Abstract
The VapBC toxin-antitoxin (TA) system, composed of VapC toxin and VapB antitoxin, has gained attention due to its relative abundance in members of the M. tuberculosis complex. Here, we have functionally characterised VapBC35 TA system from M. tuberculosis. We show that ectopic expression of VapC35 inhibits M. smegmatis growth in a bacteriostatic manner. Also, an increase in the VapB35 antitoxin to VapC35 toxin ratio results in a stronger binding affinity of the complex with the promoter-operator DNA. We show that VapBC35 is necessary for M. tuberculosis adaptation in oxidative stress conditions but is dispensable for M. tuberculosis growth in guinea pigs. Further, using a combination of co-expression studies and biophysical methods, we report that VapC35 also interacts with non-cognate antitoxin VapB3. Taken together, the present study advances our understanding of cross-interaction networks among VapBC TA systems from M. tuberculosis.
Similar content being viewed by others
Introduction
Toxin antitoxin (TA) systems are typically bicistronic and encode for a stable toxin and a labile antitoxin1,2,3,4,5,6,7. TA systems in prokaryotes have been implicated in various biological processes such as post-segregational killing, stress adaptation, phage defence, antibiotic persistence, and disease pathogenesis2,3,5,8,9,10,11,12,13,14,15,16. The antitoxin can be either an RNA or a protein. Depending on the mechanism of toxin neutralisation of the activity of the cognate toxin, TA systems have been classified into eight subfamilies2,3. In the Type I-VII subfamily, the toxin component is a protein, except for Type VIII, where the toxin is a small non-coding RNA2,3,17,18,19,20. In Type II, III and VI TA systems, antitoxins directly bind to the toxin to inhibit its activity2,3,21,22. The antitoxins belonging to Type I, V, and VIII TA systems have been reported to prevent the expression of the toxin by degrading its mRNA or sRNA. In Type IV TA systems, antitoxin and toxin compete for the same cellular targets, whereas in Type VII TA systems, the toxin is post-translational modified by the cognate antitoxin6,17,23. Studies have shown that the toxin component of the TA system inhibits bacterial growth by targeting a cellular process essential for bacterial growth. These pathways include translation, transcription, replication, membrane integrity, cell wall synthesis, ribosome assembly or cytoskeleton formation4,6,15,19,20,24,25,26. Among these subfamilies, Type II is the most abundant and well-characterised subfamily of the TA systems5,18,21,27,28.
The genome of Mycobacterium tuberculosis harbours an extensive repertoire of Type II TA systems belonging to various subfamilies such as vapBC, mazEF, relBE, higBA, parDE, hicAB, and mbcAT12,29,30,31. Among these, virulence-associated protein B and C (VapBC) is the largest, with 51 members belonging to this subfamily29,30,31. The vapBC TA system encodes for the labile antitoxin VapB and the stable toxin VapC. The PilT- amino-terminus domain of VapC toxins consists of a conserved quartet of acidic residues and a Ser/Thr amino acid residue responsible for coordinating divalent ions32. Numerous studies have demonstrated that overexpression of most VapC toxins inhibits bacterial growth in a bacteriostatic manner29,33,34. VapC toxins from M. tuberculosis recognise and cleave unique tRNAs such as tRNA10Gln, tRNA32Gln and tRNA3Leu (for VapC11, Rv1561), tRNA2Ala, tRNA26Ser, tRNA24Ser and tRNA21Cys (for VapC4, Rv0595c), tRNAfmet (for VapC2, Rv0301 and VapC21, Rv2757c), tRNA25Ser and tRNA28Ser (for VapC28, Rv0609 and VapC30, Rv0624); and tRNA7Trp (for VapC25, Rv0277c; VapC29, Rv0617; VapC33, Rv1242, VapC37, Rv2103c and VapC39, Rv2530c)34,35,36,37. In addition to the recognition of tRNAs, VapC toxins from M. tuberculosis, such as VapC20 (Rv2549c) and VapC4 (Rv0595c), show distinct endoribonuclease activities34,36,38,39,40,41. VapC4 cleaves mRNA sequences with ACGC and AC(A/U)GC motifs, while VapC20 targets the Sarcin-Ricin loop in 23S ribosomal RNA34,39,41. The three-dimensional structures for a few of VapBC TA systems, such as VapBC3 (Rv0301-Rv0300, PDB ID: 3H87), VapBC5 (Rv0626-Rv0627, PDB ID:3DBO), VapBC11 (Rv1560-Rv1561, PDB ID: 6A7V), VapBC15 (Rv2009-Rv2010, PDB ID: 4CHG), VapBC26 (Rv0581-Rv0582, PDB ID: 5X3T), VapBC30 (Rv0623-Rv0624, PDB ID: 4XGQ) and VapC toxins such as VapC20 (Rv2549c, PDB ID: 5WZ4), VapC21 (Rv2757c, PDB ID: 5SV2) from M. tuberculosis have been solved39,42,43,44,45,46,47,48. The solved structure demonstrated that the oligomeric state of the VapBC complexes can vary in solution, ranging from a heterodimer (VapBC5) to a hetero-tetramer (VapBC15, VapBC30) or a hetero-octamer (VapBC3, VapBC11, and VapBC26)42,43,44,45,46,47. Typically, the interactions between cognate toxins and antitoxins of Type II TA systems are governed by precise molecular recognition mechanisms. In addition to the interactions between cognate pairs, cross-talk has been reported between different non-cognate TA systems from M. tuberculosis and Haemophilus influenzae49,50,51,52,53. These instances of cross-talk emphasize a complex regulatory network among TA systems in bacteria.
Previously, it has been shown that a subset of TA systems is differentially expressed after exposure of M. tuberculosis to stress conditions and drugs29,33,54,55. We also reported earlier that a few TA systems are essential for M. tuberculosis to establish infection in host tissues33,44,56. In the present study, we show that compared to vector control, inducible expression of VapC35 results in cell elongation and growth inhibition of Mycobacterium smegmatis. We also report that VapBC35 is required for M. tuberculosis survival upon exposure to oxidative stress but is dispensable for growth in macrophages and aerosol-infected guinea pigs. Using various biophysical methods, we have also biochemically characterized the VapBC35 TA system from M. tuberculosis. Co-expression and binding studies revealed that VapC35 engages in cross-interactions with the non-cognate antitoxin VapB3. We have also determined the relative stabilities and oligomeric states of the toxin, antitoxin and reconstituted cognate and non-cognate TA complexes with varying ratios of both components. This is the first comprehensive study to provide insights into the biochemical and functional role of the VapBC35 TA system from M. tuberculosis.
Results
VapC35 overexpression in M. smegmatis causes growth inhibition and cell elongation
Previously, it has been reported that overexpression of VapC toxins belonging to the VapBC TA family inhibits E. coli, M. smegmatis, Mycobacterium bovis BCG, or M. tuberculosis growth in a bacteriostatic manner29,33,34,38. Here, we have functionally characterised the VapBC35 TA system from M. tuberculosis. VapC35 was purified with an N-terminal 6X-His tag for biochemical assays as per standard protocols. We have performed in vitro RNA cleavage assay using MS2 RNA to determine the ribonuclease activity of the purified VapC35. We observed that the purified VapC35 toxin possesses ribonuclease activity and was able to cleave the MS2 RNA in vitro (Fig. 1A). As expected, we did not observe ribonuclease activity of VapC35 in the presence of EDTA (Fig. 1A). Using an Atc inducible episomal vector, we noticed that compared to the vector control, overexpression of VapC35 inhibited M. smegmatis growth (Fig. 1B). As shown in Fig. 1C, in comparison to the vector control, ectopic expression of VapC35 significantly reduced the bacterial counts at 9 h and 24 h post-induction. We also observed that mutation of the glutamic acid residue of the PIN domain to alanine abrogated the ability of VapC35 to inhibit the growth of M. smegmatis upon overexpression (Fig. 1B, C). As expected, the growth defect associated with the overexpression of VapC35 was restored upon co-expression of its cognate antitoxin, VapB35, at both time points (Fig. 1D, E). We also observed that ectopic expression of VapC35 resulted in increased cell length in comparison to the strain harbouring vector control (Fig. 1F). Cell measurement studies showed that the average length of M. smegmatis harbouring pTetR-vapC35 or vector control was ~6 μm and 4 μm, respectively (Fig. 1G). In summary, our study demonstrates that VapC35 possesses ribonuclease activity, and its overexpression in M. smegmatis results in growth inhibition and cell elongation.
A Ribonuclease activity was performed by incubating MS2 RNA with 2 µM purified VapC35 protein at 37 °C for 30 min, in the absence or presence of EDTA and electrophoresed in 1.2% agarose gel. The data shown in this panel is representative of two independent experiments. B, C M. smegmatis strains harbouring pTetR or pTetR-vapC35 or pTetR-vapC35E5A were grown till OD600nm ~ 0.2–0.3. The expression of the toxin was induced by adding 50 ng/ml of Atc. The growth patterns of induced cultures were determined by measuring OD600nm (B) or bacterial counts (C) at designated time points. The data shown in (B) is individual replicates from two independent experiments performed in duplicates. The data shown in (C) is mean ± SD of log10 CFU obtained from two independent experiments performed in duplicates. D, E M. smegmatis harbouring an integrative and episomal copy of vapC35 and vapB35, respectively, were grown till OD600nm ~ 0.2–0.3. The expression of toxin and antitoxin was induced by the addition of Atc and Ac, respectively. The growth patterns of various strains were determined by measuring either OD600nm (D) or bacterial counts (E). The data shown in (D) is individual replicates obtained from three independent experiments. The data shown in (E) is mean ± SD of log10 CFU obtained from three independent experiments. F The effect of the expression of VapC35 on the cell viability of M. smegmatis was determined. For live-dead staining, Atc-induced cultures were harvested, stained using SYTO9/propidium iodide and imaged using a confocal microscope at 100× magnification. The images shown in this panel are representative of three independent experiments. G The length of M. smegmatis harbouring either pTetR or pTetR-vapC35 was determined from SYTO9/propidium iodide stained cells using Olympus CellSens software. The data shown in this panel is mean ± SD of the length obtained from ~200–225 bacilli. The data obtained was statistically analysed using paired t-test. Source data is provided in supplementary data 1.
VapBC35 TA complex binds to its native promoter in a toxin:antitoxin ratio-dependent manner
Previous studies have shown that antitoxins belonging to Type II TA systems possess a DNA binding motif at the amino-terminus and repress the transcription of the TA operon either alone or in a complex formed with the cognate toxin57,58,59. This promoter binding affinity is modulated by the toxin:antitoxin ratio, and this phenomenon is known as conditional cooperativity60. Conditional cooperativity enables bacteria to respond and control the stoichiometric ratios of toxin:antitoxin and finetune the transcriptional activity of the TA operon61,62. Through conditional cooperativity, the TA complex fine-tunes transcriptional repression, with the repression being strongest at a specific stoichiometric ratio of the two proteins61,62,63,64. This mode of gene regulation prevents random toxin activation and maintains toxin and antitoxin stoichiometry ratios. Next, we performed microscale thermophoresis (MST) experiments to determine the apparent binding of the in vitro assembled VapBC35 complexes of different stoichiometries with the vapBC35 promoter-operator region as described in ‘Methods’. The binding affinity of the VapBC35 complex to the native promoter improved with an increase in the antitoxin concentration (Fig. 2A–E). We observed that the binding affinity towards the promoter-operator regions was lowest for the VapB35:VapC35 (1:4) complex (Fig. 2A). The apparent binding affinity of the VapB35:VapC35 complex (4:1) towards the promoter was ~5.0-fold higher in comparison to the VapB35:VapC35 complex (1:4) (Fig. 2A, E, F). Taken together, this study demonstrates that the binding affinity of the VapBC35 TA complex to its native promoter is dependent on the relative ratios of VapB35 and VapC35.
A–E The dissociation constant between in vitro re-constituted VapBC35 complexes with vapBC35 promoter-operator DNA was measured using MST. The data was fitted using MO. Affinity Analysis software and the fitted data were plotted. F The KD, as a function of various ratios of the in vitro constituted VapBC35 complexes with promoter DNA. The increase in the antitoxin concentration resulted in an apparent increase in the affinity of the VapBC35 complex towards the P-O DNA. The data shown in these panels is from individual replicates and the mean fit of the values is obtained from two independent experiments performed in duplicates. Source data is provided in supplementary data 1.
Deletion of vapC35 or vapBC35 increases the susceptibility of M. tuberculosis upon exposure to oxidative stress
TA systems are differentially expressed in response to various stress conditions and contribute to bacterial stress adaptation9,13. Previous studies have shown that a subset of VapC toxins are induced in response to relevant stress conditions such as nitrosative, nutrient-limiting, and low oxygen conditions29,33. To investigate the role of the VapBC35 TA system in M. tuberculosis stress adaptation, ΔvapC35 and ΔvapBC35 mutant strains of M. tuberculosis were constructed using temperature-sensitive mycobacteriophages. The generation of toxin-deficient (ΔvapC35) or toxin-antitoxin deficient (ΔvapBC35) strains of M. tuberculosis was confirmed by Southern blot. As shown in Supplementary Fig. 1A, Supplementary Fig. 1B and Supplementary Fig. 6, the locus-specific probe hybridized with ~2.7 kb, 4.2 kb and 3.9 kb fragments of Sma I digested genomic DNA from wild type, ΔvapC35 and ΔvapBC35 M. tuberculosis strains, respectively. Next, we compared the ability of wild type, ΔvapC35 and ΔvapBC35 to survive after exposure to different in vitro stress conditions such as oxidative, nitrosative, nutritional and acidic stress as described in ‘Methods’. We observed that deletion of vapC35 increased the susceptibility of M. tuberculosis by ~20.0-fold and 64.0-fold after being exposed to oxidative stress for 6 h and 24 h, respectively (Fig. 3A). In agreement, in comparison to the parental strain, ΔvapBC35 also showed a growth defect of 24.0-fold upon exposure to oxidative stress for 24 h (Fig. 3E). As shown in Fig. 3A and Fig. 3E, the growth defect associated with these mutant strains upon exposure to oxidative stress was restored in the complemented strains. The survival of parental and complemented strains was comparable after exposure to oxidative stress. We observed that both VapC35 and VapBC35 TA systems are dispensable for M. tuberculosis adaptation to other conditions such as nitrosative for 72 h or nutritional for 14 days or acidic stress for 14 days (Fig. 3B–D, F–H). The contribution of TA systems to antibiotic persistence is debatable21,65. Nevertheless, some studies indicate that the activation of TA systems may contribute to the formation of drug-tolerant persisters66,67. It has also been reported that expression of toxins such as RelE1, RelE2, RelE3, MazF3, or VapC21 enhances the survival of M. smegmatis or M. tuberculosis after drug exposure49,68,69,70. Also, the expression of a subset of TA systems is increased in M. tuberculosis persisters54,55. In the present study, we also compared the survival of wild type, ΔvapC35 and ΔvapBC35 M. tuberculosis strains upon exposure to either rifampicin, isoniazid or levofloxacin. As shown in Supplementary Fig. 2A, B, deletion of either VapC35 or VapBC35 does not impair the survival of M. tuberculosis upon exposure to various drugs. These results suggest that VapC35 and VapBC35 are required for M. tuberculosis survival under oxidative stress conditions.
Early log phase cultures (OD600nm ~ 0.2–0.3) of various strains were subjected to either oxidative stress (A, E) or nitrosative stress (B, F) or nutritional stress (C, G) or acidic stress (D, H) as described in ‘Methods’. The values shown for the wild-type strain in (A) and (E) are similar. The data shown for time zero readings for wild type and ΔvapBC35 strain are similar in (F) and (H). The data shown in this panel is mean ± SD of log10 CFU obtained from two or three independent experiments performed in duplicates. The data obtained was statistically analysed using paired t-test. Source data is provided in supplementary data 1.
VapC35 and VapBC35 are not required for M. tuberculosis to establish disease in guinea pigs
Previous studies have shown that a subset of VapBC TA systems belonging to the type II subfamily is essential for M. tuberculosis to establish infection in guinea pigs and mice33,44,56. In addition to VapBC TA systems, toxins belonging to either MazEF or HigBA1 or MenAT TA systems are essential for M. tuberculosis pathogenesis71,72,73,74. Since we observed that both ΔvapC35 and ΔvapBC35 are susceptible to oxidative stress killing compared to the wild-type strain, we hypothesised that VapBC35 might be essential for M. tuberculosis survival in macrophages. However, we did not observe any significant differences in the growth of the mutant strains in THP-1 macrophages relative to the wild-type strain at day 0, 2, 4 and 6 post-infection (Supplementary Fig. 2C, D). Mouse and guinea pig models of TB infection have been extensively used to identify mutant strains that are attenuated for growth in vivo75. Studies have shown that in contrast to mice, the tuberculous granulomas formed in guinea pigs are hypoxic76,77. It has also been previously reported that M. tuberculosis-infected guinea pigs show the formation of caseating granulomas, as reported in humans75,78. Since in vivo models are more reliable in studying disease pathogenesis than in vitro studies, we next compared the ability of wild type, ΔvapC35 and ΔvapBC35 to establish disease in guinea pigs. We noticed that the bacterial loads in the lungs and spleens of guinea pigs infected with wild type and ΔvapC35 are comparable at 4- and 8-week post-infection. The lung bacillary loads in wild type infected guinea pigs were log10 5.28 and log10 4.90 at 4- and 8-week post-infection, respectively (Fig. 4A, B). The lung bacillary loads in guinea pigs infected with ΔvapC35 strain were log10 5.42 and log10 4.63, respectively, at 4- and 8-week post-infection (Fig. 4A, B). As shown in Fig. 4C, the growth of ΔvapBC35 was comparable relative to the wild-type strain in the lungs and spleens of aerosol-infected guinea pigs at 8-weeks post-infection. The lung bacterial loads in guinea pigs infected with wild-type and ΔvapBC35 guinea pigs were log10 5.43 and log10 5.40, respectively (Fig. 4C). In agreement with the bacterial burdens, the tissue damage seen in H & E stained sections of lung tissues infected with either wild type or ΔvapC35 or ΔvapBC35 strains was comparable at both time points (Fig. 4D–F). Overall, these findings suggest that both vapC35 and vapBC35 are dispensable for M. tuberculosis to establish infection in macrophages and guinea pigs. These observations also suggest that functional redundancy exists between the VapBC TA system, and a phenotype might only be observed upon simultaneous deletion of multiple TA systems.
A–C Guinea pigs were infected with either wild type or ΔvapC35 or ΔvapBC35 via the aerosol route. The bacterial loads were determined in the lungs and spleens of the infected animals at 4-weeks (A) or 8-weeks (B, C) post-infection. The data shown is mean ± SD of log10 CFU obtained from 6 animals per group per time point. D–F Histopathological analyses of lungs infected with various strains at 4-and 8-weeks post-infection were performed by staining the tissue sections with haematoxylin and eosin and viewed at a magnification of 10×. The images shown in this panel represent data obtained from 6 animals. Scale bar, 500 μm. Source data is provided in supplementary data 1.
VapC35 toxin interacts with non-cognate VapB3 antitoxin
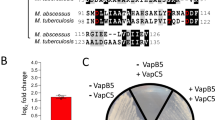
Earlier studies have shown cross-interaction between non-cognate TA systems in microorganisms such as E. coli, M. tuberculosis and H. influenzae49,52,53,79,80,81. Here, we performed experiments to determine cross-talk between VapC35 and non-cognate antitoxins. Using co-expression studies, we determined the ability of non-cognate antitoxins belonging to the Type II subfamily, such as VapBC, MazEF, RelBE or ParDE, to restore the growth defect associated with overexpression of VapC35 in M. smegmatis. The recombinant constructs were electroporated in M. smegmatis, harbouring an Atc inducible integrative copy of vapC35. The expression of antitoxin and VapC35 in early-log phase cultures of recombinant M. smegmatis strains was induced by adding Ac and Atc, respectively, and growth was monitored by measuring absorbance at regular intervals. As expected, we did not observe any growth inhibition in M. smegmatis coexpressing both VapB35 and VapC35 (Fig. 5A). We noticed that among the various VapB non-cognate antitoxins, co-expression of VapB3 was able to restore the growth defect associated with VapC35 expression. We also noticed that co-expression of non-cognate MazE, RelB or ParD antitoxins could not restore the growth defect associated with overexpression of VapC35 in M. smegmatis (Fig. 5B–D). In our repeat experiments, as expected, co-expression of VapB35 and VapB3 restored the growth defect associated with VapC35 expression in M. smegmatis in liquid cultures and spotting assays (Fig. 5E, F). We did not observe any growth restoration upon co-expression of other non-cognate VapB antitoxins such as VapB5, VapB19, VapB29, VapB38, VapB46 or VapB47 (Fig. 5E). Taken together, these results suggest that VapC35 also interacts with non-cognate antitoxin, VapB3.
A–D M. smegmatis harbouring an Atc inducible integrative and Ac inducible episomal copy of vapC35 and antitoxin (vapB, (A), mazE, (B), relB, (C) and parE, (D)) were grown till early log phase (OD600nm ~ 0.2–0.3). The expression of toxin and antitoxin was induced by the addition of Atc and Ac, respectively. The heat map represents the growth patterns of various M. smegmatis strains as determined by measuring OD600nm at regular intervals. E Growth kinetics of M. smegmatis harbouring an integrative copy of VapC35 either alone or in the presence of VapB35, VapB3, VapB5, VapB19, VapB29, VapB38, VapB46 or VapB47. The growth of various strains was determined by measuring OD600nm at regular intervals. The data shown in (A–E) are representative of two independent experiments. F The induced cultures were serially diluted at time zero, 12 h post-induction, 24 h post-induction and spotted on MB7H11 plates at 37 °C for 48 h. The images shown in this panel are representative of two independent experiments. Source data is provided in supplementary data 1.
The full-length VapBC complexes adopt diverse oligomeric states in solution
Previously, it has been shown that toxins and antitoxins belonging to the VapBC family form homodimers44,48,49,82,83. It has also been reported that toxins and antitoxins form either hetero-hexameric (T2A2T2) or hetero-octameric (T4A4) complexes49,84,85,86. Next, we purified and determined the oligomeric status of toxins (VapC3, VapC35) and antitoxins (VapB3, VapB35) by size exclusion chromatography coupled to multi-angle light scattering (SEC-MALS). For individually purified VapB35, VapC35, VapB3 and VapC3, peak 1 corresponds to a homodimer (Fig. 6A, B, H, I, Table 1). Peak 2 observed in the case of purified VapC3 corresponds to higher order aggregates (Fig. 6I). We also measured the oligomeric status of the in vitro assembled cognate TA complexes (VapB35:VapC35, VapB3:VapC3) at different stoichiometric ratios. We observed different oligomeric states of the VapBC complexes depending on the relative molar ratios of VapB to VapC. At a 1:1 ratio of cognate antitoxin and toxin as well as at a higher antitoxin to toxin ratio (VapB:VapC, 2:1; 4:1), we observed the formation of a hetero octameric complex (T4A4) (Fig. 6E, F, G, K, L, Table 1). However, at a higher VapC to VapB ratio (VapB:VapC, 1:2; 1:4), we observed that a hetero-hexameric complex (T2A2T2) was formed (Fig. 6C, D, J, Table 1). We also noticed two additional peaks in cases where the complexes were reconstituted in vitro. As shown in Fig. 6C, D, L, peak 1 corresponds to free antitoxin or toxin. Notably, peak 2, observed in Fig. 6J, K corresponds to higher-order oligomers and peak 3, observed in Fig. 6J corresponds to aggregates. Further, we also determined the oligomeric status of the in vitro constituted non-cognate TA complex (VapB3:VapC35) at different stoichiometric ratios of 1:1 and 2:1. We observed distinct hetero-oligomeric states in non-cognate TA complex, where peak 1, peak 2 and peak 3 correspond to the formation of hetero hexamer (T2A2T2) or hetero octamer (T4A4) or higher order aggregates, respectively (Fig. 6M, N, Table 1). In summary, our analysis has revealed a range of oligomeric states of the in vitro-constituted VapBC complexes. The formation of these oligomeric states varies significantly depending on the stoichiometric ratios of antitoxin and toxin.
Traces for refractive index are shown in blue. The molar mass and fits of all traces are plotted as a function of elution time as black dots and red lines, respectively. Elution profiles for purified toxins, antitoxins and in vitro constituted VapB:VapC complexes at different stoichiometric ratios are shown. A VapB35, (B) VapC35, in vitro assembled VapB35:VapC35, complexes at different ratios of VapB:VapC (C) VapB35:VapC35::1:4, (D) VapB35:VapC35::1:2, (E) VapB35:VapC35::1:1, (F) VapB35:VapC35::2:1, (G) VapB35:VapC35::4:1. Purified (H) VapB3, (I) VapC3, in vitro assembled VapB3:VapC3 complexes at different ratios of VapB:VapC (J) VapB3:VapC3::1:2, (K) VapB3:VapC3::1:1, (L) VapB3:VapC3::2:1, in vitro assembled VapB3:VapC35, complexes at different ratios of VapB:VapC (M) VapB3:VapC35::1:1, (N) VapB3:VapC35::2:1. Data is representative of two independent experiments. Source data is provided in supplementary data 1.
Thermal stability and binding studies of cognate and non-cognate TA systems
Next, purified proteins VapB3, VapB35, VapC3, and VapC35, and the in vitro assembled VapBC35, VapBC3, VapB3:VapC35 complexes of different stoichiometries were subjected to thermal denaturation as described in ‘Methods’. Protein unfolding was monitored using the intrinsic fluorescence of tryptophan and tyrosine residues with temperature, and the apparent Tm was calculated. We observed that VapB35 and VapC35 displayed Tm of 51 °C and 58 °C, respectively (Fig. 7A). In agreement with previous studies, we also observed higher thermal stability of the in vitro assembled VapBC35 complexes in comparison to free toxin or antitoxin (Fig. 7A)49,81,87,88,89,90,91. These observations indicate stabilization of both proteins upon complex formation. As shown in Fig. 7B, the stability of the complex increased (in the range of 71–73 °C), with an increase in the antitoxin concentration. As shown in Fig. 7C, VapC3 and VapBC3 displayed Tm of 56 °C and 86 °C, respectively (Fig. 7C). Further, we also determined the stabilities of in vitro assembled VapB3:VapC35 complex of different stoichiometries (Fig. 7C). We observed that non-cognate complex (Tm in the range of 70–76 °C) had higher thermal stability as compared to toxins VapC35 (Tm 58 °C) and VapC3 (Tm 56 °C), but similar thermal stability as compared to the cognate VapBC35 complex (Tm in the range of 71–73 °C) or lower thermal stability as compared to the cognate VapBC3 complex (Tm 86 °C) (Fig. 7C). We next determined the binding of VapC35 with the full-length antitoxins VapB3, VapB9, VapB15, VapB34 and VapB35, respectively, using MST as described in ‘Methods’. We observed that VapC35 binds with VapB35 antitoxin with an affinity of 1 nM (Fig. 7D). The binding of VapC3 with VapB3 antitoxin displayed a KD value of ~7 nM (Supplementary Fig. 3). Among various non-cognate antitoxins, we observed that VapB3 binds to VapC35 with a KD value of ~14 nM (Fig. 7D). As expected, we did not observe any interaction between VapC35 with other non-cognate antitoxins VapB9, VapB15, and VapB34.
A Thermal unfolding profiles of 10 μM of purified VapB35, VapC35, and different ratios of in vitro assembled VapB35:VapC35 complexes are shown. B The stability of the VapBC35 complex (Tm) increases as the VapB35:VapC35 ratio increases. C Thermal unfolding profiles of 10 μM of purified VapC3, in vitro assembled full-length VapB3:VapC3 complex (1:1), and different ratios of in vitro assembled VapB3 and VapC35 complexes are shown in this panel. D The dissociation constant between VapC35 and various antitoxins such as VapB35 (red), VapB3 (blue), VapB9 (yellow), VapB15 (green) and VapB34 (magenta) was measured using MST. The data shown in (A) and (C) is representative of two independent experiments. The data shown in (B) is mean ± SD of Tm obtained from two experiments performed in duplicates. The data shown in (D) is from individual replicates and the mean fit of the values is obtained from two independent experiments performed in duplicates. Source data is provided in supplementary data 1.
Discussion
TA systems were initially reported to be important for plasmid maintenance in prokaryotes92. However, later studies revealed that chromosomally encoded TA systems in prokaryotes are essential for stress adaptation, persistence, biofilm formation and establishment of successful infection in the host18,20,93,94,95,96,97. M. tuberculosis possesses a remarkably large number of TA systems, most of which belong to the VapBC family29,30,31. Despite significant advances in the field of TA biology, the contribution of most of these TA systems to the pathophysiology of M. tuberculosis is still not understood. We have earlier functionally characterized TA systems such as VapBC3, VapBC4, VapBC22, VapBC28, VapBC21, HigB1, MazEF3, MazEF6, MazEF9, MenAT2, MenAT3 and MenAT4 from M. tuberculosis33,44,49,56,71,72,73,74. Here, we have functionally characterized the VapBC35 TA system from M. tuberculosis. As reported in the case of other VapC toxins, we also show that ectopic expression of VapC35 inhibits M. smegmatis growth in a bacteriostatic manner33,34. As expected, this growth defect was restored upon co-expression of its cognate antitoxin VapB35. Previous studies have shown that toxins belonging to Type II TA systems inhibit bacterial growth by targeting essential cellular processes such as translation, DNA replication, cell-wall synthesis, transcription, replication, membrane integrity, ribosome assembly, cytoskeleton formation and cell-wall synthesis4,19,24,25. VapC toxins are characterized by the presence of the PIN domain, and most of these proteins possess ribonuclease activity98. In agreement, we also show that the VapC35 toxin possesses ribonuclease activity and can cleave MS2 RNA. We observed that compared to vector control, overexpression of VapC35 in M. smegmatis resulted in growth inhibition and increased cell length. We speculate that this increase in bacterial length might be associated with the interference of the activity of cell division proteins in the VapC35 overexpression strain.
Previously, it has been shown that a subset of toxins and antitoxins are differentially expressed in conditions that M. tuberculosis might encounter in host tissues, such as nitrosative, oxidative, nutrient-limiting or low oxygen conditions29,33. These observations suggest that TA systems might contribute either alone or in a cumulative manner to the ability of M. tuberculosis to adapt to different stress conditions. It has been reported that deletion of vapC22 or overexpression of VapB22 increases the susceptibility of M. tuberculosis to oxidative stress56. Previously, we have shown that VapBC3, VapBC4, VapBC11 and VapC22 are essential for M. tuberculosis pathogenesis33,44,56. In the present study, we also determined the contribution of the VapC35 toxin and VapBC35 TA system in stress adaptation and pathogenesis of M. tuberculosis. The results revealed that both VapC35 and VapBC35 are required for M. tuberculosis to survive upon exposure to oxidative stress. We also observed that VapC35 and VapBC35 are dispensable for M. tuberculosis growth after exposure to other stress conditions, such as nitrosative, acidic and nutrient-limiting growth conditions. Previously, it has been reported that, similar to humans, oxidative stress conditions exist in the guinea pig model of TB and that M. tuberculosis strains more susceptible to oxidative stress show attenuated phenotype in guinea pigs73,74,99,100,101. Since both ΔvapC35 and ΔvapBC35 strains are susceptible to oxidative stress, we speculated that compared to the wild-type strain, these strains might be defective in their ability to establish disease in guinea pigs. Surprisingly, our macrophage and guinea pig infection studies revealed that neither VapC35 nor VapBC35 are required by M. tuberculosis to establish infection in host tissues. The absence of an attenuated phenotype for ΔvapC35 and ΔvapBC35 in guinea pigs might be attributed to the following reasons. Firstly, the extent and duration of oxidative stress encountered by M. tuberculosis might vary in our in vitro experiments and in vivo. Secondly, M. tuberculosis encounters multiple stress conditions such as oxidative, nitrosative, acidic, low oxygen or nutritional stress inside host tissues. Since VapBC35 is dispensable for survival in other stress conditions, there is a possibility that the deletion of vapC35 or vapBC35 does not affect the survival of M. tuberculosis in an environment where the bacteria is simultaneously exposed to multiple stress conditions. Lastly, the lack of attenuated phenotype in vivo could be attributed to functional redundancy between TA systems in M. tuberculosis, which could potentially compensate for the lack of VapC35 and VapBC35 under in vivo conditions.
Among the VapBC TA systems from M. tuberculosis, three-dimensional structures of VapBC3, VapBC5, VapBC11, VapBC15, VapBC26, and VapBC30 have been determined42,43,44,45,46,47. These structures have provided insights into the oligomeric states and interactions within these complexes, revealing the binding interfaces between cognate toxin and antitoxin components. The backbone root-mean-square error of superposition (RMS) between the solved structures of the VapBC TA complexes ranges between 1.86 and 4.79 Å (Fig. S4)102. In comparison, the toxins (VapC20, VapC21) exist as dimers in solution39,48. The toxin structures are highly conserved with a backbone RMS of 2.21 Å. The composition of residues at the interface among different TA systems is highly conserved (Fig. S5A–F). The interfaces show a consistently high prevalence of amide and aromatic residues, followed by basic and acidic residues (Fig. S5G). In addition, mutational scanning approaches such as aspartate or cysteine scanning have been performed to elucidate the secondary structure of antitoxins and functional residues within TA systems81,103. These studies have resulted in the mapping of binding interfaces and the identification of crucial residues that are important for TA complex formation. Since neither the crystal structure nor mutational scanning data of the VapC35 toxin or the VapBC35 complex are available, we performed a detailed biophysical characterisation of the VapBC35 TA system. SEC-MALS studies revealed that depending upon the stoichiometric ratios of antitoxins and toxins, the VapBC35 complex adapts hetero-hexameric and hetero-octameric states. These observations are consistent with previous findings for other TA systems, such as MazEF3, MazEF6, MazEF9, VapBC11, VapBC21, VapC20 and VapC2244,48,49,56,81,82,91. As expected, the VapBC35 TA complex was thermally more stable than the VapC35 toxin and VapB35 antitoxin. In agreement with SEC-MALS data, higher antitoxin concentrations led to increased complex stability, possibly due to the formation of higher-order oligomers.
The autoregulation of TA operons by cognate TA complexes underscores the importance of maintaining a precise balance between toxin and antitoxin levels to regulate bacterial growth and responses to stress conditions57,104,105,106. In agreement with earlier reports, we observed that the binding affinity of the VapBC35 complex to the native promoter is contingent on the antitoxin-to-toxin ratios, with higher antitoxin concentrations enhancing promoter binding. This observed autoregulation is governed by conditional cooperativity, which has also been reported for other TA systems61,62,63. We have not performed promoter binding assays at VapB35:VapC35 ratios of greater than 1:4 because, at these ratios, excess antitoxin might outcompete the TA complex, thereby resulting in unreliable KD estimations. Furthermore, increasing the concentration of these complexes resulted in visible precipitation, which compromises the accuracy of protein concentration measurements that are important for the accurate determination of KD values. In addition to autoregulation, cross-interaction between TA systems has been reported in various microorganisms such as E. coli, M. tuberculosis, and H. influenzae49,52,53,79,80. Using co-expression studies in M. smegmatis, we report that co-expression of VapB3 was able to restore the growth defect associated with the inducible expression of VapC35. SEC-MALS and MST experiments also confirmed interactions between non-cognate VapC35 and VapB3 proteins. In agreement with the co-expression study, MST studies revealed that VapC35 binds with a higher affinity with VapB3 in comparison to other non-cognate antitoxins. Based on these observations, we speculate that regulatory networks exist between TA systems, and these networks are important for their functioning.
This is the first study reporting a detailed functional and biophysical characterization of the VapBC35 TA system from M. tuberculosis. We show that inducible expression of VapC35 exhibits a bacteriostatic effect upon expression in M. smegmatis, which is reversed by co-expression of cognate antitoxin, VapB35 and non-cognate antitoxin, VapB3. We also report that despite being necessary for the adaptation of M. tuberculosis to oxidative stress, VapBC35 is dispensable for in vivo growth. These observations suggest functional redundancy exists between TA systems, and these modules might function cumulatively in vivo. Future studies would involve the construction of multiple deletion mutants to understand the cumulative role of TA systems in M. tuberculosis pathogenesis. The extensive biophysical characterization revealed multiple oligomeric states of the VapBC35 complex, with higher antitoxin concentrations promoting complex stability and tighter autoregulation. Future studies would involve identifying the residues involved in cognate (VapB35:VapC35) and non-cognate (VapB3:VapC35) TA systems using a combination of various methodologies such as high-throughput display methods, screening mutagenesis libraries, flow cytometry and next-generation sequencing as reported earlier81,103,107,108,109,110,111. This study also expands our understanding of the regulatory networks that exist between non-cognate TA systems from M. tuberculosis.
Methods
Bacterial strains and culture conditions
The strains and plasmids used in the study are listed in Table 2. Escherichia coli strains (XL-1 blue, (λDE3, plysE) or Rosetta (DE3), HB-101) used in the study were grown in Luria-Bertani (LB) broth and Luria-Bertani agar plates. Various mycobacterial strains were grown in 7H9 medium with 0.2% glycerol, 0.05% Tween-80 with 1x ADS and on 7H11 agar with 1x OADS as per standard protocols112. All cultures were grown by incubating at 37 °C, 150 rpm unless specified. The media were supplemented with antibiotics, as appropriate. All chemicals, unless specified, were procured from Sigma Aldrich.
Generation of mutant and complemented strain of M. tuberculosis
The sequences of primers used in the study are listed in Supplementary Table 1. M. tuberculosis H37Rv was used as the parental strain for constructing mutant and complemented strains. M. tuberculosis strain lacking either vapC35 (Rv1962c) or vapBC35 (Rv1962a-Rv1962c) was constructed using temperature-sensitive mycobacteriophages113. To construct mutant strains, approximately 800 bp flanking upstream and downstream regions of either vapC35 or vapBC35 were PCR amplified using gene-specific primers and cloned into pYUB854 cosmid flanking the hygromycin cassette. The resulting cosmids pYUB854-ΔvapC35 or pYUB854-ΔvapBC35 were Pac I digested and packaged into pYUB159 phagemid using MaxPlax™ lambda packaging extract. The resulting phagemids pYUB159-ΔvapC35 or pYUB159-ΔvapBC35 were electroporated into M. smegmatis mc2155 strain to generate high-titre temperature-sensitive mycobacteriophages. Subsequently, mid-log phase cultures of M. tuberculosis (OD600nm ~ 0.8–1.0) were transduced with high titre of mycobacteriophages and transformants were selected on MB7H11 plates supplemented with hygromycin. The replacement of either vapC35 or vapBC35 in the respective mutant strains with the hygromycin resistance gene was confirmed by Southern blot using a locus-specific probe. To construct the complemented strain, the vapBC35 locus was PCR amplified and cloned into pJEB402, resulting in pJEB402-vapBC35. The recombinant DNA was electroporated in electrocompetent cells of ΔvapC35 and ΔvapBC35 to generate complemented strains, and the transformants were selected on MB7H11 plates supplemented with kanamycin and hygromycin.
Growth inhibition studies
For growth inhibition studies in M. smegmatis, vapC35 was PCR amplified and cloned into either an anhydrotetracycline (Atc) inducible episomal (pTetR) or integrative vector (pTetR-int) resulting in pTetR-vapC35 and pTetR-int-vapC3533. Also, vapC35E5A harbouring a mutation in glutamic acid residue at the 5th position of the PIN domain was PCR amplified and cloned into pTetR, resulting in pTetR-vapC35E5A. The resulting plasmids were electroporated in M. smegmatis, and transformants were selected on MB7H11 plates supplemented with either hygromycin (for pTetR constructs) or kanamycin (for pTetR-int constructs). The expression of vapC35 or vapC35E5A in early-log phase cultures of M. smegmatis (OD600nm ~ 0.2–0.3) was induced by adding 50 ng/ml Atc. For co-expression studies, various antitoxins were cloned into an acetamide (Ac) inducible vector, pLam12 and electroporated into M. smegmatis harbouring an integrative copy of VapC35 as reported earlier49. The addition of Atc and Ac induced the expression of toxin and antitoxin, respectively, in early-log phase cultures of recombinant M. smegmatis strains. The growth patterns of various recombinant strains were determined by measuring OD600nm or bacterial counts or by spotting at different time points post-induction.
Live-dead staining and cell length measurement
To determine the effect of overexpression of VapC35 on cell length and viability of M. smegmatis, a live dead staining assay was performed as previously reported49. For live dead staining, induced cultures were harvested, washed with 1x phosphate-buffered saline (1x PBS) and single-cell suspension was prepared as per standard protocols. The single-cell suspension of washed bacilli was stained with SYTO9/Propidium iodide and viewed under confocal microscope (Olympus FV3000, Japan). The length of approximately 220–250 individual bacilli in Atc-induced cultures was measured using Olympus cellSens software.
In vitro growth, stress and macrophage experiments
The growth patterns of wild type, ΔvapC35 and ΔvapBC35 strains of M. tuberculosis were determined by measuring OD600nm till the stationary phase was attained. To study the role of vapC35 and vapBC35 in stress adaptation, early-log phase cultures (OD600nm ~ 0.2–0.3) of various strains were exposed to different stress conditions such as oxidative (5 mM H2O2 in MB7H9 medium for 6 or 24 h), nutritional (1x tris-buffered saline with 0.05% Tween-80 for either 7 or 14 days), nitrosative (MB7H9 medium pH 5.2, 5 mM NaNO2 for either 24 or 72 h) and acidic stress (MB7H9 medium, pH 5.2 for either 7 or 14 days) as previously reported49,112. For drug tolerance experiments, mid-log phase cultures (OD600nm ~ 0.8–1.0) of various strains were exposed to either 0.4 µg/mL of rifampicin or 10 µg/mL of levofloxacin or 10 µg/mL of isoniazid for either 7 or 14 days as reported earlier49,112. The samples were collected at designated time points, 10.0-fold serial dilutions were prepared and plated on MB7H11 agar at 37 °C for 3–4 weeks. For macrophage infection studies, THP-1 monocytes (National Centre for Cell Science, Pune) were differentiated into macrophages using 20 ng/mL of phorbol 12-myristate 13-acetate for 48 h. The differentiated macrophages were washed once with 1x PBS and supplemented with RPMI containing 10% FBS. After 24 h, the cells were infected with either wild type or mutant strains at a multiplicity of infection of 1:10. At 4 h post-infection, macrophages were overlaid with RPMI medium containing amikacin (200 μg/mL) to remove extracellular bacteria. After amikacin treatment for 2 h, macrophages were washed twice with 1x PBS and overlaid with complete RPMI medium. At designated time points, infected macrophages were lysed using 1x PBS containing 0.1% Triton-100. For bacterial enumeration, at designated time points, 10.0-fold serial dilutions were prepared and plated on MB7H11 agar at 37 °C for 3–4 weeks.
In vivo infection experiments
The animal experiments were approved by the animal ethical committee of the Translational Health Science and Technology Institute (THSTI) and performed as per the guidelines of the committee for the purpose of control and supervision of experiments on animals (CCSEA). The performed animal experiments complied with all relevant ethical regulations. 6–8-week-old female Duncan Hartley Guinea pigs (250–300 g) were procured from Lala Lajpat Rai University of Veterinary and Animal Sciences, Hisar, India. The guinea pigs were housed in individually ventilated cages in group of 3 in a biosafety level 3 facility. The guinea pigs were housed at a room temperature of 22 ± 3°C, relative humidity of 30 to 70%, 15–20 air changes/hr, light intensity of 325–350 lux with a 14 h light/10 h dark cycle and noise intensity of <85 db. For infection studies, the guinea pigs were infected using a Glas-Col aerosol inhalation exposure chamber with approximately 50–100 bacteria of either wild type or ΔvapC35 or ΔvapBC35 strains of M. tuberculosis. For bacterial enumeration at 4- and/or 8-week post-infection, lungs and spleen tissues were harvested, homogenised, serially diluted and plated on MB7H11 plates at 37 °C for 3–4 weeks. To determine the extent of tissue damage, a portion of lung tissue was fixed with 10% formalin and stained with haematoxylin and eosin112.
Bioinformatic analysis of the solved VapBC TA complex structures
To determine the structural conservation among different VapBC complex structures, the backbone root mean square error of superposition (RMS) was calculated using TopMatch-web, a pairwise structure alignment tool102. The amino acid propensities at the interfaces of the TA complexes with solved structures were determined using PDBsum114,115. In addition, the abundance of different functional groups of amino acids (e.g., amide, aromatic, basic and acidic) at the interface of the TA complex was also determined.
Cloning, protein expression, and purification
For protein purification, individual toxins (vapC3, Rv0549c or vapC35, Rv1962c) and antitoxins (vapB3, Rv0550c, or vapB35, Rv1962a, or vapB9, Rv0959a, or vapB15, Rv2009, or vapB34, Rv1740) were PCR amplified, cloned into either pET15b or pET28b and transformed intro either BL21 (λDE3, plysE) or Rosetta (DE3). The recombinant BL21 (λDE3, plysE for purification of VapB3 and VapC3) and Rosetta (λDE3, for purification of VapB35, VapC35, VapB9, VapB15 and VapB34) strains were cultured in TB medium until an OD600nm ~ 0.8 was attained. The expression of recombinant proteins was induced by adding 1.0 mM isopropyl β-D-1-thiogalactopyranoside overnight at 20 °C. The induced cultures were harvested by centrifugation, and bacterial pellets were resuspended and sonicated in lysis buffer (10 mM HEPES, pH-8.0, 100 mM NaCl, 100 mM L-arginine, 10% glycerol containing protease inhibitor). The clarified lysates were prepared by centrifugation and incubated with Ni-NTA Sepharose 6 Fast Flow for 4 h at 4 °C. Subsequently, the protein-bound resin was washed with twenty-five column volumes of wash buffer (10 mM HEPES, pH-8.0, 100 mM NaCl, 100 mM L-arginine, 50 mM imidazole, 10% glycerol). The recombinant protein fractions were eluted with elution buffer (10 mM HEPES, pH-8.0, 100 mM NaCl, 100 mM L-arginine, 10% glycerol and a gradient of imidazole (100–900 mM)). The eluted fractions were subjected to 15% tricine SDS-PAGE, and the protein concentration was determined using densitometric analysis. The purified fractions were dialysed, concentrated and stored in storage buffer (10 mM HEPES, pH-8.0, 100 mM NaCl, 100 mM L-arginine, 500 mM imidazole, 10% glycerol, protease inhibitor) at −80 °C till further use.
Ribonuclease assays
For ribonuclease assays, 0.5 μg of MS2 RNA was incubated with 2 μM purified VapC35 protein at 37 °C in the absence or presence of 50 mM EDTA. After incubation for 30 min, the reactions were electrophoresed on 1.2% agarose at 60 V, and images were captured using the gel documentation system (Bio-Rad ChemiDoc MP imaging system).
nanoDSF experiments
Thermal unfolding of purified toxins, antitoxins and in vitro assembled TA complexes at different stoichiometric ratios was performed using nanoDSF (Prometheus NT.48). The sample was filled into capillaries and placed into the instrument as reported earlier86,116. All the thermal unfolding experiments were performed at 10 µM of purified protein. Briefly, the samples were excited at 280 nm, and emission was detected at 330 nm and 350 nm to derive the F350/F330 ratio for data analysis, with a bandwidth of measurement set at 5 nm.
The first derivative of the ratio (\({F}^{\prime}=\frac{d\frac{F350}{F330}}{{dT}}\)) is normalised using Eq. 1:
Where y is the first derivative of the observed fluorescence (F350/F330 ratio), ymin is the value of the first derivative minimum, and ymax is the value of the first derivative maximum.
SEC-MALS experiments
100 μg of the purified toxins, antitoxins and TA complexes at different stoichiometric ratios were analysed on a Superdex-200 analytical gel filtration column (GE Healthcare) equilibrated in elution buffer (10 mM HEPES, pH-8.0, 100 mM NaCl, 100 mM L-arginine, 500 mM imidazole) with in-line UV (SHIMADZU), MALS (mini DAWN TREOS, Wyatt Technology Corp.) and refractive index detectors (WATERS24614) for molecular weight analysis. UV, MALS and RI data were collected and analysed using ASTRATM software (Wyatt Technology) as reported earlier49.
MST experiments
For MST experiments, purified toxins were equilibrated in 10 mM HEPES, pH-8.4 and labelled using the Monolith™ Protein Labelling Kit RED-NHS (NanoTemper Technologies) according to the manufacturer’s instructions. The labelled toxins were used at a dimeric concentration of 500 nM and 20 nM for VapC3 and VapC35, respectively, and titrated with different concentrations of unlabelled full-length cognate and non-cognate antitoxins ranging from 1 pM to 5 µM in 10 mM HEPES, pH-8.4 at 25 °C. For promoter binding studies, labelled VapC35 was used at a dimeric concentration of 40 nM and was incubated with 160, 80, 40, 20 and 10 nM of cognate antitoxin VapB35 to form 4:1, 2:1, 1:1, 1:2 and 1:4 (A:T) complexes. Subsequently, serially diluted cognate VapBC35 promoter-operator DNA (concentrations from 10 nM to 25 µM) was added. The measurement was performed using MST power settings, medium and high, at LED/excitation power settings of 40–60%. Data was analysed using MO. Affinity Analysis software (version 2.2.5, NanoTemper Technologies)86,117.
Statistical analysis and data reproducibility
The statistical analyses and the graphs were plotted using GraphPad Prism version 9.5.1 (GraphPad Software Inc., CA, United States). Groups with a P value < 0.05 were considered statistically significant. The respective figure legends mention the number of replicates and the statistical tests used.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The published article and its supplementary information include all the data generated and analysed in this study. All the source values underlying all figures and the uncropped and unedited blots are available in supplementary data 1.
References
Harms, A., Brodersen, D. E., Mitarai, N. & Gerdes, K. Toxins, targets, and triggers: an overview of toxin-antitoxin biology. Mol. Cell 70, 768–784 (2018).
Qiu, J., Zhai, Y., Wei, M., Zheng, C. & Jiao, X. Toxin-antitoxin systems: classification, biological roles, and applications. Microbiol. Res. 264, 127159 (2022).
Jurenas, D., Fraikin, N., Goormaghtigh, F. & Van Melderen, L. Biology and evolution of bacterial toxin-antitoxin systems. Nat. Rev. Microbiol. 20, 335–350 (2022).
Yamaguchi, Y., Park, J. H. & Inouye, M. Toxin-antitoxin systems in bacteria and archaea. Annu. Rev. Genet. 45, 61–79 (2011).
Sonika, S., Singh, S., Mishra, S. & Verma, S. Toxin-antitoxin systems in bacterial pathogenesis. Heliyon 9, e14220 (2023).
Lobato-Marquez, D., Diaz-Orejas, R. & Garcia-Del Portillo, F. Toxin-antitoxins and bacterial virulence. FEMS Microbiol. Rev. 40, 592–609 (2016).
Page, R. & Peti, W. Toxin-antitoxin systems in bacterial growth arrest and persistence. Nat. Chem. Biol. 12, 208–214 (2016).
LeRoux, M. & Laub, M. T. Toxin-antitoxin systems as phage defense elements. Annu. Rev. Microbiol. 76, 21–43 (2022).
Zhang, L. Y. et al. Toxin-antitoxin systems alter adaptation of Mycobacterium smegmatis to environmental stress. Microbiol. Spectr. 10, e0281522 (2022).
Pizzolato-Cezar, L. R., Spira, B. & Machini, M. T. Bacterial toxin-antitoxin systems: Novel insights on toxin activation across populations and experimental shortcomings. Curr. Res. Micro. Sci. 5, 100204 (2023).
Yang, Q. E. & Walsh, T. R. Toxin-antitoxin systems and their role in disseminating and maintaining antimicrobial resistance. FEMS Microbiol. Rev. 41, 343–353 (2017).
Pandey, D. P. Gerdes K. Toxin-antitoxin loci are highly abundant in free-living but lost from host-associated prokaryotes. Nucleic Acids Res. 33, 966–976 (2005).
Khan, S. et al. Toxin-Antitoxin system of Mycobacterium tuberculosis: Roles beyond stress sensor and growth regulator. Tuberculosis143, 102395 (2023).
Buts, L., Lah, J., Dao-Thi, M. H., Wyns, L. & Loris, R. Toxin-antitoxin modules as bacterial metabolic stress managers. Trends Biochem. Sci. 30, 672–679 (2005).
Slayden, R. A., Dawson, C. C. & Cummings, J. E. Toxin-antitoxin systems and regulatory mechanisms in Mycobacterium tuberculosis. Pathog. Dis. 76, 1–12 (2018).
Wang, X. & Wood, T. K. Toxin-antitoxin systems influence biofilm and persister cell formation and the general stress response. Appl Environ. Microbiol 77, 5577–5583 (2011).
Choi, J. S. et al. The small RNA, SdsR, acts as a novel type of toxin in Escherichia coli. RNA Biol. 15, 1319–1335 (2018).
Kamruzzaman, M., Wu, A. Y. & Iredell, J. R. Biological functions of type II toxin-antitoxin systems in bacteria. Microorganisms 9, 1276, 1–23 (2021).
Unterholzner, S. J., Poppenberger, B. & Rozhon, W. Toxin-antitoxin systems: Biology, identification, and application. Mob. Genet Elem. 3, e26219 (2013).
Singh, G., Yadav, M., Ghosh, C. & Rathore, J. S. Bacterial toxin-antitoxin modules: classification, functions, and association with persistence. Curr. Res. Micro. Sci. 2, 100047 (2021).
Holden, D. W. & Errington, J. Type II toxin-antitoxin systems and persister cells. mBio 9, e01574-18, 1–2 (2018).
Kang, S. M., Kim, D. H., Jin, C. & Lee, B. J. A systematic overview of type II and III toxin-antitoxin systems with a focus on druggability. Toxins 10, 515, 1–21 (2018).
Wang, X., Yao, J., Sun, Y. C. & Wood, T. K. Type VII toxin/antitoxin classification system for antitoxins that enzymatically neutralize toxins. Trends Microbiol. 29, 388–393 (2021).
Schuster, C. F. & Bertram, R. Toxin-antitoxin systems are ubiquitous and versatile modulators of prokaryotic cell fate. FEMS Microbiol. Lett. 340, 73–85 (2013).
Van Melderen, L. Toxin-antitoxin systems: why so many, what for? Curr. Opin. Microbiol. 13, 781–785 (2010).
Prysak, M. H. et al. Bacterial toxin YafQ is an endoribonuclease that associates with the ribosome and blocks translation elongation through sequence-specific and frame-dependent mRNA cleavage. Mol. Microbiol. 71, 1071–1087 (2009).
Fraikin, N., Goormaghtigh, F. & Van Melderen, L. Type II toxin-antitoxin systems: evolution and revolutions. J. Bacteriol. 202, 1–14 (2020).
Ramisetty, B. C. & Santhosh, R. S. Horizontal gene transfer of chromosomal Type II toxin-antitoxin systems of Escherichia coli. FEMS Microbiol. Lett. 363, fnv238, 1–7 (2016).
Ramage, H. R., Connolly, L. E. & Cox, J. S. Comprehensive functional analysis of Mycobacterium tuberculosis toxin-antitoxin systems: implications for pathogenesis, stress responses, and evolution. PLoS Genet. 5, e1000767 (2009).
Akarsu, H. et al. TASmania: a bacterial toxin-antitoxin systems database. PLoS Comput. Biol. 15, e1006946 (2019).
Tandon, H. et al. Bioinformatic and mutational studies of related toxin-antitoxin pairs in Mycobacterium tuberculosis predict and identify key functional residues. J. Biol. Chem. 294, 9048–9063 (2019).
Arcus, V. L., McKenzie, J. L., Robson, J. & Cook, G. M. The PIN-domain ribonucleases and the prokaryotic VapBC toxin-antitoxin array. Protein Eng. Des. Sel. 24, 33–40 (2011).
Agarwal, S. et al. System-wide analysis unravels the differential regulation and in vivo essentiality of virulence-associated proteins B and C toxin-antitoxin systems of Mycobacterium tuberculosis. J. Infect. Dis. 217, 1809–1820 (2018).
Winther, K., Tree, J. J., Tollervey, D. & Gerdes, K. VapCs of Mycobacterium tuberculosis cleave RNAs essential for translation. Nucleic Acids Res. 44, 9860–9871 (2016).
Cintron, M. et al. Accurate target identification for Mycobacterium tuberculosis endoribonuclease toxins requires expression in their native host. Sci. Rep. 9, 5949 (2019).
Cruz, J. W. et al. Growth-regulating Mycobacterium tuberculosis VapC-mt4 toxin is an isoacceptor-specific tRNase. Nat. Commun. 6, 7480 (2015).
Chauhan, U., Barth, V. C. & Woychik, N. A. tRNA(fMet) inactivating Mycobacterium tuberculosis VapBC toxin-antitoxin systems as therapeutic targets. Antimicrob. Agents Chemother. 66, e0189621 (2022).
Ahidjo, B. A. et al. VapC toxins from Mycobacterium tuberculosis are ribonucleases that differentially inhibit growth and are neutralized by cognate VapB antitoxins. PLoS ONE6, e21738 (2011).
Winther, K. S., Brodersen, D. E., Brown, A. K. & Gerdes, K. VapC20 of Mycobacterium tuberculosis cleaves the sarcin-ricin loop of 23S rRNA. Nat. Commun. 4, 2796 (2013).
Barth, V. C. et al. Mycobacterium tuberculosis VapC4 toxin engages small ORFs to initiate an integrated oxidative and copper stress response. Proc. Natl Acad. Sci. USA 118, 1–12 (2021).
Sharp, J. D. et al. Growth and translation inhibition through sequence-specific RNA binding by Mycobacterium tuberculosis VapC toxin. J. Biol. Chem. 287, 12835–12847 (2012).
Min, A. B. et al. The crystal structure of the Rv0301-Rv0300 VapBC-3 toxin-antitoxin complex from M. tuberculosis reveals a Mg(2)(+) ion in the active site and a putative RNA-binding site. Protein Sci. 21, 1754–1767 (2012).
Miallau, L. et al. Structure and proposed activity of a member of the VapBC family of toxin-antitoxin systems: VapBC-5 from Mycobacterium tuberculosis. J. Biol. Chem. 284, 276–283 (2009).
Deep, A. et al. Structural, functional and biological insights into the role of Mycobacterium tuberculosis VapBC11 toxin-antitoxin system: targeting a tRNase to tackle mycobacterial adaptation. Nucleic Acids Res. 46, 11639–11655 (2018).
Das, U. et al. Crystal structure of the VapBC-15 complex from Mycobacterium tuberculosis reveals a two-metal ion dependent PIN-domain ribonuclease and a variable mode of toxin-antitoxin assembly. J. Struct. Biol. 188, 249–258 (2014).
Kang, S. M. et al. Functional details of the Mycobacterium tuberculosis VapBC26 toxin-antitoxin system based on a structural study: insights into unique binding and antibiotic peptides. Nucleic Acids Res 45, 8564–8580 (2017).
Lee, I. G. et al. Structural and functional studies of the Mycobacterium tuberculosis VapBC30 toxin-antitoxin system: implications for the design of novel antimicrobial peptides. Nucleic Acids Res. 43, 7624–7637 (2015).
Jardim, P., Santos, I. C., Barbosa, J. A., de Freitas, S. M. & Valadares, N. F. Crystal structure of VapC21 from Mycobacterium tuberculosis at 1.31 A resolution. Biochem. Biophys. Res. Commun. 478, 1370–1375 (2016).
Sharma, A. et al. VapC21 toxin contributes to drug-tolerance and interacts with non-cognate VapB32 antitoxin in Mycobacterium tuberculosis. Front Microbiol. 11, 2037 (2020).
Tu, C. H., Holt, M., Ruan, S. & Bourne, C. Evaluating the potential for cross-interactions of antitoxins in type II TA systems. Toxins 12, 422, 1–25 (2020).
Boss, L. & Kedzierska, B. Bacterial toxin-antitoxin systems’ cross-interactions-implications for practical use in medicine and biotechnology. Toxins 15, 380, 1–30 (2023).
Yang, M., Gao, C., Wang, Y., Zhang, H. & He, Z. G. Characterization of the interaction and cross-regulation of three Mycobacterium tuberculosis RelBE modules. PLoS ONE5, e10672 (2010).
Zhu, L., Sharp, J. D., Kobayashi, H., Woychik, N. A. & Inouye, M. Noncognate Mycobacterium tuberculosis toxin-antitoxins can physically and functionally interact. J. Biol. Chem. 285, 39732–39738 (2010).
Gupta, A., Venkataraman, B., Vasudevan, M. & Gopinath Bankar, K. Co-expression network analysis of toxin-antitoxin loci in Mycobacterium tuberculosis reveals key modulators of cellular stress. Sci. Rep. 7, 5868 (2017).
Keren, I., Minami, S., Rubin, E. & Lewis, K. Characterization and transcriptome analysis of Mycobacterium tuberculosis persisters. mBio 2, e00100–e00111 (2011).
Agarwal, S. et al. VapBC22 toxin-antitoxin system from Mycobacterium tuberculosis is required for pathogenesis and modulation of host immune response. Sci. Adv. 6, eaba6944 (2020).
Robson, J., McKenzie, J. L., Cursons, R., Cook, G. M. & Arcus, V. L. The vapBC operon from Mycobacterium smegmatis is an autoregulated toxin-antitoxin module that controls growth via inhibition of translation. J. Mol. Biol. 390, 353–367 (2009).
Bendtsen, K. L. & Brodersen, D. E. Higher-order structure in bacterial VapBC toxin-antitoxin complexes. Subcell. Biochem. 83, 381–412 (2017).
Kumari, K. & Sarma, S. P. Structural and mutational analysis of MazE6-operator DNA complex provide insights into autoregulation of toxin-antitoxin systems. Commun. Biol. 5, 963 (2022).
Overgaard, M., Borch, J., Jorgensen, M. G. & Gerdes, K. Messenger RNA interferase RelE controls relBE transcription by conditional cooperativity. Mol. Microbiol 69, 841–857 (2008).
Cataudella, I., Trusina, A., Sneppen, K., Gerdes, K. & Mitarai, N. Conditional cooperativity in toxin-antitoxin regulation prevents random toxin activation and promotes fast translational recovery. Nucleic Acids Res. 40, 6424–6434 (2012).
Garcia-Pino, A. et al. Allostery and intrinsic disorder mediate transcription regulation by conditional cooperativity. Cell 142, 101–111 (2010).
Overgaard, M., Borch, J. & Gerdes, K. RelB and RelE of Escherichia coli form a tight complex that represses transcription via the ribbon-helix-helix motif in RelB. J. Mol. Biol. 394, 183–196 (2009).
Magnuson, R. & Yarmolinsky, M. B. Corepression of the P1 addiction operon by Phd and Doc. J. Bacteriol. 180, 6342–6351 (1998).
Ronneau, S. & Helaine, S. Clarifying the link between toxin-antitoxin modules and bacterial persistence. J. Mol. Biol. 431, 3462–3471 (2019).
Lemma, A. S. & Brynildsen, M. P. Toxin induction or inhibition of transcription or translation posttreatment increases persistence to fluoroquinolones. mBio 12, e0198321 (2021).
Shah, D. et al. Persisters: a distinct physiological state of E. coli. BMC Microbiol. 6, 53 (2006).
Singh, R., Barry, C. E. 3rd & Boshoff, H. I. The three RelE homologs of Mycobacterium tuberculosis have individual, drug-specific effects on bacterial antibiotic tolerance. J. Bacteriol. 192, 1279–1291 (2010).
Miallau, L. et al. Comparative proteomics identifies the cell-associated lethality of M. tuberculosis RelBE-like toxin-antitoxin complexes. Structure 21, 627–637 (2013).
Han, J. S. et al. Characterization of a chromosomal toxin-antitoxin, Rv1102c-Rv1103c system in Mycobacterium tuberculosis. Biochem. Biophys. Res. Commun. 400, 293–298 (2010).
Gosain, T. P., Singh, M., Singh, C., Thakur, K. G. & Singh, R. Disruption of MenT2 toxin impairs the growth of Mycobacterium tuberculosis in guinea pigs. Microbiology 168, 1–15 (2022).
Sharma, A. et al. HigB1 toxin in Mycobacterium tuberculosis is upregulated during stress and required to establish infection in guinea pigs. Front. Microbiol. 12, 748890 (2021).
Tiwari, P. et al. MazF ribonucleases promote Mycobacterium tuberculosis drug tolerance and virulence in guinea pigs. Nat. Commun. 6, 6059 (2015).
Gosain, T. P. et al. Mycobacterium tuberculosis strain with deletions in menT3 and menT4 is attenuated and confers protection in mice and guinea pigs. Nat. Commun. 15, 5467 (2024).
Orme, I. M. & Ordway, D. J. Mouse and guinea pig models of tuberculosis. Microbiol. Spectr. 4, 1–20 (2016).
Via, L. E. et al. Tuberculous granulomas are hypoxic in guinea pigs, rabbits, and nonhuman primates. Infect. Immun. 76, 2333–2340 (2008).
Aly, S. et al. Oxygen status of lung granulomas in Mycobacterium tuberculosis-infected mice. J. Pathol. 210, 298–305 (2006).
Clark, S., Hall, Y. & Williams, A. Animal models of tuberculosis: guinea pigs. Cold Spring Harb. Perspect. Med. 5, a018572 (2014).
Walling, L. R. & Butler, J. S. Structural determinants for antitoxin identity and insulation of cross talk between homologous toxin-antitoxin systems. J. Bacteriol. 198, 3287–3295 (2016).
Kamphuis, M. B. et al. Interactions between the toxin Kid of the bacterial parD system and the antitoxins Kis and MazE. Proteins 67, 219–231 (2007).
Ahmed, S. et al. Combining cysteine scanning with chemical labeling to map protein-protein interactions and infer bound structure in an intrinsically disordered region. Front Mol. Biosci. 9, 997653 (2022).
Deep, A., Kaundal, S., Agarwal, S., Singh, R. & Thakur, K. G. Crystal structure of Mycobacterium tuberculosis VapC20 toxin and its interactions with cognate antitoxin, VapB20, suggest a model for toxin-antitoxin assembly. FEBS J. 284, 4066–4082 (2017).
Park, D. et al. Crystal structure of proteolyzed VapBC and DNA-bound VapBC from Salmonella enterica Typhimurium LT2 and VapC as a putative Ca(2+)-dependent ribonuclease. FASEB J. 34, 3051–3068 (2020).
Simanshu, D. K., Yamaguchi, Y., Park, J. H., Inouye, M. & Patel, D. J. Structural basis of mRNA recognition and cleavage by toxin MazF and its regulation by antitoxin MazE in Bacillus subtilis. Mol. Cell 52, 447–458 (2013).
Kamada, K., Hanaoka, F. & Burley, S. K. Crystal structure of the MazE/MazF complex: molecular bases of antidote-toxin recognition. Mol. Cell 11, 875–884 (2003).
Chattopadhyay, G. et al. Mechanistic insights into global suppressors of protein folding defects. PLoS Genet. 18, e1010334 (2022).
Cherny, I. et al. Structural and thermodynamic characterization of the Escherichia coli RelBE toxin-antitoxin system: indication for a functional role of differential stability. Biochemistry 46, 12152–12163 (2007).
Cherny, I., Rockah, L. & Gazit, E. The YoeB toxin is a folded protein that forms a physical complex with the unfolded YefM antitoxin. Implications for a structural-based differential stability of toxin-antitoxin systems. J. Biol. Chem. 280, 30063–30072 (2005).
Nieto, C. et al. The yefM-yoeB toxin-antitoxin systems of Escherichia coli and Streptococcus pneumoniae: functional and structural correlation. J. Bacteriol. 189, 1266–1278 (2007).
Sahoo, A., Khare, S., Devanarayanan, S., Jain, P. C. & Varadarajan, R. Residue proximity information and protein model discrimination using saturation-suppressor mutagenesis. Elife 4, e09532, 1–29 (2015).
Chattopadhyay, G. et al. Functional and Biochemical Characterization of the MazEF6 Toxin-Antitoxin System of Mycobacterium tuberculosis. J. Bacteriol. 204, e0005822 (2022).
Gerdes, K., Rasmussen, P. B. & Molin, S. Unique type of plasmid maintenance function: postsegregational killing of plasmid-free cells. Proc. Natl. Acad. Sci. USA 83, 3116–3120 (1986).
Berne, C., Zappa, S. & Brun, Y. V. eDNA-stimulated cell dispersion from Caulobacter crescentus biofilms upon oxygen limitation is dependent on a toxin-antitoxin system. Elife 11, e80808, 1–25 (2023).
Ma, D. et al. The toxin-antitoxin MazEF drives Staphylococcus aureus biofilm formation, antibiotic tolerance, and chronic infection. mBio 10, e01658-19, 1–15 (2019).
Rycroft, J. A. et al. Activity of acetyltransferase toxins involved in Salmonella persister formation during macrophage infection. Nat. Commun. 9, 1993 (2018).
Lobato-Marquez, D., Moreno-Cordoba, I., Figueroa, V., Diaz-Orejas, R. & Garcia-del Portillo, F. Distinct type I and type II toxin-antitoxin modules control Salmonella lifestyle inside eukaryotic cells. Sci. Rep. 5, 9374 (2015).
Wang, Y. et al. Functional RelBE-Family Toxin-Antitoxin Pairs Affect Biofilm Maturation and Intestine Colonization in Vibrio cholerae. PLoS ONE10, e0135696 (2015).
Sharrock, A., Ruthe, A., Andrews, E. S. V., Arcus, V. A. & Hicks, J. L. VapC proteins from Mycobacterium tuberculosis share ribonuclease sequence specificity but differ in regulation and toxicity. PLoS ONE13, e0203412 (2018).
Palanisamy, G. S. et al. Evidence for oxidative stress and defective antioxidant response in guinea pigs with tuberculosis. PLoS ONE6, e26254 (2011).
Edwards, K. M. et al. Iron-cofactored superoxide dismutase inhibits host responses to Mycobacterium tuberculosis. Am. J. Respir. Crit. Care Med. 164, 2213–2219 (2001).
Arora, G., Chaudhary, D., Kidwai, S., Sharma, D. & Singh, R. CitE enzymes are essential for Mycobacterium tuberculosis to establish infection in macrophages and guinea pigs. Front. Cell Infect. Microbiol. 8, 385 (2018).
Wiederstein, M. & Sippl, M. J. TopMatch-web: pairwise matching of large assemblies of protein and nucleic acid chains in 3D. Nucleic Acids Res. 48, W31–W35 (2020).
Chandra, S., Chattopadhyay, G. & Varadarajan, R. Rapid identification of secondary structure and binding site residues in an intrinsically disordered protein segment. Front. Genet. 12, 755292 (2021).
Wilbur, J. S. et al. Neisseria gonorrhoeae FitA interacts with FitB to bind DNA through its ribbon-helix-helix motif. Biochemistry 44, 12515–12524 (2005).
Gu, Q. et al. An auto-regulating type II toxin-antitoxin system modulates drug resistance and virulence in Streptococcus suis. Front. Microbiol. 12, 671706 (2021).
Beck, I. N., Usher, B., Hampton, H. G., Fineran, P. C. & Blower, T. R. Antitoxin autoregulation of M. tuberculosis toxin-antitoxin expression through negative cooperativity arising from multiple inverted repeat sequences. Biochem. J. 477, 2401–2419 (2020).
Aakre, C. D. et al. Evolving new protein-protein interaction specificity through promiscuous intermediates. Cell 163, 594–606 (2015).
Lite, T. V. et al. Uncovering the basis of protein-protein interaction specificity with a combinatorially complete library. Elife 9, e60924, 1–22 (2020).
Snead, K. J., Moore, L. L. & Bourne, C. R. ParD antitoxin hotspot alters a disorder-to-order transition upon binding to its cognate ParE toxin, lessening its interaction affinity and increasing its protease degradation kinetics. Biochemistry 61, 34–45 (2022).
Ding, D. et al. Co-evolution of interacting proteins through non-contacting and non-specific mutations. Nat. Ecol. Evol. 6, 590–603 (2022).
Chattopadhyay, G. et al. High-throughput method to stabilize transient ternary complexes and measure associated kinetics. Protein Sci. 32, e4514 (2023).
Singh, R. et al. Polyphosphate deficiency in Mycobacterium tuberculosis is associated with enhanced drug susceptibility and impaired growth in guinea pigs. J. Bacteriol. 195, 2839–2851 (2013).
Bardarov, S. et al. Specialized transduction: an efficient method for generating marked and unmarked targeted gene disruptions in Mycobacterium tuberculosis, M. bovis BCG and M. smegmatis. Microbiology148, 3007–3017 (2002).
Laskowski, R. A. et al. PDBsum: a Web-based database of summaries and analyses of all PDB structures. Trends Biochem. Sci. 22, 488–490 (1997).
Laskowski, R. A., Jablonska, J., Pravda, L., Varekova, R. S. & Thornton, J. M. PDBsum: Structural summaries of PDB entries. Protein Sci. 27, 129–134 (2018).
Chattopadhyay, G. & Varadarajan, R. Facile measurement of protein stability and folding kinetics using a nano differential scanning fluorimeter. Protein Sci. 28, 1127–1134 (2019).
Seidel, S. A. et al. Label-free microscale thermophoresis discriminates sites and affinity of protein-ligand binding. Angew. Chem. Int. Ed. Engl. 51, 10656–10659 (2012).
Ehrt, S. et al. Controlling gene expression in mycobacteria with anhydrotetracycline and Tet repressor. Nucleic Acids Res. 33, e21 (2005).
Acknowledgements
R.S. acknowledges the financial support received from Wellcome Trust-DBT India Alliance (IA/S/19/2/504646). R.S. is a recipient of the National Bioscience Award from the Department of Biotechnology and Ramalingaswami fellowship. N.S. and G.C. acknowledge the Department of Biotechnology and the Ministry of Education (MOE), respectively, for their research fellowship. N.S.S. is thankful to the Department of Biotechnology for the M.K. Bhan fellowship. The authors thank staff members of the biosafety level 3 facility and experimental animal facility at the Translational Health Science and Technology Institute for their technical help. The authors are also thankful to the staff members of the National Liver Disease Biobank at the Institute of Liver and Biliary Diseases for histopathology analysis. We also acknowledge the nanoDSF and MST facility at the Biological Sciences Division, IISc, for acquiring the thermal stability data and binding affinity characterisation studies. The authors thank Dr Saurabh Chugh, Dr Shikha Singh, Mr Arun Sharma, and Ms Neha Khan for critical manuscript reading. The authors thank Mr Saqib Kidwai for his technical assistance. We acknowledge Dr Deepika Chaudhary for help with animal experiments. The authors sincerely acknowledge the help of Mr Ashish Kumar, Mr Rajesh and Mr Sher Singh in performing experiments.
Author information
Authors and Affiliations
Contributions
R.S. conceived the idea and designed the work plan. N.S. and N.S.S. performed the microbiology and molecular biology experiments, N.S. performed the animal experiments, G.C. performed biophysical characterization experiments. R.S. and R.V. supervised the experiments. R.S., N.S., G.C. and N.S.S. analysed and interpreted the data. R.S., G.C. and N.S.S. wrote the manuscript with inputs from other authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks the anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editors: Karthika Rajeeve and Johannes Stortz. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Singh, N., Chattopadhyay, G., Sundaramoorthy, N.S. et al. Understanding the physiological role and cross-interaction network of VapBC35 toxin-antitoxin system from Mycobacterium tuberculosis. Commun Biol 8, 327 (2025). https://doi.org/10.1038/s42003-025-07663-2
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s42003-025-07663-2