Abstract
A complex interplay of genetic and environmental factors influences bacterial growth. Understanding these interactions is crucial for insights into complex living systems. This study employs a data-driven approach to uncover the principles governing bacterial growth changes due to genetic and environmental variation. A pilot survey is conducted across 115 Escherichia coli strains and 135 synthetic media comprising 45 chemicals, generating 13,944 growth profiles. Machine learning analyzes this dataset to predict the chemicals’ priorities for bacterial growth. The primary gene-chemical networks are structured hierarchically, with glucose playing a pivotal role. Offset in bacterial growth changes is frequently observed across 1,445,840 combinations of strains and media, with its magnitude correlating to individual alterations in strains or media. This counterbalance in the gene-chemical interplay is supposed to be a general feature beneficial for bacterial population growth.
Similar content being viewed by others
Introduction
The interplay between genetic and environmental factors significantly influences population dynamics, adaptive evolution, and survival strategies across various contexts. Genotypes and environments determine bacterial growth. Escherichia coli (E. coli) is a model organism, providing comprehensive knowledge on cell growth1,2,3. At the genetic level, studies on single-gene knockout4 and genome reduction5,6 have shown that bacterial growth rates were correlated with genome size7, transcriptome periodicity8,9, mutation rates10,11, and evolutionary changes8,12. Environmental factors impacting bacterial growth include physical variables (e.g., temperature, pH, osmotic pressure)13,14,15,16, biological interactions (e.g., interspecific competition, symbiosis)17,18,19,20, and chemical conditions (e.g., resource availability, chemical composition)21,22,23,24. Despite extensive knowledge of these biological mechanisms and theoretical simulations25,26, predicting bacterial growth remains challenging due to insufficient experimental and analytical observations linking genetic and environmental factors to growth outcomes. This gap underscores the need for more integrated studies to understand and predict bacterial growth dynamics.
The genetic and environmental combinations are critically important in determining bacterial growth. Extensive studies on bacterial auxotrophy27,28, conditional essentiality29, medium optimization for the minimal cell30, and related topics have demonstrated that culture medium composition significantly impacts bacterial growth in addition to genetic features. Furthermore, this environmental dependence has been observed in the context of antibiotic resistance31 and gene-gene interactions32,33,34. These findings indicated that genetic and environmental crosstalk played a strategic role in regulating bacterial growth. Understanding how genetic and environmental combinations function in bacterial growth remained challenging. Recent studies have incorporated genetic and environmental variations35,36 to address this issue, revealing the underlying mechanism of negative epistasis36,37,38,39. This approach has been used to understand transcriptome reorganization40,41, antimicrobial interactions42, and interspecies interactions43. Despite these advances, the precise dynamics of how these combinations influenced bacterial growth were required for further integrated studies.
This study employed a data-driven approach to elucidate the underlying mechanism of bacterial growth changes resulting from genetic and environmental variations. Since vitamin B (VB) has been used in studying bacterial auxotrophy since the 1950s44,45, the genes involved in VB metabolic pathways were selected as the genetic variations for studying bacterial growth. According to our previous studies24,46, the chemical components of the culture media were employed as the environmental variation. Single-gene knockout strains of E. coli were combined with various chemical compositions of the culture media to assess bacterial growth. A comprehensive dataset was experimentally collected, encompassing the growth profiles of diverse single-gene knockout E. coli strains across a wide range of synthetic media. The resultant dataset was subjected to machine learning (ML), a data-driven approach successfully used to analyze bacterial growth dynamics47,48. To identify the chemical impacts on bacterial growth, ML considering regression of chemical composition to growth was conducted. Further analysis considering the interplay between genetic and environmental factors in bacterial growth was performed using an additive model36,37,38,39,42,43 for easy understanding. Our findings provided data-driven insights into how these factors impacted bacterial growth and offered principles for future investigation and application of microbial systems.
Results
Bacterial growth profiles across genetic and environmental diversities
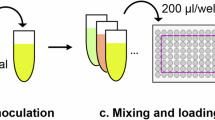
A large dataset linking bacterial growth to the combinations of E. coli strains and culture media was experimentally obtained to investigate the impact of genetic and environmental factors on bacterial growth. Genetic variation was achieved using 115 E. coli strains, comprising 114 single-gene knockout mutants and their wild-type BW25113 (Fig. 1Ai and Supplementary Data 1). These knockout genes participated in nine pathways related to VB metabolism in the KEGG database49 (details in Supplementary Table 1) and were nonessential4 when growing on either rich or poor nutritional media9. These strains grew differentially between the two media of LB and M63 (Supplementary Fig. 1), demonstrating that genetic variation was responsible for the growth change in response to chemical environments. Environmental diversity was attained by preparing 135 synthetic media using 48 pure compounds (Fig. 1Aii and Supplementary Data 2). As known, some pure compounds would ionize in solution, resulting in 45 chemical components when considering their ionic forms. Finally, the synthetic media consisted of 45 chemical components of concentration gradients altering logarithmic order (Fig. 1B and Supplementary Fig. 2). The chemical compositions of the 135 media were randomly distributed in principal component analysis (PCA) (Fig. 1C). Their concentrations were more or less irrelevant to each other (Supplementary Fig. 3), indicating an unbiased environmental diversity. Subsequently, a high-throughput growth assay was conducted (Supplementary Fig. 4) to obtain the temporal changes in OD600 of 115 strains grown under 135 media for 72 h. The preliminary tests confirmed that the E. coli growth reached saturation within 72 h (Supplementary Fig. 5). Finally, a total of 15,525 growth profiles (four biological replicates per profile) linking to the combinations of strains and media were obtained (Supplementary Data 3), in which bacterial growth (saturated population density, K) was evaluated (Fig. 1Aiii). The mean of biological replicates was subjected to data mining to discover the gene–chemical interplay contributing to K (Fig. 1Aiv).
A Study overview. Genetic (i) and environmental (ii) variations were achieved by an assortment of E. coli strains and synthetic media, respectively. A high-throughput growth assay was performed to acquire the bacterial growth profiles linked to the genetic and environmental diversity (iii). The maximum OD600 (K), highlighted in red, was designated as the saturated population density and used for the following data mining (iv). B Chemical concentration of synthetic media. Color variation indicates the categories of the 45 chemical components. Black dots indicate the experimentally tested concentrations. Concentrations are shown in the logarithmic scale, and zero is not indicated. C PCA of medium compositions. Two main PCs of all medium compositions used in the study are shown.
A large dataset connecting K to strains (genes) and media was obtained (Fig. 2A). Of the 15,525 growth profiles, 13,944 growth profiles with a standard deviation of K between replicates less than 0.1 were used for the following analyses (Fig. 2B). The mean K values of 115 strains considerably differed across varied media (Fig. 2C and Supplementary Fig. 6A), whereas that of 135 media remained roughly similar regardless of the different strains (Fig. 2D and Supplementary Fig. 6B). The changes in K were more significant across 135 media than those across 115 strains. It might be due to the broad distributions of K detected in individual strains (Supplementary Fig. 7) compared to the narrow ones in individual media (Supplementary Fig. 8). The variation in K due to genetic or environmental diversity was represented by the standard deviation (Ksd). Its distribution was broader caused by media (Fig. 2E) than by strains (Fig. 2F). These findings indicated that the environmental alteration caused more significant changes in K than the genetic variation. It was probably because the knockout genes (strains) were within a limited metabolism in the present study.
A Growth (K) of 115 strains under 135 media. The blue gradation from dark to light indicates the value of K from high to low. B Distribution of K. The number of strain and medium combinations (N) tested in the study is indicated. C K of 115 strains grown under individual media. All 135 media were arrayed in a decent order of the median of K, highlighted in yellow. The mean of K is indicated in magenta. D K in 135 media of individual strains. All 115 strains were arrayed in a decent order of the median of K, highlighted in yellow. The mean of K is indicated in magenta. E Distribution of the standard deviations of 115 strains’ K in each medium. F Distribution of each strain’s standard deviations of the K in 135 media.
Gene clusters categorized according to the impact of chemicals on bacterial growth
Gradient-boosting decision tree (GBDT) was used to construct 115 machine learning (ML) models, which were trained with the corresponding 115 datasets linking the chemical compositions to K of each strain (Fig. 3A). The GBDT models were constructed, in which regression of explanatory variables (concentrations of 45 chemicals) to the objective variable (K) was performed to build a series of decision trees for increased accuracy. Consequently, the feature importance of each chemical was predicted for each strain by the GBDT models, which was interpreted as the quantitative contribution of the chemical component to K (Supplementary Data 4). The chemicals most impacted bacterial growth were specified. Accordingly, hierarchical clustering divided 115 strains (genes) into four clusters, comprising 11, 60, 7, and 37 genes, respectively (Fig. 3B, C, C1–C4). Enrichment analysis was conducted to determine the correspondence between the four identified gene clusters and the VB metabolic pathways. As a result, no significantly enriched gene clusters were identified in the nine pathways related to VB metabolism (Fig. 3D). For instance, the WT and ten knockout strains were categorized in C1 (Supplementary Data 4), in which the knockout genes participated in six out of the nine pathways (Fig. 3D). The results suggested either the functional insignificance of these knockout genes or the nonspecific gene–chemical interactions involved broadly across the metabolic pathways.
A Machine learning model construction and prediction for clustering the strains. The feature importance of each chemical in individual strain was predicted by the GBDT model, which was constructed using the corresponding growth profiles. B Clustering dendrogram of the strains. The heatmap indicates the feature importance of each chemical in each strain. Chemicals representing the medium components comprised in 135 media are arrayed vertically. Four clusters (C1–C4) are shown in green and orange. The number of strains in each cluster is bracketed. C Chemicals of high priority in the four clusters. The top five chemicals are shown. Boxplots of the genes (indicated by gray dots) that participated in the cluster are indicated. D Clusters participated in the nine pathways. The number of 114 knockout genes assigned in the four clusters participating in each pathway is shown. E Gene–chemical network constructed upon their contribution to bacterial growth (K). A total of 204 combinations of genes and chemicals are used to create the network, in which the chemicals of high importance to determine the growth of 115 strains are indicated. The large nodes in transparent yellow represent the determinative chemicals, whose sizes are the sum of the feature importance to all linked strains (genes). The small nodes in green and orange indicate the knockout genes categorized in the four clusters. The thickness of the edges reflects the magnitude of the feature importance.
Although 115 strains (genes) were categorized into different gene clusters, the chemicals of high priority determining the growth (K) of the strains in the four clusters were in common: glucose (Glc), isoleucine (Ile), and valine (Val) (Fig. 3B, asterisks). They were in charge of the gene clusters, i.e., Ile and Val were of great importance for C1; Val showed the highest priority for C2; Glc, Ile, and Val were of equal importance for C3; and Glc was the most crucial for C4 (Fig. 3B, C). Three core gene–chemical networks were created (Fig. 3E) using the primary feature importance across the 114 genes and 45 chemicals. The gene clusters were determined by the chemicals for bacterial growth, revealing the diverse patterns of gene–chemical interplay. Note that applying an alternative clustering algorithm (i.e., gaussian mixture) or changing the threshold of the standard deviation of K between replicates did not change the chemicals of high impact on K, regardless of the changes in the number of clusters (Supplementary Figs. 9 and 10).
Hierarchical gene–chemical networks
The chemical deciding the variation in K of 115 strains (Ksd) upon media (Fig. 2E) was further evaluated by SHapley Additive exPlanations (SHAP)50,51 to the ML model (Fig. 3A). SHAP was used to explain which factors (i.e., chemicals) were most important in determining objective (i.e., Ksd). The highest priority chemical determining Ksd was glucose (Glc), of which the high concentration caused the large Ksd (Fig. 4A), indicating the Glc abundance differentiated the gene–chemical networks. Accordingly, the growth profiles were divided into three groups dependent on Glc, i.e., low (Glc_l), medium (Glc_m), and high (Glc_h) concentrations (Supplementary Fig. 11, note that four concentrations of Glc were used, and the middle two were designated “medium”), which comprised 6005, 4192, and 3747 growth profilings, linking 115 strains to 57, 40, and 38 media, respectively (Supplementary Fig. 11). The ML models were reconstructed using the three datasets separately. Varied patterns of chemical feature importance to K were observed, in which discriminated gene clusters varying from one to three (Fig. 4B–D, note that Glc_l, Glc_m, and Glc_h were analyzed with the same threshold; see “Methods”). The determinative chemicals for K were altered from two to three in the three groups, i.e., Val and Ile for Glc_l, NH4, Fe, and VB6 for Glc_m, and Val and VB9 for Glc_h (Fig. 4B–D, asterisks). Note that applying an alternative clustering algorithm (i.e., gaussian mixture) or changing the threshold of the standard deviation of K between replicates did not change the chemicals of high impact on K, regardless of the changes in the number of clusters (Supplementary Fig. 9, Supplementary Fig. 10). The gene–chemical networks using the primary feature importance (Supplementary Fig. 11) showed that they were multiply separated in Glc_m and Glc_h and highly overlapped in Glc_l (Fig. 4E–G). Fe and NH4 were in charge of the primary clusters in Glc_m (Fig. 4F), consistent with our previous finding of the decision-making chemicals for the saturated population density of a genome-reduced E. coli strain52. VB6 and VB9 were responsible for the primary clusters in Glc_m and Glc_h, respectively (Fig. 4F, G), suggesting the task divergent of vitamin B metabolites in response to gene–chemical networks. The sole gene cluster determined by Val and Ile in Glc_l (Fig. 4B, E) indicated that BCAA metabolism became a priority under insufficient glucose. The results revealed that the abundance of glucose structured the gene–chemical networks. The gene–chemical networks were somehow layered hierarchically.
A Evaluating the ML model of Ksd by SHAP analysis. The SHAP value represents the chemical contribution to the variation of Ksd. The color gradation indicates the relative concentration of the chemical that normalized its concentrations across 135 media. The chemicals of significant contributions are shown in order. B–D Clustering dendrograms of the strains at varied glucose levels. Glc_l, Glc_m, and Glc_h indicate low, medium, and high glucose levels, comprising varied media and resultant gene–chemical combinations. The heatmaps indicate the feature importance of each chemical in each strain. Chemicals are arrayed vertically. The clusters are shown in green and orange. E–G Gene–chemical networks at varied glucose levels. 208, 216, and 218 combinations of genes and chemicals are used to create the networks of Glc_l, Glc_m, and Glc_h, respectively. The chemicals of high importance in determining the growth of 115 strains are indicated. The large nodes in transparent yellow represent the determinative chemicals, whose sizes are the sum of the feature importance to all linked strains (genes). The small nodes in green and orange indicate the knockout genes categorized in the four clusters. The thickness of the edges reflects the magnitude of the feature importance.
Intriguingly, the determinative chemicals were completely discriminated in these gene clusters. The chemicals of the highest priority to determine the growth of the strains in each cluster were Ile for the sole cluster in Glc_l (Fig. 5A), NH4, Fe, and VB6 for each cluster in Glc_m (Fig. 5B), and Val, VB9, and K for each cluster in Glc_h (Fig. 5C). As the gene clusters comprised different strains, the significant differentiation in the determinative chemicals indicated the divergence in gene–chemical sub-networks. Further analysis showed no significantly enriched gene clusters in the nine pathways (Fig. 5D). However, a pathway was somehow significantly enriched in C2 if without multiple test corrections (Fig. 5D, asterisks). The global network (Fig. 3E) and the hierarchical sub-networks (Fig. 4E–G) failed to enrich any specific pathways, suggesting the underlying mechanism was independent of genetic function but related to the general working principle.
A–C Chemicals of high priority in the clusters. The top five chemicals are shown. Boxplots of the genes (indicated by gray dots) that participated in the cluster are indicated. Glc_l (A), Glc_m (B), and Glc_h (C) indicate low, medium, and high glucose levels. The number of strains in each cluster is indicated. D Composition of clusters in each pathway. The colors are corresponding to the clusters shown in (A–C). Asterisks indicate significantly enriched clusters or pathways without multiple test corrections (P < 0.05).
Directional changes in the gene–chemical interplay
The changes in K were theoretically evaluated to investigate any interactions of gene–chemical combinations. Changes in K caused by the alterations in strains (Gi, i = 1–114) and media (Ej, j = 1–134) from the WT at any medium (Base) were designed as ΔGi and ΔEj, respectively, and that caused by the simultaneous alterations was ΔGEij (Fig. 6A). As an example, when the Base was the WT at a particular medium (ID: C0003) out of 135 media, a total of 114, 116, and 12,778 Gi, Ej, and GEij were attained, resulting in an equal number of ΔGi, ΔEj, and ΔGEij, respectively. Note that the experimentally or statistically invalidated GEij was removed from the analysis, so its number could be smaller than the theoretical calculation. Their distributions ranged broadly from negative to positive and were of varied shapes (Fig. 6B), independent of Base (Supplementary Fig. 12). Simultaneous alterations of strains (ΔGi) and media (ΔEj) widened the distributions of ΔGEij, which was likely that combining the monomodal ΔGi and the multimodal ΔEj turning out to be the bimodal ΔGEij. It indicated the crosstalk of genetic and environmental contributions in bacterial growth. i.e., the changes in K.
A Evaluating the changes in K triggered by either genes or chemicals. Base, Gi, Ej, and GEij indicate the K values of the WT at any medium, of any single-gene knockout strain at the same medium, of the WT at any other medium, and of any single-gene knockout strain at any other medium, respectively. ΔGi, ΔEj, and ΔGEij indicate the changes in K of the corresponding pairs. The details are described in “Methods”. B Histograms of the changes in K caused by strains, media, and their combinations. The total number of changes is indicated. C Definition of the directional changes in K. P and N represent positive and negative changes, respectively. NN, PN, and PP indicate the three patterns of the directional changes of ΔGi and ΔEj. D An example of a NN, PN, and PP scatter plot. 2865, 2524, and 7389 combinations of NN, PP, and PN are shown, respectively, while the base is the WT at medium C0003. E Relative frequencies of NN, PN, and PP at each Base. Color variation indicates PP, PN, and NN. F. Distributions of relative frequencies of PP, PN, and NN. The number of all combinations evaluated in PP, PN, and NN is indicated.
To understand the relations among ΔGEij, ΔGi, and ΔEj, the combinations of ΔGi and ΔEj were divided into three categories, i.e., NN, PP, and PN, representing that ΔGi and ∆Ej were negative, positive, and negative or positive, respectively (Fig. 6C). In the present case (WT at C0003), more than half of the total 12,778 combinations presented reverse changes between ∆Gi and ΔEj (PN), and the remaining combinations showed the equivalent frequency of identical directional changes (NN and PP) (Fig. 6D). Difference Base led to the varied numbers of Gi, Ej, and GEij, and the differentiated ΔGi, ΔEj, and ΔGEij. More than ten thousand combinations were evaluated in response to each Base. Consequently, the changes in Base led to the changes in the ratios of PN, NN, and PP (Fig. 6E), i.e., different Base resulted in the broad distributions of the ratios (Fig. 6F). It demonstrated that the directions of the changes in K, i.e., increase or decrease, depended on both strains (genes) and media without a general trend.
Counterbalance in the gene–chemical interplay
The additive model36,37,38,39,42,43 was applied to predict the K (GEijExpected) of any combinations of Gi and Ej according to Base, ΔGi, and ΔEj (Fig. 6A). Comparing GEijExpected to the experimentally obtained GEij resulted in any of the three consequences, i.e., Additive, Strengthen, and Offset, representing GEijExpected was equal to, larger, and smaller than GEij, respectively (Fig. 7A). For instance, 75.6%, 60.3%, and 77.5% were Offset in NN, PN, and PP, respectively, leading to an overall ratio of 67.1% across all combinations in the base of WT at C0003 (Fig. 7B). The high proportion of Offset was independent of Base (Supplementary Fig. 13). Evaluating the alternative Base showed that over half of the combinations were Offset, regardless of NN, PN, and PP (Fig. 7C). The ratio of Offset was highly significant than that of Strenghen (Fig. 7D), indicating the counterbalance occurred more often when the genetic and environmental alterations caused the changes in K (ΔGi and ΔEj) of the same direction (PP and NN). In comparison, when the changes (ΔGi and ΔEj) were in a reverse direction (PN), more combinations of Strengthen were allowed, which probably avoided too strong the canceling effect caused by opposite changes to maintain bacterial growth. The finding was common even if the definition of Additive was modified to allow for a range of errors, as experimental errors and calculation bias might occur (Supplementary Fig. 14).
A Evaluating the changes in K triggered by gene–chemical combinations. ΔGEijExpected is the sum of Base, ΔGi, and ΔEj; all other labels are described in Fig. 6A. Red, blue, and gray represent Offset, Strengthen, and Additive mechanisms in gene–chemical crosstalk. Three examples of Additive in NN, Strengthen in PN, and Offset in PP are illustrated. B An example diagram of the relative frequency of the three mechanisms. PP, PN, and NN combinations shown in Fig. 6D are presented. C Relative frequencies of Offset in NN, PN, and PP at each Base. The broken lines indicate 50%. D Distributions of the Offset and Strengthen relative frequencies in NN, PN, and PP. The statistical significance of the Mann–Whitney U test is shown. E Comparing the Offset to the additive changes. The magnitudes of the Offset and the additive changes are illustrated as the black arrow lines of GEij - GEijExpected and ΔGi + ΔEj. F An example of the correlation between the magnitude of Offset and the additive changes. Correlation coefficients and the p-value are indicated. G Correlation between the magnitude of Offset and the additive changes in all combinations in PP and NN. Color gradations indicate the correlation coefficients in the linear scale and the corresponding p-values in the logarithmic scale.
The significance of the Offset in NN and PP was additionally analyzed, as the changes in identical directions might enhance the damage. The subtraction between GEijExpected and GEij positively correlated to the sum of ΔGi and ΔEj (Fig. 7E, F). A more substantial Offset resulted in more extensive changes in K, of which significantly correlated combinations were frequently observed in NN and PP (Fig. 7G). It demonstrated that the magnitude of Offset depended on the magnitudes of genetic and environmental changes, likely the negative epistasis commonly proposed in genetics53. In the present study, 296,747 and 485,343 pairs of PP and NN were identified in a total of 1,445,840 combinations across 45 medium components and 115 strains, of which 78.70% and 75.40% were in the relationship of Offset, respectively (Supplementary Data 5). Surprisingly, a significantly large proportion of Offset showed even more repressed changes in growth (K) than any of that caused by genetic or environmental alterations in both PP and NN (Fig. 8A), i.e., the changes caused by GEij (ΔGEij) were milder than those caused by either Gi or Ej (ΔGi or ΔEj). It indicated that the Offset in gene–chemical interplay prevented more extensive changes than those triggered by genetic and environmental factors, frequently happening in biological systems.
A Ratio of highly repressed changes in the Offset. Green and blue indicate NN and PP, respectively. ΔGi, ΔEj, and ΔGEij indicate the changes in K of the corresponding pairs, as described in the main text. B Benefits and pains. The canceling and compensating effects are caused by the genetic and environmental alterations in synthetic biology and homeostasis of living systems, respectively. Black lines indicate the observed consequences of genetic or environmental alteration. Solid and dotted lines in green and blue indicate the observed and expected consequences of simultaneous alterations in genes and environments, respectively.
Discussion
Our study demonstrated that the magnitude of growth changes was limited within a limited range, which was reasonable for the living cell as a homeostatic system. Genetic and environmental alterations could either increase or decrease bacterial growth. As the representative cases, i.e., PP and NN, although the genetic and environmental alterations increased or decreased bacterial growth in identical directions, their simultaneous alterations favored repressed growth changes as if the canceling effect participated (Fig. 8B, green). The so-called Offset might benefit bacterial survival from genetic and environmental interruptions. Although its underlying global mechanisms remained unclear, the Offset in gene–chemical crosstalk should be essential for living organisms to maintain homeostasis. It well explained some conflicting biological findings. For instance, the amino acids or the biosynthetic genes were beneficial for bacterial growth; however, both presences were unbeneficial, reported as less was more in amino acid biosynthesis54. The addition of antibiotics or the expression of its resistance gene was disadvantageous for bacterial growth; however, both presences were beneficial, as observed in the ampC production for Salmonella55. As representative cases of PP and NN, these studies were explained well by the Offset mechanism observed here. Nevertheless, whether the Offset was a universal phenomenon in living cells required further demonstration, as it was identified in the gene–chemical interactions highly related to VB metabolism in bacteria.
The Offset could explain why the culturing output of genetically engineered cells did not always meet the synthetic design, in addition to the mechanisms of gene expression noise and biological fluctuation56,57. The canceling effect might disturb the genetic circuit (synthetic pathway) to function precisely as its design principle relied on the regulatory mechanisms of the target gene and chemical without considering Offset (Fig. 8B, blue). Synthetic biology requires data-driven design principles for predictable output and advanced application46,58. In addition, the case of Strengthen also happened but was considerably restricted within specific gene–chemical combinations, such as the combinations between the knockout gene ilvN and the medium components of Ile and Val (Supplementary Fig. 15). In the presence of ilvN (wild-type strain), a high concentration of Val inhibited bacterial growth59, which could be rescued by adding the high concentration of Ile60. Without ilvN (single-gene knockout strain), bacterial cells lost their sensitivity to Val61. Consequently, the growth suppression caused by Val was released, allowing the additional change derived from Ile. Strengthen was probably an alternative approach for living cells to maintain homeostasis as if the chemical was supplementary to enhance the gene function.
Glucose was identified as the primary decision-making chemical for bacterial growth, consistent with the findings of its coordination with amino acids to regulate bacterial growth62,63. Here, glucose might serve as a switch of microbial life history strategies: high yield (Y), resource acquisition (A), and stress tolerance (S), the so-called Y-A-S strategies64. The gene–chemical networks structured in Glc_m, Glc_l, and Glc_h tended to Y, A, and S strategies, respectively. Firstly, as glucose starvation might occur in Glc_l, the chemicals of Val and Ile became the growth determinators for resource acquisition (A-strategy). The stringent response, induced by glucose depletion65, could be regulated by branched-chain amino acids66, i.e., Val, Leu, and Ile. Since Val and Ile shared the metabolic pathways downstream to glucose67,68, they could also trigger the stringent response69. Val could cause Ile starvation67, and Ile was found to be highly sensitive to bacterial growth24. It might be why Val and Ile were also identified as the primary decision-making chemicals here. Therefore, global metabolic reorganization under carbon downshift70,71 might happen for resource acquisition or allocation independent of genetic variation. Secondly, multiple core gene–chemical networks were formed in Glc_m, indicating sufficient glucose allowed varied chemicals in charge of yield (Y-strategy) of 115 strains. Finally, since accessive amounts of glucose were stressors for cells72, stress tolerance (S-strategy) might be mediated by the chemicals of Val and VB9 in Glc_h. The underlying regulatory mechanism or pathway was unknown.
The present study linking genes and chemicals to bacterial growth provided evidence of the proposals on the adaptation and evolution of life on Earth. For example, the growth distributions (K) of 115 strains were similar in two different media, which could be considered genetic diversity within a single population as proposed in the population genetics73. Two strains of WT and ΔacpT presented different K values in the same media (Supplementary Fig. 16) as if they were genetic variations in the population of varied growth features under identical environments. Although ΔacpT showed lower fitness (K) than WT in medium C0058, it was better when grown in medium C0037 (Supplementary Fig. 16). It was likely the preadaptation strategy74, holding cryptic genetic variation at the population level for adapting to the unpredictive environment and keeping robustness75,76.
As a pilot survey, this study linked bacterial growth to a broad genetic and chemical environmental space. The biological limitation of the study was that only the single-gene knockout strains related to VB metabolism were used. Considering the genes as the smallest genetic unit, discovering the single-gene knockout strains was supposed to be a straightforward way to understand the genetic and environmental interactions to bacterial growth. VB was a precursor for various cofactors and participated in various metabolisms, including its biosynthesis and carbon fixation77. VB exchange was reported to be highly essential for the microbes that could not synthesize VB78 for maintenance79. Due to its essentiality, the counterbalance effect and hierarchical gene–chemical networks reported in the present study might provide hints and insights for analyzing the ecological and microbiome interactions. The analytical limitation remained as only the saturated population density (K) under static culture conditions was analyzed due to the experimental restriction. Future investigation on the growth rate, an alternative parameter critical for understanding bacterial growth, may provide in-depth and differentiated findings in the genetic and environmental contributions to bacterial population dynamics. In addition, as the first experimental assays across media and strains to discover the genetic and environmental interactions on bacterial growth, the additive model benefited from the easy understanding of complex interactions. However, as biological interactions are assumed to be highly complex, other theoretical models, in addition to the additive model, such as the non-additive model for bacterial competitive interactions43, might be considered in the future. In particular, the question of why the chemical impact on bacterial growth was more significant than the genetic one remained to be further investigated. Whether the phenomenon was restricted to the genes that participated in VB metabolism or globally was intriguing.
In summary, this study provided an extensive quantitative dataset connecting genetic and environmental diversity to bacterial growth and discovered the gene–chemical interplay participating in the growth. Whether the hierarchically determined core gene–chemical networks and the Offset were universal should be further verified, as the genetic variation was somewhat lower than the environmental variety, according to the magnitude of growth changes. The findings provided valuable insights to fill the gap in gene–environment interactions that participated in survival strategies for bacterial growth in nature. The counterbalance effect in gene–chemical interactions might also reflect a working principle for synthetic biology: over-optimizing the genetic construct might decrease its performance while culturing the cell carrying it. Applying the working principle to synthetic design and metabolic optimization required more effort. The present study tried to bridge the gap between computational prediction and biological significance; nevertheless, linking the data-driven analytical findings to the biological context remained challenging. Further experimental verification was required to clarify the underlying biological mechanisms and functions.
Methods
E. coli strains and culture stock preparation
The wild-type E. coli strain BW25113 and its derivative single-gene knockout strains, i.e., 114 strains associated with vitamin B metabolism in the Keio collection4, were obtained from the National BioResource Project (NBRP), National Institute of Genetics (Shizuoka, Japan). The gene names and IDs and the associated pathways of these strains were summarized in Supplementary Data 1 Common stocks were prepared beforehand for all E. coli strains used in the study to reduce the experimental errors of the repeated growth assay on different days. E. coli cells were cultured in 5 ml of M63 minimal medium in a bioshaker (BR-23FP, TAITEC) at 200 rpm rotation speed and 37 °C. The cell cultures were stopped during the exponential phase and were dispensed into 1.5-mL microtubes (Watson) at 750 µL each, added with 250 µL of 60% glycerol solution. They were subsequently divided into new 1.5 mL microtubes at 50 µL per tube and stored at −80 °C. More than 20 aliquots (culture stocks) were prepared at once and disposably used to avoid the biases caused by repeated freezing-and-thawing or pre-culture; aliquots were used only once, and the remaining cultures were discarded.
Chemical compounds
A total of 48 pure chemicals were used in this study, of which 44 were previously determined24, and nine were related to vitamin B pathways. All chemical compounds were commercially available and purchased from Wako or Sigma, as summarized in Supplementary Data 2. The concentrations of the nine chemicals related to vitamin B pathways were determined according to the Difco Manual of Microbiological Culture Media 11th Edition80, as summarized in Supplementary Data 6. The concentration gradients of these nine chemicals varied from zero to 20-fold their amounts in the commonly used natural media, which were estimated according to their amounts used in these natural media (details in Supplementary Table 2). The maximum concentrations were experimentally verified to cause growth changes in various ranges in the 114 single-gene knockout strains when added to the minimal medium (Supplementary Fig. 17). The concentration gradients of the other 39 chemicals were equivalent to those previously reported24,46. Briefly, the minimum concentration was usually set to zero, and the maximum concentration was determined individually according to literature and manuals and also from experimental verification24. The maximal and minimal concentrations of a total of 48 pure compounds were summarized in Supplementary Data 2. The following analyses used logarithmical values of the chemical concentrations, where zero was transformed to 1% of the minimum concentrations and excluded water ions (H+, OH−).
Media preparation
Stock solutions of 48 chemicals were prepared in advance as previously described24. In brief, the pure compounds were dissolved in highly pure water (Direct-Q UV, Merck) at high concentrations according to their maximal concentrations used in the growth assay (Supplementary Data 2). These solutions were subsequently sterilized by filtration with syringes equipped with 0.22 µm pore size (hydrophilic PVDF membrane, Merck) or autoclaving at 121 °C for 20 min (TOMY). The sterilized chemical solutions were divided into 1050 µL aliquots in 1.5-mL microtubes (Watson) or 4050 µL aliquots in 5-mL microtubes (Watson) and stored at −30 °C for future use. Every 10–100 stock solutions were prepared at once for the 48 chemicals. Aliquots were used only once to avoid repeated thawing and freezing of the stock solutions. The media were prepared by mixing the stock solutions (aliquots) before use. The chemical concentrations varied on a logarithmic scale, and only one chemical was altered for each assay. A total of 135 media were tested, of which the chemical concentrations were calculated considering their ionized forms in water (Supplementary Data 3).
Growth assay
The growth assay was performed in the following steps (Supplementary Fig. 4). The media were first prepared by mixing the 48 stock solutions in 50 ml tubes (Watson) and dispensed into the inner 60 wells on 96-well plates (Coster), 180 µL per well. Every eight 96-well plates were used for each medium, leading to 4800 wells per medium, to meet the assay of 115 E. coli strains with four biological replicates each. Every 10 µL of culture stocks were inoculated to the inner 60 wells on 96-deep well plates (BM Bio), in which 990 µL of M63 medium was dispensed per well in advance. It resulted in a 100-fold dilution of the culture stocks. Every 20 µL of the diluted culture stocks was loaded into the wells containing 180 µL media on 96-well plates. The 96-well plates were placed in a plate reader (Epoch2, BioTek) and shaken with 731 rpm rotation speed at 37 °C for 15 s; followingly, the initial cell culture density was detected at an absorbance of 600 nm (OD600). The 96-well plates were then incubated at 37 °C statically in the incubators (BR-23FP, TAITEC or IC602, YAMATO SCIENTIFIC). Shake and absorbance measurements were performed at 24, 48, and 72 h, as was conducted initially. A total of 13,944 growth profiles were obtained, of which the biological replicates (N = 4) were conducted per profile, i.e., per combination of strain and medium. Note that biological replications’ variance (noise) was considerably smaller than the variation among the genetic or environmental changes (Supplementary Fig. 18).
Data processing and calculation
Data processing of OD600 records and calculations were performed with Python. Unreliable OD600 records, caused mainly by experimental errors, were removed from the analysis. These experimental errors were identified by eyes, e.g., precipitated media, water droplets on the microplate’s lid, reduced culture volume due to evaporation, etc. The optical background was subtracted according to the initially recorded OD600 at 0 h, performed for each well at 24, 48, and 72 h. The Smirnoff–Grubbs test with a significant difference of 0.05 determined the outliers in the four biological replicates, considering that OD600 of biological replicates followed a normal distribution. The mean OD600 of biological replicates was calculated without the outliers. The maximum of the mean OD600 at 24, 48, and 72 h was defined as the saturated density, K Note that a unique value was used for the standard deviation threshold, regardless of the mean OD600, to avoid potential experimental errors that might cause unreliable analytical results. Finally, a total of 13,944 K values, comprising 115 strains grown under 135 media, were reliable and used in the following data mining.
Principal component analysis
Principal component analysis (PCA)81,82 was used to reduce the dimensions of the large dataset by comparison of the variance in the dataset. The present study used the logarithmic values of chemical concentrations for the analysis, as the concentration gradients were changed on a logarithmical scale. Note that the absence of the chemical (zero concentration) was transformed to 1% of its minimum concentration. PCA was performed with Python using “decomposition.PCA” in the scikit-learn library83. The chemical concentrations were normalized within one unit and subjected to PCA. A total of 135 medium compositions were subjected to the analysis.
Machine learning (ML) model construction
The gradient-boosting decision tree (GBDT) was used as a white box ML algorithm to benefit from its high interpretability. It was demonstrated in our previous study of successful prediction of the determinative chemicals for bacterial growth24. The high accuracy of GBDT compared to other ML models was verified using the experimental data acquired in the present study (Supplementary Fig. 19). GBDT regression was performed with “ensemble.GradientBoostingRegressor” in the scikit-learn library. In all, 115 ML models were constructed to predict K for 115 strains at any defined medium compositions; the objective variables were K, and the explanatory variables were the chemical concentrations. An additional ML model was constructed to predict the variation in K due to genetic diversity, assigned as Ksd, calculated as the standard deviation of 115 K. For model construction, a grid search was used for searching the hyperparameters, which were explored in the following ranges by a supercomputer (Cygnus system, NEC LX 124Rh-4G): learning_rate from 0.05 to 0.5 in 0.05 increments, max_depth among 2, 3, 4, and 5, n_estimators among 100, 200, and 300, min_samples_leaf among 0.05 and 0.1, and random_state was fixed at 0. All other hyperparameters were used as default. The mean squared error (MSE) was calculated using “metrics.mean_squared_error” in the scikit-learn library to evaluate the reconstructed ML models. Fivefold non-nested cross-validation was performed to explore the hyperparameters and evaluate the ML models. The dataset was firstly divided into five subsets, of which four were used for training and one for validation. Then, five combinations of training and validation were performed. The hyperparameters in each combination were explored by grid search, a prediction model was trained using the training data, and the prediction accuracy was evaluated using the validation data. The hyperparameters with the highest average prediction accuracy were obtained after five rounds of training and validation, which were used to construct the final prediction model. The best hyperparameters in searching and the MSEs in model verification were summarized in Supplementary Data 7 (115 Kgene models) and Supplementary Table 3 (Ksd model). The best hyperparameters were used to reconstruct the ML models with the data used for searching the hyperparameter and model verification. Random forest, support vector machine (SVM), and multiple regression were performed using “ensemble.RandomForestRegressor”, “svm.SVR”, and “liner_model.LinearRegression”, respectively. All these modules were in the “scikit-learn” library. The hyperparameters were explored as follows. In SVM, “C” was among 0.1, 1, 10, and 100, “epsilon” was among 0.01, 0.1, 0.2, and 0.3, and the kernel was fixed at “rbf”. In random forest, “max_depth” was among 2, 3, 4, and 5, “n_estimators” was among 100, 200, and 300, “min_samples_leaf” was between 0.05 and 0.1, and “random_state” was fixed at zero. All other hyperparameters were used as default.
SHAP
Feature importances were calculated using the “feature_importances_“ attribute included in “GradientBoostingRegressor” to estimate the influence of individual chemical components in the constructed ML model. SHAP (SHapley Additive exPlanations)50,51 was used to explain the output of the ML model predicting Ksd. The constructed ML model predicting Ksd was applied to “Explainer” in the “shap” library, and the SHAP values were calculated by inputting the explanatory variables, i.e., the chemical concentrations in the 135 media, into “Explainer”.
Clustering and enrichment analyses
Hierarchical clustering was performed with Python using the “linkage” function of the “cluster.hierarchy” package in the scipy library84. Ward’s linkage method was used in common. The feature importance was used to cluster the genes (strains) overall and separately. The thresholds for determining the gene (strain) clusters were as follows: half of the maximum distance using all 135 media for overall clustering, half of the maximum distance using 38 media for Glc_h, ~1.10-fold of the maximum distance using 57 media for Glc_l, ~0.51-fold of the maximum distance using 40 media for Glc_m. The threshold for clustering the genes (strains) based on K was 0.15-fold of the maximum distance. Non-hierarchical clustering was performed with Python using the “mixture.GaussianMixture” from the scikit-learn library. Bayesian information criterion (BIC) and Akaike information criterion (AIC) were calculated using the “bic” and “aic” methods for between one and ten clusters, and the number of clusters with the lowest overall BIC and AIC was judged to be optimal. Enrichment analysis was performed with Python with “stats.binomtest” package in the scipy library. The null hypothesis was the overall proportion of pathways to which the 114 genes belonged in the case of pathway enrichment and the overall proportion of clusters to which the 114 genes belonged in the case of cluster enrichment. A binomial test was performed with an upper one-tailed test, and the significance level was set at 0.05. Multiple testing corrections were performed using the Bonferroni or Benjamini/Hochberg method.
Network construction and visualization
The gene–chemical network was visualized using the NetworkX library in Python85. The chemicals of the accumulated feature importance higher than the threshold, determined according to the 0.5% of the total 5175 feature importance (Supplementary Fig. 11), were extracted for each strain and linked by edges to its corresponding gene. The overall and separated (Glc_l, Glc_m, and Gld_h) gene–chemical networks were formed by 115 genes linking to 8, 11, 23, and 23 chemicals, respectively. The thickness of the edges was correlated to the chemical’s feature importance to the linked gene (strain), and the node’s size was correlated to the sum of the chemical’s feature importance to all linked genes (strains).
Evaluation of gene–chemical interactions
The effect of genetic (i.e., single-gene knockout) and environmental (i.e., medium) changes on bacterial growth, K, was evaluated according to the additive model36,37,38,39,42,43. Base indicated the wild-type strain (WT) at any of the tested media, except 18 media (Supplemantry Data 5), which failed to obtain the valid K of WT. The changes in K caused by the alteration of strains (genes) and media, represented by \(\varDelta {G}_{i}\) and \(\varDelta {E}_{j}\), were evaluated by the following equations, where Base, Gi, and Ei were the K values of the WT at any of the defined media, any other gene (strain) i, and any other medium j, respectively.
If the changes in K caused by “G” and “E” were additive, then the simultaneous alteration of strain/gene and medium (GEi,jExpected) were supposed to be equivalent to the sum of individual changes (\(\varDelta {G}_{i}\) and \(\varDelta {E}_{j}\)), as follows.
According to the following criteria, the gene–chemical interactions were categorized into three categories, i.e., NN, PP, and PN, indicating were negative, positive, and negative or positive.
The effects of gene–chemical interactions were then defined as follows.
NN
PP
PN
Here, max(Gi, Ej) and min(Gi, Ej) indicated the maximum and minimum of gi and ej, respectively. Offset and Strengthen represented the gene–chemical interactions in a negative and positive epistatic manner, respectively. The effect of Additive was determined to allow for a range of errors as follows, where sd is the standard deviation of \(G{E}_{i,j}\).
Statistics and reproducibility
Four biological replicates of the growth assay of 115 strains under 135 media were performed, and the Smirnoff–Grubbs test was used to identify the outlier. Five cross-validations were used in the machine learning. A binomial test was used for enrichment analysis with multiple testing corrections. The detailed descriptions can be found in the corresponding “Methods” sections.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All data are provided in the paper as Supplementary Data.
Code availability
ML code is available at https://github.com/aida288571/genetic-and-environmental-interplay.
References
Monk, J. M. et al. iML1515, a knowledgebase that computes Escherichia coli traits. Nat. Biotechnol. 35, 904–908 (2017).
Karp, P. D. et al. The EcoCyc database (2023). EcoSal 11, eesp00022023 (2023).
Tong, M. et al. Gene dispensability in Escherichia coli grown in thirty different carbon environments. mBio https://doi.org/10.1128/mBio.02259-20 (2020).
Baba, T. et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2, 2006.0008 (2006).
Pósfai, G. et al. Emergent properties of reduced-genome Escherichia coli. Science 312, 1044–1046 (2006).
Hashimoto, M. et al. Cell size and nucleoid organization of engineered Escherichia coli cells with a reduced genome. Mol. Microbiol. 55, 137–149 (2005).
Kurokawa, M., Seno, S., Matsuda, H. & Ying, B.-W. Correlation between genome reduction and bacterial growth. DNA Res. 23, 517–525 (2016).
Hitomi, K., Ishii, Y. & Ying, B.-W. Experimental evolution for the recovery of growth loss due to genome reduction. eLife https://doi.org/10.7554/elife.93520.3 (2024).
Liu, L., Kurokawa, M., Nagai, M., Seno, S. & Ying, B.-W. Correlated chromosomal periodicities according to the growth rate and gene expression. Sci. Rep. 10, 15531 (2020).
Lao, Z., Matsui, Y., Ijichi, S. & Ying, B. W. Global coordination of the mutation and growth rates across the genetic and nutritional variety in Escherichia coli. Front. Microbiol. 13, 990969 (2022).
Nishimura, I., Kurokawa, M., Liu, L. & Ying, B. W. Coordinated changes in mutation and growth rates induced by genome reduction. mBio https://doi.org/10.1128/mBio.00676-17 (2017).
Kurokawa, M., Nishimura, I. & Ying, B. W. Experimental evolution expands the breadth of adaptation to an environmental gradient correlated with genome reduction. Front. Microbiol. 13, 826894 (2022).
Baig, I. A. & Hopton, J. W. Psychrophilic properties and the temperature characteristic of growth of bacteria. J. Bacteriol. 100, 552–553 (1969).
Ratkowsky, D. A., Olley, J., McMeekin, T. A. & Ball, A. Relationship between temperature and growth rate of bacterial cultures. J. Bacteriol. 149, 1–5 (1982).
Pietikainen, J., Pettersson, M. & Baath, E. Comparison of temperature effects on soil respiration and bacterial and fungal growth rates. FEMS Microbiol. Ecol. 52, 49–58 (2005).
Record, M. T. Jr., Courtenay, E. S., Cayley, D. S. & Guttman, H. J. Responses of E. coli to osmotic stress: large changes in amounts of cytoplasmic solutes and water. Trends Biochem. Sci. 23, 143–148 (1998).
Fredrickson, A. G. & Stephanopoulos, G. Microbial competition. Science 213, 972–979 (1981).
Pekkonen, M., Ketola, T. & Laakso, J. T. Resource availability and competition shape the evolution of survival and growth ability in a bacterial community. PLoS ONE 8, e76471 (2013).
Minter, E. J. A. et al. Variation and asymmetry in host-symbiont dependence in a microbial symbiosis. BMC Evol. Biol. 18, 108 (2018).
Kehe, J. et al. Positive interactions are common among culturable bacteria. Sci. Adv. 7, eabi7159 (2021).
Egli, T. & Zinn, M. The concept of multiple-nutrient-limited growth of microorganisms and its application in biotechnological processes. Biotechnol. Adv. 22, 35–43 (2003).
Bren, A., Hart, Y., Dekel, E., Koster, D. & Alon, U. The last generation of bacterial growth in limiting nutrient. BMC Syst. Biol. 7, 27 (2013).
Ehrenberg, M., Bremer, H. & Dennis, P. P. Medium-dependent control of the bacterial growth rate. Biochimie 95, 643–658 (2013).
Aida, H., Hashizume, T., Ashino, K. & Ying, B.-W. Machine learning-assisted discovery of growth decision elements by relating bacterial population dynamics to environmental diversity. eLife 11, e76846 (2022).
Grishkevich, V. & Yanai, I. The genomic determinants of genotype × environment interactions in gene expression. Trends Genet 29, 479–487 (2013).
Martin, G. & Lenormand, T. The fitness effect of mutations across environments: Fisher’s geometrical model with multiple optima. Evolution 69, 1433–1447 (2015).
Lundgren, D. G. & Bott, K. F. Growth and sporulation characteristics of an organic sulfur-requiring auxotroph of Bacillus cereus. J. Bacteriol. 86, 462–472 (1963).
Rossi, J. J. & Berg, C. M. Effect of enrichment procedure upon auxotroph recovery in Escherichia coli K-12. Antimicrob. Agents Chemother. 7, 110–112 (1975).
Joyce, A. R. et al. Experimental and computational assessment of conditionally essential genes in Escherichia coli. J. Bacteriol. 188, 8259–8271 (2006).
Lachance, J.-C., Rodrigue, S. & Palsson, B. O. Minimal cells, maximal knowledge. eLife 8, e45379 (2019).
Soley, J. K. et al. Pervasive genotype-by-environment interactions shape the fitness effects of antibiotic resistance mutations. Proc. Biol. Sci. 290, 20231030 (2023).
Flynn, K. M., Cooper, T. F., Moore, F. B. G. & Cooper, V. S. The environment affects epistatic interactions to alter the topology of an empirical fitness landscape. PLoS Genet. 9, e1003426 (2013).
Hall, A. E. et al. Environment changes epistasis to alter trade-offs along alternative evolutionary paths. Evolution 73, 2094–2105 (2019).
Bank, C. Epistasis and adaptation on fitness landscapes. Annu. Rev. Ecol. Evol. Syst. 53, 457–479 (2022).
Rodriguez-Gijon, A. et al. Linking prokaryotic genome size variation to metabolic potential and environment. ISME Commun. 3, 25 (2023).
Baier, F., Gauye, F., Perez-Carrasco, R., Payne, J. L. & Schaerli, Y. Environment-dependent epistasis increases phenotypic diversity in gene regulatory networks. Sci. Adv. 9, eadf1773 (2023).
Li, X., Lalić, J., Baeza-Centurion, P., Dhar, R. & Lehner, B. Changes in gene expression predictably shift and switch genetic interactions. Nat. Commun. 10, 3886 (2019).
Poelwijk, F. J., Socolich, M. & Ranganathan, R. Learning the pattern of epistasis linking genotype and phenotype in a protein. Nat. Commun. 10, 4213 (2019).
Mani, R., St.Onge, R. P., Hartman, J. L., Giaever, G. & Roth, F. P. Defining genetic interaction. Proc. Natl. Acad. Sci. USA 105, 3461–3466 (2008).
Matsui, Y., Nagai, M. & Ying, B. W. Growth rate-associated transcriptome reorganization in response to genomic, environmental, and evolutionary interruptions. Front. Microbiol. 14, 1145673 (2023).
Van Driessche, N. et al. Epistasis analysis with global transcriptional phenotypes. Nat. Genet. 37, 471–477 (2005).
Bollenbach, T. Antimicrobial interactions: mechanisms and implications for drug discovery and resistance evolution. Curr. Opin. Microbiol. 27, 1–9 (2015).
Baichman-Kass, A., Song, T. & Friedman, J. Competitive interactions between culturable bacteria are highly non-additive. eLife 12, e83398 (2023).
Alimchandani, H. R. & Sreenivasan, A. Reversal of sulphonamide action in Escherichia coli (B12 auxotroph) by vitamin B12. Biochim. Biophys. Acta 18, 567 (1955).
Dickerman, H. & Weissbach, H. Altered folate metabolism in a vitamin B 12-methionine auxotroph. Biochem. Biophys. Res. Commun. 16, 593–599 (1964).
Aida, H. et al. Machine learning-assisted medium optimization revealed the discriminated strategies for improved production of the foreign and native metabolites. Comput. Struct. Biotechnol. J. 21, 2654–2663 (2023).
Baig, Y., Ma, H. R., Xu, H. & You, L. Autoencoder neural networks enable low dimensional structure analyses of microbial growth dynamics. Nat. Commun. 14, 7937 (2023).
Zhang, C. et al. Temporal encoding of bacterial identity and traits in growth dynamics. Proc. Natl. Acad. Sci. USA 117, 20202–20210 (2020).
Kanehisa, M. & Goto, S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30 (2000).
Lundberg, S. M. & Lee, S.-I. A unified approach to interpreting model predictions. Adv. Neural Inf. Process. Syst. 30, arXiv:1705.07874 (2017).
Lundberg, S. M. et al. From local explanations to global understanding with explainable AI for trees. Nat. Mach. Intell. 2, 56–67 (2020).
Ashino, K., Sugano, K., Amagasa, T. & Ying, B. W. Predicting the decision making chemicals used for bacterial growth. Sci. Rep. 9, 7251 (2019).
Khan, A. I., Dinh, D. M., Schneider, D., Lenski, R. E. & Cooper, T. F. Negative epistasis between beneficial mutations in an evolving bacterial population. Science 332, 1193–1196 (2011).
D’Souza, G. et al. Less is more: selective advantages can explain the prevalent loss of biosynthetic genes in bacteria. Evolution 68, 2559–2570 (2014).
Morosini, M. I., Ayala, J. A., Baquero, F., Martínez, J. L. & Blázquez, J. Biological cost of AmpC production for serotype Typhimurium. Antimicrob. Agents Chemother. 44, 3137–3143 (2000).
Ying, B. W., Seno, S., Matsuda, H. & Yomo, T. A simple comparison of the extrinsic noise in gene expression between native and foreign regulations in Escherichia coli. Biochem. Biophys. Res. Commun. 486, 852–857 (2017).
Bar-Even, A. et al. Noise in protein expression scales with natural protein abundance. Nat. Genet. 38, 636–643 (2006).
Radivojevic, T., Costello, Z., Workman, K. & Garcia Martin, H. A machine learning automated recommendation tool for synthetic biology. Nat. Commun. 11, 4879 (2020).
Vonderhaar, R. A. & Umbarger, H. E. Isoleucine and valine metabolism in Escherichia-coli K-12-detection and measurement of Ilv-specific messenger ribonucleic-acid. J. Bacteriol. 120, 687–696 (1974).
Umbarger, H. E. & Brown, B. Isoleucine and valine metabolism in Escherichia-coli .5. antagonism between isoleucine and valine. J. Bacteriol. 70, 241–248 (1955).
Eoyang, L. & Silverman, P. M. Role of small subunit (Ilvn polypeptide) of acetohydroxyacid synthase-I from Escherichia-coli-K-12 in sensitivity of the enzyme to valine inhibition. J. Bacteriol. 166, 901–904 (1986).
Zampieri, M., Hörl, M., Hotz, F., Müller, N. F. & Sauer, U. Regulatory mechanisms underlying coordination of amino acid and glucose catabolism in Escherichia coli. Nat. Commun. https://doi.org/10.1038/s41467-019-11331-5 (2019).
Shao, D. et al. Glucose promotes cell growth by suppressing branched-chain amino acid degradation. Nat. Commun. 9, 2935 (2018).
Malik, A. A. et al. Defining trait-based microbial strategies with consequences for soil carbon cycling under climate change. ISME J. 14, 1–9 (2020).
Irving, S. E., Choudhury, N. R. & Corrigan, R. M. The stringent response and physiological roles of (pp)pGpp in bacteria. Nat. Rev. Microbiol. 19, 256–271 (2021).
Fang, M. X. & Bauer, C. E. Regulation of stringent factor by branched-chain amino acids. Proc. Natl. Acad. Sci. USA 115, 6446–6451 (2018).
De Felice, M., Levinthal, M., Iaccarino, M. & Guardiola, J. Growth inhibition as a consequence of antagonism between related amino acids: effect of valine in Escherichia coli K-12. Microbiol. Rev. 43, 42–58 (1979).
Kanehisa, M., Sato, Y., Kawashima, M., Furumichi, M. & Tanabe, M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 44, D457–D462 (2016).
Gummesson, B. et al. Valine-induced isoleucine starvation in Escherichia coli K-12 studied by spike-in normalized RNA sequencing. Front. Genet. 11, 144 (2020).
Zhu, M. & Dai, X. Stringent response ensures the timely adaptation of bacterial growth to nutrient downshift. Nat. Commun. 14, 467 (2023).
Traxler, M. F., Chang, D. E. & Conway, T. Guanosine 3’,5’-bispyrophosphate coordinates global gene expression during glucose-lactose diauxie in Escherichia coli. Proc. Natl. Acad. Sci. USA 103, 2374–2379 (2006).
Mizzi, L. et al. Assessing the individual microbial inhibitory capacity of different sugars against pathogens commonly found in food systems. Lett. Appl. Microbiol. 71, 251–258 (2020).
Ellegren, H. & Galtier, N. Determinants of genetic diversity. Nat. Rev. Genet. 17, 422–433 (2016).
Masel, J. Cryptic genetic variation is enriched for potential adaptations. Genetics 172, 1985–1991 (2006).
Masel, J. & Siegal, M. L. Robustness: mechanisms and consequences. Trends Genet. 25, 395–403 (2009).
Masel, J. & Trotter, M. V. Robustness and evolvability. Trends Genet. 26, 406–414 (2010).
Monteverde, D. R., Gomez-Consarnau, L., Suffridge, C. & Sanudo-Wilhelmy, S. A. Life’s utilization of B vitamins on early Earth. Geobiology 15, 3–18 (2017).
Rodionov, D. A. et al. Micronutrient requirements and sharing capabilities of the human gut microbiome. Front. Microbiol. 10, 1316 (2019).
Sharma, V. et al. B-vitamin sharing promotes stability of gut microbial communities. Front Microbiol 10, 1485 (2019).
Difco, L. DIFCO Manual. 11th edn (Difco Laboratories, 1998).
Ringnér, M. What is principal component analysis? Nat. Biotechnol. 26, 303–304 (2008).
Abdi, H. & Williams, L. J. Principal component analysis. Wiley Interdiscip. Rev.: Comput. Stat. 2, 433–459 (2010).
Pedregosa, F. et al. Scikit-learn: machine learning in Python. J. Mach. Learn. Res. 12, 2825–2830 (2011).
Virtanen, P. et al. SciPy 1.0: fundamental algorithms for scientific computing in Python. Nat. Methods 17, 261–272 (2020).
Hagberg, A., Swart, P. & Chult, D. In Proceedings of the 7th Python in Science Conference (SciPy2008) (Pasadena, CA, 2008).
Acknowledgements
The authors thank Jialin Xu for the experimental support and NBRP for the E. coli strains (BW25113 and Keio collection). This work was supported by JST SPRING (JPMJSP2124 to H.A.), JSPS KAKENHI Grant-in-Aid for Scientific Research (B) (19H03215 to B.W.Y.), and Challenging Exploratory Research (21K19815 to B.W.Y.).
Author information
Authors and Affiliations
Contributions
H.A. conducted the experiments and data analyses and drafted the manuscript; B.W.Y. conceived the research, designed the experiments and analyses, and rewrote the manuscript; all authors approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks the anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editors: Aylin Bircan and Tobias Goris. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Aida, H., Ying, BW. Data-driven discovery of the interplay between genetic and environmental factors in bacterial growth. Commun Biol 7, 1691 (2024). https://doi.org/10.1038/s42003-024-07347-3
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s42003-024-07347-3
This article is cited by
-
Non-model bacteria as platforms for endogenous gene expression in synthetic biology
Nature Reviews Bioengineering (2025)
-
Data-driven analysis reveals distinct genomic and environmental contributions to bacterial growth curves
Scientific Reports (2025)
-
Population Dynamics of Escherichia coli Growing under Chemically Defined Media
Scientific Data (2025)
-
Experimental mapping of bacterial fitness landscapes reveals eco-evolutionary fingerprints
Scientific Reports (2025)