosylation sites on YAP1 was predicted via the NetNGlyc1.0 online tool (Fig. 6C). To further validate the role of fucosylation in YAP1, cells were treated with 2F-Peracetyl-Fucose (2F-PAF), a specific inhibitor of fucosylation. Results showed that fucosylation of YAP1 was significantly decreased by 2F-PAF treatment (Fig. 6B). Meanwhile, the phosphorylation of YAP1 was increased and the expression of CTGF was decreased by 2F-PAF treatment (Fig. 6D). These results suggest that the effect of YAP1 was regulated by FUT2-mediated fucosylation.
A Co-immunoprecipitation assay to determine the interaction between FUT2 and YAP1/TAZ in SW480 cells, indicating direct protein-protein interactions of FUT2 with YAP1. B SW480 cells were transfected with FUT2-shRNA or treated with 1 μM 2F-PAF for 24 h. Following the treatment, western blot analysis of YAP1 expressions were assessed after UEA-1 pull-down. C Visualization of the glycosylation sites on YAP1 and SREBP-1, showing the potential sites of modification. D SW480 cells were transfected with FUT2-shRNA. After transfection, the cells were treated with 1 μM 2F-PAF for 24 h, and then western blot analysis of CTGF and p-YAP1 expressions were determined. The relative density of protein bands was quantified, normalized to β-actin, and fold changes were shown in histograms from three independent experiments. E SW480 cells were transfected with FUT2-shRNA and YAP1-siRNA. After transfection, cells were analyzed for western blot analysis of indicated proteins. The relative density of protein bands was quantified, normalized to β-actin, and fold changes were shown in histograms from three independent experiments. F SW480 cells were transfected with YAP1-siRNA, and then the mRNA levels of YAP1, FASN, and CPT1A were assessed by qRT-PCR. Data shown are mean ± SD; n = 3 samples. G, H SW480 cells were transfected with FUT2/Lv105. After transfection, cells were treated with verteporfin (500 nM) for 24 h, and then and then stained with Nile red (G) or phalloidin (H) to evaluate intracellular cytoskeleton organization. Green or red fluorescence signals were captured and visualized through a fluorescent microscope using constant fluorescence parameters explained in the methods section: scale bar, 50 μm. I Design of the tumor-establishment studies. BALB/c nude mice were subcutaneously implanted with the indicated FUT2-knockdown (sh-FUT2 SW480) or control SW480 (shRNAcont) cells. J After 30 days of transplantation, western blot analysis of ACC1, FASN, YAP1, and TAZ expression in isolated tumors (n = 6 mice/group) from the SW480 xenograft model were determined. The relative density of protein bands was quantified, normalized to β-actin, and fold changes were shown in histograms from three independent experiments. Statistical significance was determined using one-way ANOVA using the Bonferroni post-hoc test, with *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, ns: not significant.
FUT2 promotes fatty acid synthesis via YAP/TAZ signaling
To investigate whether FUT2 regulates lipid metabolism via YAP/TAZ signaling, we assessed the expression of key enzymes involved in lipid metabolism. Western blot analysis revealed that YAP1 knockdown led to a decrease in the expression of FASN and CPT1A, whereas ATGL expression remained unaffected (Fig. 6E). Quantitative real-time PCR (qRT-PCR) results showed that the mRNA expression of FASN and CPT1A was significantly decreased by YAP1 knockdown (Fig. 6F). Furthermore, the functional impact of YAP/TAZ signaling on lipid synthesis and cytoskeleton formation was investigated. Treated with verteporfin, the lipid synthesis and cytoskeleton formation of CRC cells were significantly impaired (Fig. 6G, H). Overexpression of FUT2 promoted both lipid synthesis and cytoskeleton formation in SW480 cells, and these effects were suppressed by verteporfin treatment. Furthermore, the results showed that the expression of FASN, ACC1, and TAZ was decreased, and the p-YAP1 was increased in knockdown FUT2 nude mice xenografts models (Fig. 6I, J). These results suggest that FUT2 regulates fatty acid synthesis via YAP/TAZ signaling.
FUT2 promotes fatty acid synthesis by increasing glucose uptake in CRC cells
The main raw material for lipid synthesis is derived from the products of cellular glucose metabolism11. Research has shown that the expression of GULT1, a major glucose transporter, is regulated by the Hippo pathway31. Here, we found that YAP1 (or FUT2) knockdown significantly decreased the mRNA expression of GLUT1 in SW480 cells (Fig. 7A, B). The western blot results revealed that FUT2 knockdown significantly decreased GLUT1 expression in CRC cells (Fig. 7C). To further verify the regulatory effect of FUT2 on GLUT1, the total cellular levels of glucose (Glu) were examined. The results revealed that the level of glucose was markedly decreased by FUT2 knockdown in CRC cells (Fig. 7D). We therefore hypothesized that FUT2 affects glucose uptake capacity, which in turn affects lipid synthesis. To determine whether glucose restriction affects fatty acid synthesis, we treated SW480 cells with medium containing either 25 mM or 5 mM glucose. The results revealed that the levels of TG and glucose were markedly lower in FUT2-knockdown cells treated with 25 mM or 5 mM glucose than in the corresponding control cells. High glucose treatment significantly increased the level of TG; however, there was no significant difference in the level of TG between 25 mM and 5 mM glucose treated in FUT2 knockdown SW480 cells (Fig. 7E). Interestingly, compared with the corresponding controls, FUT2 knockdown markedly reduced the intracellular levels of ATP under both 25 mM and 5 mM glucose conditions, whereas compared with FUT2 knockdown, the level of ATP was no significant different under different concentrations of glucose (Fig. 7E). Moreover, FASN expression was significantly decreased with FUT2 knockdown under glucose limitation (Fig. 7F). The results indicated that the conversion of glucose to TG was inhibited by FUT2 knockdown in SW480 cells.
SW480 cells were transfected with YAP1-siRNA (A) or FUT2-shRNA (B), and then GLUT1 mRNA levels were assessed by qRT-PCR. Data shown are mean ± SD; n = 3 samples. C SW480 and DLD-1 cells were transfected with FUT2-shRNA, and then western blot analysis of GLUT1 expressions were determined. The relative density of protein bands was quantified, normalized to GAPDH, and fold changes were shown in histograms from three independent experiments. D SW480 and DLD-1 cells were transfected with FUT2-shRNA, and then intracellular glucose (Glu) levels were assessed with detection kit. Data shown are mean ± SD; n = 3 samples. E SW480 cells were transfected with FUT2-shRNA. After transfection, SW480 cells were cultured in 5 mM or 25 mM glucose media for 48 h, and then intracellular ATP, TG and Glu levels were assessed with detection kits. Data shown are mean ± SD; n = 3 samples. F SW480 cells were transfected with FUT2-shRNA. After transfection, SW480 cells were cultured in 5 mM or 25 mM glucose media for 48 h, and then western blot analysis of FASN was determined. The relative density of protein bands was quantified, normalized to α-tubulin, and fold changes were shown in histograms from three independent experiments. G SW480 cells were cultured in 5 mM or 25 mM glucose media for 48 h, and then the mRNA levels of ACC1, FASN, SCD, SREBP-1, CHREBP, MLX and FUT2 were assessed by qRT-PCR. Data shown are mean ± SD; n = 3 samples. Statistical significance was determined using one-way ANOVA using the Bonferroni post-hoc test, with *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, ns: not significant.
Changes in gene expression are coordinately regulated at the level of the transcriptional pathway by transcription factors, where the transcription factors carbohydrate response element-binding protein (ChREBP) and max-like protein X (MLX) bind and activate triglyceride synthesis genes in a glucose-dependent manner32,33, among which, SREBP-1 (sterol regulatory element binding protein 1) predominantly regulates fatty acid de novo synthesis genes34. By further examining the mRNA levels of the above transcription factors and fatty acid synthesis genes in cells under high glucose conditions, we found that FUT2 knockdown mainly passively affected the expression of SREBP-1 under glucose limitation (Fig. 7G). Taken together, these results indicate that FUT2 enhances the transcription of GLUT1 via YAP/TAZ signaling and thereby enhances the glucose uptake capacity of CRC cells, which in turn promotes fatty acid synthesis.
Fucosylation of mSREBP-1 regulated by FUT2 enhances mSREBP-1 stability
The SREBP-1 precursor (pSREBP-1) located in the endoplasmic reticulum is activated to enter the Golgi apparatus for shear processing to form mature SREBP-1 (mSREBP-1), which is translocated to the nucleus and promotes the transcription of genes involved in de novo lipogenesis by binding to specific sterol regulatory elements (SREs)35. As shown in Fig. 8A, FUT2 knockdown did not affect the expression of pSREBP-1, whereas the level of mSREBP-1 was markedly decreased by FUT2 knockdown in SW480 cells. The mRNA levels of SREBP-1 downstream factors, including ACC1, FASN, SCD, HMGCR, HMGCS, and INSIG1, were markedly decreased by FUT2 knockdown (Fig. 8B). Co-immunoprecipitation results showed that there is an interaction between FUT2 and mSREBP-1 (Fig. 8C). Lectin UEA-1 assays revealed that both FUT2 knockdown or fucosylation inhibitor treatment significantly reduced the fucosylation of mSREBP-1 (Fig. 8D), and NetNGlyc1.0 online predicted the presence of glycosylation sites of SREBP-1 (Fig. 6C). To further determine how FUT2 stabilizes the mSREBP-1 protein, the protein synthesis inhibitor cycloheximide (CHX) and the proteasome inhibitor MG-132 were used to test the degradation pathway of mSREBP-1. The results revealed that the mSREBP-1 content was markedly decreased by FUT2 knockdown with CHX treatment (Fig. 8E). MG132 treatment can prevent the degradation of mSREBP-1 by FUT2 knockdown (Fig. 8F), indicating that FUT2 stabilizes mSREBP-1 by protecting it from proteasome degradation.
A SW480 and DLD-1 cells were transfected with FUT2-shRNA, and then western blot analysis of pSREBP-1 and mSREBP-1 was determined. The relative density of protein bands was quantified, normalized to GAPDH, and fold changes were shown in histograms from three independent experiments. B SW480 cells were transfected with FUT2-shRNA, and then the mRNA levels of HMGCS, HMGCR, INSIG1, SCD, ACC1, FASN and FUT2 were assessed by qRT-PCR. Data shown are mean ± SD; n = 3 samples. C Co-immunoprecipitation assay was applied to determine the interaction of FUT2 and mSREBP-1 in SW480 cells. D SW480 cells were transfected with FUT2-shRNA or treated with 1 μM 2F-PAF for 24 h. Following the treatment, western blot analysis of mSREBP-1 was assessed after UEA-1 pull-down. E SW480 cells were transfected with FUT2-shRNA. After transfection, the cells were treated with 50 µM CHX for the indicated time, and then western blot analysis of mSREBP-1 expression were determined. The relative density of protein bands was quantified, normalized to β-actin, and fold changes were shown in histograms from three independent experiments. F SW480 cells were transfected with FUT2-shRNA. After transfection, the cells pre-treated with CHX (50 µM) were treated with MG132 (20 µM) for 4 h. Following the treatment, cells were analyzed for Western blot. The relative density of protein bands was quantified, normalized to GAPDH, and fold changes were shown in histograms from three independent experiments. G Schematic representation of the predicted TEADs family and SREBP-1 in the FASN gene promoter. H Co-immunoprecipitation assay was applied to determine the interaction of mSREBP-1 and YAP1 in SW480 cells. I Kaplan-Meier survival curves stratified TCGA-CRC patients into high and low risk groups based on the mRNA level of FASN, SREBP-1, YAP1, and FUT2, displaying differential survival rates. J Time-dependent ROC curves assessed prognostic model’s prognostic accuracy for CRC over 1, 3, and 5 years, presenting the AUC for each interval. Statistical significance was determined using one-way ANOVA using the Bonferroni post-hoc test, with *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, ns: not significant.
Furthermore, JASPER analysis revealed that TEADs and SREBP-1 have close binding sites in the FASN promoter region (Fig. 8G). Co-IP experiments revealed an interaction between YAP1 and mSREBP-1 (Fig. 8H). These results suggest that the interaction of YAP1 and SREBP-1 can promote the transcription of FASN.
Prognostic value of FUT2, FASN, SREBP-1 and YAP1 in CRC
Owing to the diversity in clinical presentation and individual differences in disease progression in colorectal cancer, accurate prognostic prediction for CRC patients is crucial for clinical management. Precise prognostic assessment not only aids in formulating personalized treatment, but also optimizes resource allocation, enhances treatment efficacy, and improves the quality of patients’ life. By identifying key genes and signaling pathways associated with disease progression and prognosis, and integrating the expression information of multiple biomarkers, a more accurate prognostic model can be constructed. When individual genes such as YAP1 and SREBP-1 were evaluated as prognostic markers for CRC patients, no significant difference in overall survival (OS) was observed between the high- and low-expression groups (Supplementary Fig. 3A–D). However, by combining YAP1, SREBP-1, FUT2, and FASN into a prognostic model via LSASSO regression, distinct risk categories were established. The OS of patients in the high-risk group was significantly shorter than that of patients in the low-risk group (Fig. 8I). The model demonstrated prognostic accuracy, with concordance indices of 0.57, 0.57, and 0.71 for predicting OS at 1, 3, and 5 years, respectively (Fig. 8J). These results provide a potential model for CRC prognostic diagnosis and offer a valuable reference for further development and application of molecular biomarkers in assessing CRC prognosis.
Discussion
CRC is highly lethal due to distant metastases; 20% of CRC patients present with distant metastases at initial diagnosis and more than 30% of patients eventually develop metastases, with a 5-year survival rate of less than 20%36. Glycosylation has emerged as an important regulator in the process of metastasis37. Accordingly, the dysregulation of glycosyltransferases has been extensively studied in CRC, and altered gene expression of glycosyltransferases, as well as mutations, has been described for CRC tumor tissues and cell lines38. Our previous study revealed that FUT2 was upregulated in CRC and acted as an oncogene in the progression of CRC, and knockdown of FUT2 markedly suppressed cell proliferation, colony formation, migration and invasion18. Surprisingly, it was reported that FUT2 inhibited cell proliferation, invasion and tumor metastasis of CRC39. Here, we found that high expression of FUT2 in CRC can significantly distinguish between cancerous and non-cancerous tissues, and is linked to increased stromal and ESTIMATE scores, indicating that FUT2 is closely related to the metastasis of CRC.
Metabolic reprogramming is a common feature of cancer cells, among which, lipid metabolic reprogramming has received increasing attention because of its critical role in the development and progression of cancer40. The ability of cancer cells to rewire their lipid metabolism provides them with fatty acids for the building blocks of membranes, energy sources and signal transduction7. Like many other tumors, CRC shows a lipogenic phenotype with profound changes in synthesis of fatty acids41. In the present study, we found that there was a clear correlation between enhanced lipid metabolism and poorer outcomes, and this correlation was associated with the expression of FUT2. FUT2 knockdown markedly reduced the intracellular levels of TG, T-CHO, and FFA and the content of neutral lipids. Studies have shown that the dysregulation of lipid metabolism in many cancers is linked to elevated de novo fatty acid synthesis, and related enzymes, such as FASN, ACC1 and ACLY, are abnormally overexpressed7,25. FASN, in particular, is crucial for catalyzing the de novo synthesis of fatty acids and has emerged as a key target for understanding the molecular pathways controlling CRC metastasis42,43. The upregulated expression of ACC1 could promote cancer cell proliferation and metastasis25. Consistently, our study revealed that FUT2 knockdown significantly inhibited de novo fatty acid synthesis by decreasing the expression of FASN, ACC1 and ACLY, thus repressing CRC cell growth and metastasis. CPT1A, a subtype of the CPT1 transport system, plays a critical role in fatty acid oxidation, controlling the transport of fatty acids into the mitochondria for oxidation, which provides intermediate metabolites and energy to support cell proliferation44,45. Here, we found that the knockdown of FUT2 led to reduction in the expression of CPT1A, which might lead to the attenuation of fatty acid utilization and elimination of the proliferation of CRC cells. Furthermore, FUT2 knockdown enhances lipid peroxidation and ferroptosis in CRC cells. Ferroptosis sensitivity in cells hinges on peroxidized polyunsaturated fatty acid levels and related enzymes, which are essential for lipid metabolism homeostasis46, whereas the role of FUT2 in regulation of CRC cell ferroptosis is unexplored.
The regulation of lipid metabolism is complex in cancer cells. Research has shown that oncogenic pathways can directly regulate lipid metabolism programming11. Recent studies have revealed that YAP/TAZ can reform intracellular metabolic programs via transcriptional regulation and accelerate lipid accumulation in cancer47. In the present study, we found a strong link between FUT2 and the Hippo signaling pathway in CRC via bioinformatic analysis. The present study revealed that FUT2-meditated fucosylation promotes the translocation of YAP1 to nucleus by inhibiting phosphorylation of YAP1. Meanwhile, the expression of TAZ, a homolog of YAP and interacted with YAP as a functional unit, was decreased by FUT2 knockdown in CRC cells. The activated YAP/TAZ promotes the fatty acid synthesis and cytoskeleton formation in CRC cells, enhancing their migration and invasion of CRC cells. The conversion of glucose into fatty acids is crucial for both energy storage and metabolism regulation48. Recently, studies have shown that active YAP increases the glucose utilization by promoting the transcription of glucose transporter 3 (GLUT3)47. Our study revealed that the transcription of GLUT1 was upregulated by FUT2 via the YAP signaling pathway, which increased glucose uptake to support fatty acid synthesis and accumulation. SREBP-1, an important transcription factor in the novo synthesis of FAs, can promote the transcriptional expression of multiple enzymes, including FASN, ACC1 and SCD40. SREBP-1 also serves as a critical glucose sensor, playing a vital role in bridging glucose metabolism and de novo fatty acid synthesis49. Our study revealed that SREBP-1 expression is not affected by FUT2 knockdown in CRC cells; however, mSREBP-1 expression was significantly reduced by FUT2 knockdown. Further study revealed that FUT2 stabilizes mSREBP-1 by fucosylation, which enhances de novo fatty acid synthesis.
Over the past decade, many strategies have been evaluated for targeting glycosylation to control cancer progression and prevent metastasis50. In our study, FUT2 acted as an oncogene in CRC, enhancing de novo fatty acid biosynthesis which in turn promoted CRC cell proliferation and metastasis through YAP1/TAZ signaling and SREBP-1. Furthermore, a prognostic model for CRC was constructed on the basis of FUT2, SREBP-1, FASN, and YAP1, and showed a high accuracy for predicting the OS of patients with CRC. Overall, these findings also imply that FUT2 might be a potential therapeutic target for CRC.
Materials and methods
Data collection and analysis
We acquired genomic data for colorectal adenocarcinoma (COAD) and rectum adenocarcinoma (READ) along with corresponding clinical information from the TCGA through UCSC Xena (http://xena.ucsc.edu/). The genomic data of colorectal tissue from 384 healthy people were obtained from the GTEx database. After elimination of expression, missing survival data, and different batches of data from the same patient, 975 samples were collected, including 616 tumor samples and 359 normal samples. The “tidyverse” and “dplyr” packages were employed for efficient data cleaning and ID conversion in R (version 4.3.2). To assess the tumor environment, the ESTIMATE algorithm was used to estimate tumor purity and quantify the abundance of infiltrating stromal and immune cells. We utilized the “limma” package51 to identify DEGs between the high- and low-FUT2 expression groups. The criteria for determining DEGs were a |fold change | > 1.3 and a P < 0.05. Survival analysis, which employs Cox regression analysis through the survival and “timeROC” packages, facilitated the construction of ROC curves to assess model accuracy. For a nuanced understanding of the biological pathways associated with the DEGs, we employed the “clusterProfiler” package for GSEA.
Acquisition and processing of single-cell data
The single-cell data utilized in this study were derived from an article published by Zhang L and other researchers titled “Single-Cell Analyses Inform Mechanisms of Myeloid-Targeted Therapies in Colon Cancer”52. Single-cell analysis was conducted via “Seurat v5.0.3”53. In the quality control step, cells with detected gene counts greater than 300, ribosomal protein-encoding gene expression proportions greater than 3%, mitochondrial gene expression proportions less than 20%, and haemoglobin gene expression proportions less than 0.1% were retained. The “NormalizeData” function was employed for standardizing the single-cell dataset, whereas the “FindVariableFeatures” function identified 3000 variable features within the dataset. The “ScaleData” function was utilized to scale the data, mitigating the effects of varying sequencing depths across different cells. Cell clustering and cell type identification were performed via the “FindClusters” function. Cell type annotation genes were obtained from Cell Marker 2.0 (http://117.50.127.228/CellMarker/).
Cell culture
HEK293T, SW480 and DLD-1 cells were obtained from the Cell Bank of the Chinese Academy of Sciences. They were maintained in DMEM (or RPMI-1640) (Gibco, USA) supplemented with 10% FBS (Clark Bioscience, USA) and 1% penicillin/streptomycin (Gibco). Regular monitoring and passage ensured optimal cell growth and viability.
Transfection and plasmid construction
SW480 and DLD-1 cells were transduced with hU6-MCS-Ubiquitin-EGFP-IRES-puro-FUT2 vector (Addgene plasmid#227423) (Genechem, China). FUT2-overexpressing cells and negative control cells were obtained via the Lv105-FUT2 plasmid (Addgene plasmid#227439) or empty vector Lv105 (Genechem). Lentiviruses were produced in HEK293T cells, and the viral supernatant was collected after 48 h. The supernatant was then used to infect SW480 and DLD-1 cells. Stable clones were established through selection with puromycin (Sigma, USA) for following experiments. YAP1- and TAZ-knockdown cells were generated via transient transfection of cells with 100 pM siRNA (Genechem) via the Lipofectamine 3000 transfection system (Thermo Fisher, USA). For details of the sequences used please see Supplementary Tab. 2.
Quantitative real-time PCR
Total RNA was isolated from CRC cell lines via TRIzol reagent (Takara, Japan). The cDNA was obtained via PrimeScript RT reagent (Vazyme,China), and qRT-PCR was performed via SYBR Green Master Mix (Vazyme). The results were normalized to GAPDH and are presented as the fold change in mRNA expression (2-△△CT). The sequences of primer used are listed in Supplementary Tab.2.
Western blot analysis
In brief, the lysis of CRC cells was accomplished by adding 1% phenylmethanesulfonyl fluoride (PMSF) (Beyotime, China) and 2% phosphatase inhibitor (Beyotime) to RIPA buffer (Beyotime). Following separation via sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), the proteins were transferred onto a polyvinylidene fluoride membrane. Rapid blocking solution (Epizyme, China) was used in TBS containing 0.1% Tween-20 to block the membranes for 1 h, which were than incubated with specific primary antibodies overnight at 4 °C and subsequently probed with a secondary antibody at room temperature for 1 h. Finally, enhanced chemiluminescence (ECL) (NCM Biotech, China) reagents were used to visualize the results on the Gel DocXR (Bio-Rad,USA), which was analyzed via Image J software. The primary antibodies used are listed in Supplementary Tab. 3.
Measurement of intracellular components
According to the manufacturer’s instruction, the levels of intracellular components, including Glu (Beyotime), FFAs (Solarbio, China), TG (Solarbio), T-CHO (Solarbio), ATP (Thermo Fisher), MDA (Solarbio), GSSG (Solarbio), and GSH (Solarbio), were meticulously quantified via specialized kits.
For the RSL3 treatment, SW480 cells were pretreated with 2 μmol/L RSL3 (MedChemExpress, USA) for 24 h, followed by treatment with or without 1 μmol/L ferrostatin (MedChemExpress) for another 24 h. Finally, the cells were collected for MDA assays.
ROS assay
For the assessment of ROS, the cells were treated with 100 μmol/L DCFH-DA (Beyotime) in the absence of light for 30 min. Then, the cells were digested and collected via trypsin. Employing on-line flow cytometry (Agilent, China), the average fluorescence intensity of 2 × 104 cells was measured as a representative indicator of ROS levels. The data were processed via NovoExpress software.
Cell viability assay
SW480 cells (3 × 103) or DLD-1 cells (5 × 103) were seeded in 96-well plates in 200 μl of medium containing 5% FBS with or without 50 μmol/L palmitic acid (Sigma). After 24, 48 and 72 h, 10 μl of CCK 8 (Dojindo, Japan) reagent was added to 100 μl of the mixture, which was subsequently cultured at 37 °C for 2 h. The absorbance of each sample was assessed at 450 nm via an automated plate reader (Thermo Fisher).
Colony formation assay
CRC cells were cultured in 6-well plates and treated according to the experimental protocols. Two weeks later, the cells were fixed with 4% paraformaldehyde (Phygene, China), stained with crystal violet staining solution (Beyotime) for 30 min and photographed. The colony number of crystal violet stained cells was quantified using Image J software v1.6.0.
Transwell migration and invasion assays
The invasion or migration capacity of CRC cells was evaluated via Transwell membranes (Corning, USA) with or without Matrigel. In briefly, 5 × 104 cells were seeded in the upper chamber in 200 μl of serum-free medium. Concurrently, 500 μl of medium supplemented with 10% FBS was placed in the lower chamber. Following a 24 h incubation at 37 °C, the cells were immobilized and photographed after they were stained with crystal violet in the chamber.
For the orlistat treatment assay, SW480 and DLD-1 cells were treated with orlistat (MedChemExpress) for 48 hours (50 μmol/L or 25 μmol/L), then the cells were subjected to migration and invasion assays. Similarly, the cells were treated with the verteporfin (MedChemExpress) for 24 h (500 nmol/L).
Immunofluorescence
For Nile red staining, the cells were treated according to the experimental instructions and stained with 0.05 μg/mL Nile red (Beyotime) at 37 °C for 10 min to visualize the lipid droplets. The cells were restrained with 1 μg/mL DAPI (Beyotime) mixed with fluorescence quencher for 1 min and then imaged under a confocal laser scanning microscope (Nikon Corporation, Japan) using a 20x objective with DAPI or Texas Red channel.
For phalloidin staining, the cells were treated according to the experimental instructions and fixed with 0.5% Triton X-100. The cells were incubated in 5 μmol/L phalloidin (Yeasen, China) for 30 min in the dark and then imaged under a confocal laser scanning microscope using a 20x objective with DAPI or FITC channel.
Co-immunoprecipitation
Proteins obtained after cell lysis were subjected to protein A + G agarose beads (Beyotime). The mixture was then incubated with the target-specific primary antibody or with rabbit or mouse IgG as a negative control, at 4 °C for 16 h. Subsequently, protein-antibody complexes were captured using Protein A + G agarose beads pre-blocked with BSA. Following three washes with PBS, the bound proteins were collected and subsequently analyzed by Western blot.
Lectin pull-down assay
The cell lysates were incubated with UEA-1-conjugated beads (Vector Laboratories, USA) and rotated at 4 °C for 16 h. After washing several times with PBS to remove any unbound proteins, the UEA-1 enriched proteins were collected and analyzed via western blotting.
For the 2F-PAF treatment assay, SW480 cells were treated with 1 μmol/L 2F-PAF (MedChemExpress) for 24 h, then the cells were extracted for WB analysis.
Animal experiments
Animal experiments were approved by the Laboratory Animal Ethics Committee of Wenzhou Medical University and the Laboratory Animal Centre of Wenzhou Medical University (NO. WYDW2019-0245), and were performed in accordance with all relevant ethical regulations for animal use. BALB/c nude mice (female, eight weeks old) were purchased from Beijing Vital River Laboratory Animal Technology Co., Ltd. SW480-NC and SW480-sh-FUT2 cells (5 × 107) were subcutaneously injected into the right flank of BALB/c nude mice. Tumor lengths were measured with calipers every 3 days. When the maximum tumor volume reached humane endpoints (750 mm3), euthanasia was performed on the mice (with an overdose of pentobarbital), and the tumors were then excised for imaging and weighing. A portion of the excised transplants was used for TG and T-CHO measurements, and the remaining xenografts were stored in liquid nitrogen for further experiments.
Statistical analysis
All the data were analyzed and reproduced at least three times for robustness and reproducibility. Statistical analyses were performed via R-4.3.2 and GraphPad Prism v8.0. The Wilcoxon rank test was used to analyze the distribution and expression of risk scores, stromal scores, immune scores, and gene expression across different groups. Student’s t-test was used to compare two independent groups, whereas one-way ANOVA followed by Tukey’s post hoc test was used for multiple comparisons. Bar graphs represent the mean values±standard deviation (SD) from three independent experiments. Statistical significance was set at P values < 0.05.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
References
Bray, F. et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 68, 394–424 (2018).
O’Sullivan, D. E. et al. Risk Factors for Early-Onset Colorectal Cancer: A Systematic Review and Meta-analysis. Clin. Gastroenterol. Hepatol. 20, 1229–1240.e1225 (2022).
Rawla, P., Sunkara, T. & Barsouk, A. Epidemiology of colorectal cancer: incidence, mortality, survival, and risk factors. Gastroenterol. Rev./Przegląd Gastroenterologiczny. 14, 89–103 (2019).
Ciardiello, F. et al. Clinical management of metastatic colorectal cancer in the era of precision medicine. CA: A Cancer J. Clinicians. 72, 372–401 (2022).
Zhang, C. et al. Cancer-derived exosomal HSPC111 promotes colorectal cancer liver metastasis by reprogramming lipid metabolism in cancer-associated fibroblasts. Cell Death Dis. 13, 57 (2022).
Siegel, R. L., Wagle, N. S., Cercek, A., Smith, R. A. & Jemal, A. Colorectal cancer statistics, 2023. CA Cancer J. Clin. 73, 233–254 (2023).
Yang, Y. et al. SLC25A1 promotes tumor growth and survival by reprogramming energy metabolism in colorectal cancer. Cell Death Dis. 12, 1108 (2021).
Luo, X. et al. Emerging roles of lipid metabolism in cancer metastasis. Mol. cancer 16, 76 (2017).
Yoshida, G. J. Metabolic reprogramming: the emerging concept and associated therapeutic strategies. J. Exp. Clin. Cancer Res. 34, 111 (2015).
Sedlak, J. C., Yilmaz, Ö. H. & Roper, J. Metabolism and Colorectal Cancer. Annu Rev. Pathol. 18, 467–492 (2023).
Koundouros, N. & Poulogiannis, G. Reprogramming of fatty acid metabolism in cancer. Br. J. Cancer 122, 4–22 (2020).
La Vecchia, S. & Sebastián, C. Metabolic pathways regulating colorectal cancer initiation and progression. Semin Cell Dev. Biol. 98, 63–70 (2020).
Reily, C., Stewart, T. J., Renfrow, M. B. & Novak, J. Glycosylation in health and disease. Nat. Rev. Nephrol. 15, 346–366 (2019).
Madunić, K. et al. Colorectal cancer cell lines show striking diversity of their O-glycome reflecting the cellular differentiation phenotype. Cell Mol. Life Sci. 78, 337–350 (2021).
Singh, J. P., Zhang, K., Wu, J. & Yang, X. O-GlcNAc signaling in cancer metabolism and epigenetics. Cancer Lett. 356, 244–250 (2015).
Sun, R., Kim, A. M. J. & Lim, S.-O. Glycosylation of immune receptors in cancer. Cells 10, 1100 (2021).
Lv, Y., Zhang, Z., Tian, S., Wang, W. & Li, H. Therapeutic potential of fucosyltransferases in cancer and recent development of targeted inhibitors. Drug Discov. Today 28, 103394 (2023).
Liu, P. et al. FUT2 promotes the tumorigenicity and metastasis of colorectal cancer cells via the Wnt/β‑catenin pathway. Int. J. Oncol. 62, 35 (2023).
Koo, J. H. & Guan, K. L. Interplay between YAP/TAZ and Metabolism. Cell Metab. 28, 196–206 (2018).
Dey, A., Varelas, X. & Guan, K.-L. Targeting the Hippo pathway in cancer, fibrosis, wound healing and regenerative medicine. Nat. Rev. Drug Discov. 19, 480–494 (2020).
Meng, Z., Moroishi, T. & Guan, K.-L. Mechanisms of Hippo pathway regulation. Genes Dev. 30, 1–17 (2016).
Zou, R. et al. YAP nuclear-cytoplasmic translocation is regulated by mechanical signaling, protein modification, and metabolism. Cell Biol. Int. 44, 1416–1425 (2020).
Li, X. et al. OGT regulated O-GlcNAcylation promotes papillary thyroid cancer malignancy via activating YAP. Oncogene 40, 4859–4871 (2021).
Yu, S. et al. GALNT12 promotes fibrosarcoma growth by accelerating YAP1 nuclear localization. Oncol. Lett. 26, 1–11 (2023).
Dai, W. et al. PTPRO represses colorectal cancer tumorigenesis and progression by reprogramming fatty acid metabolism. Cancer Commun. 42, 848–867 (2022).
Seetharaman, S. & Etienne-Manneville, S. Cytoskeletal Crosstalk in Cell Migration. Trends Cell Biol. 30, 720–735 (2020).
Mollinedo, F. & Gajate, C. Lipid rafts as signaling hubs in cancer cell survival/death and invasion: implications in tumor progression and therapy: thematic review series: biology of lipid rafts. J. Lipid Res. 61, 611–635 (2020).
Zheng, J. & Conrad, M. The Metabolic Underpinnings of Ferroptosis. Cell Metab. 32, 920–937 (2020).
Jin, X. et al. Ferroptosis: Emerging mechanisms, biological function, and therapeutic potential in cancer and inflammation. Cell Death Discov. 10, 45 (2024).
Guo, J. et al. Ferroptosis: a novel anti-tumor action for cisplatin. Cancer research and treatment. Off. J. Korean Cancer Assoc. 50, 445 (2018).
Kashihara, T. et al. YAP mediates compensatory cardiac hypertrophy through aerobic glycolysis in response to pressure overload. J. Clinical Investig. 132, e150595 (2022).
Yu, A. et al. Glucose-induced and ChREBP: MLX-mediated lipogenic program promotes hepatocellular carcinoma development. Oncogene 42, 3182–3193 (2023).
Iizuka, K., Takao, K. & Yabe, D. ChREBP-Mediated Regulation of Lipid Metabolism: Involvement of the Gut Microbiota, Liver, and Adipose Tissue. Front Endocrinol. (Lausanne). 11, 587189 (2020).
Shimano, H. & Sato, R. SREBP-regulated lipid metabolism: convergent physiology - divergent pathophysiology. Nat. Rev. Endocrinol. 13, 710–730 (2017).
Jeon, T. I. & Osborne, T. F. SREBPs: metabolic integrators in physiology and metabolism. Trends Endocrinol. Metab. 23, 65–72 (2012).
Lu, W. et al. Reprogramming immunosuppressive myeloid cells facilitates immunotherapy for colorectal cancer. EMBO Mol. Med. 13, e12798 (2021).
Bindeman, W. E. & Fingleton, B. Glycosylation as a regulator of site-specific metastasis. Cancer Metastasis Rev. 41, 107–129 (2022).
Fernández-Ponce, C. et al. The role of glycosyltransferases in colorectal cancer. Int. J. Mol. Sci. 22, 5822 (2021).
Wang, W. et al. Intestinal epithelium-specific Fut2 deficiency promotes colorectal cancer through down-regulating fucosylation of MCAM. J. Transl. Med. 21, 82 (2023).
Pham, D. V., Tilija Pun, N. & Park, P. H. Autophagy activation and SREBP‐1 induction contribute to fatty acid metabolic reprogramming by leptin in breast cancer cells. Mol. Oncol. 15, 657–678 (2021).
Currie, E., Schulze, A., Zechner, R., Walther, T. C. & Farese, R. V. Cellular fatty acid metabolism and cancer. Cell Metab. 18, 153–161 (2013).
Menendez, J. A. & Lupu, R. Fatty acid synthase and the lipogenic phenotype in cancer pathogenesis. Nat. Rev. Cancer 7, 763–777 (2007).
Jiang, T. et al. The m6A modification mediated-lncRNA POU6F2-AS1 reprograms fatty acid metabolism and facilitates the growth of colorectal cancer via upregulation of FASN. Mol. Cancer 23, 55 (2024).
Ma, Y. et al. Fatty acid oxidation: An emerging facet of metabolic transformation in cancer. Cancer Lett. 435, 92–100 (2018).
Tang, M. et al. CPT1A-mediated fatty acid oxidation promotes cell proliferation via nucleoside metabolism in nasopharyngeal carcinoma. Cell Death Dis. 13, 331 (2022).
Yang, W. S. & Stockwell, B. R. Ferroptosis: death by lipid peroxidation. Trends Cell Biol. 26, 165–176 (2016).
Koo, J. H. & Guan, K.-L. Interplay between YAP/TAZ and Metabolism. Cell Metab. 28, 196–206 (2018).
Koppenol, W. H., Bounds, P. L. & Dang, C. V. Otto Warburg’s contributions to current concepts of cancer metabolism. Nat. Rev. Cancer 11, 325–337 (2011).
Cheng, C., Geng, F., Cheng, X. & Guo, D. Lipid metabolism reprogramming and its potential targets in cancer. Cancer Commun. 38, 1–14 (2018).
Briggs, M. T. et al. Translating N‐glycan analytical applications into clinical strategies for ovarian cancer. PROTEOMICS–Clin. Appl. 13, 1800099 (2019).
Ritchie, M. E. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47–e47 (2015).
Zhang, L. et al. Single-cell analyses inform mechanisms of myeloid-targeted therapies in colon cancer. Cell 181, 442–459. e429 (2020).
Butler, A., Hoffman, P., Smibert, P., Papalexi, E. & Satija, R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420 (2018).
Acknowledgements
The work was supported by the Key Discipline of Zhejiang Province in Medical Technology (First Class, Category A).
Author information
Authors and Affiliations
Contributions
C.D., Y.Z., J.Z., S.C., Y.L., and Z.B. performed the experiments and analyzed the data. C.D. and Y.Z. performed most of the experiments. C.D., S.F., and X.C. conceived the ideas and supervised the study. C.D. wrote the manuscript. S.F. and X.C. revised the manuscript. All the authors have read and approved the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks Matthias Lochner, Gurdeep Singh and the other, anonymous, reviewer for their contribution to the peer review of this work. Primary Handling Editor: Christina Karlsson Rosenthal. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Dong, C., Zhang, Y., Zeng, J. et al. FUT2 promotes colorectal cancer metastasis by reprogramming fatty acid metabolism via YAP/TAZ signaling and SREBP-1. Commun Biol 7, 1297 (2024). https://doi.org/10.1038/s42003-024-06993-x
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s42003-024-06993-x
This article is cited by
-
l-Fucose: a dietary sugar with multifaceted potential in the biology and therapy of cancer
Nature Reviews Cancer (2026)
-
The interplay between extracellular matrix remodeling and cellular lipid metabolic reprogramming in cancer: a review
Molecular Biology Reports (2026)
-
Structure, function, and implications of fucosyltransferases in health and disease
Nature Communications (2025)
-
Liquid-liquid phase separation of GPS2-LATS1 promotes colorectal cancer progression by reprogramming lipid metabolism
Oncogene (2025)
-
The pathogenesis and therapeutic implications of metabolic reprogramming in renal cell carcinoma
Cell Death Discovery (2025)