Abstract
Different glioblastoma (GBM) subtypes have been identified based on the tumor microenvironment (TME). The discovery of new therapies for these hard-to-treat tumors requires a thorough characterization of preclinical models, including their TME, to apply preclinical results to the most similar GBM subtype. Using single-nucleus RNA sequencing (snRNA-seq), we characterized the tumor and TME in an immunocompetent mouse model with intracranially implanted GBM stem cells at different stages and treatments. Visium spatial transcriptomics confirmed the location of annotated cells. This model exhibits GBM targets related to integration into neural circuits - Grik2, Nlgn3, Gap43 or Kcnn4-, immunoevasion - Nt5e, Cd274 or Irf8- and immunosuppression - Csf1r, Arg1, Mrc1 and Tgfb1. The landscape of cytokines, checkpoint ligands and receptors uncovered Mrc1, PD-L1, TIM-3 or B7-H3, among the immunotherapy targets that can be addressed in this model. The comparison with human GBMs unveiled crucial similarities with TMEMed GBM, the most frequent subtype.
Similar content being viewed by others
Introduction
Glioblastoma (GBM) is the most aggressive and frequent form of primary brain tumor and, despite continuous effort to find an effective treatment, is considered as one of the deadliest types of cancer, with a median survival of only 16 months. GBMs are characterized by fast and aggressive growth, high infiltrative capacity, and resistance to current treatments. The main challenges underlying therapeutic failure are derived from its cellular and molecular heterogeneity. Fortunately, increasing knowledge of cellular and molecular alterations in brain tumors has improved their classification1 with benefits in personalized treatments. For instance, inhibitors of mutant IDH1 and IDH2 enzymes have shown positive results in clinical trials for IDH-mutant gliomas2. Regarding IDHwt GBM, recent studies have identified three novel subtypes with significantly different tumor microenvironment (TME) compositions and different response to immunotherapy3, which supports that deciphering GBM heterogeneity may improve their treatment. High-throughput studies, such as single-cell RNA sequencing (scRNA-Seq) studies have helped to clarify intra-tumor and inter-tumor heterogeneity4,5,6. scRNA-Seq has emerged as a key method to characterize cellular states and is continuously used to analyze tumor samples7; however, it requires a quick dissociation of fresh tissue and enzymatic digestion that can damage sensitive cell types such as neurons8. Conversely, single-nucleus RNA sequencing (snRNA-Seq) avoids the aggressive enzymatic digestion, preserving the information from most cell types9,10 in the brain TME.
Unfortunately, many experimental treatments that were successful in preclinical models have failed in subsequent clinical trials11. Among the causes of this failure are the differences between preclinical models and human GBM. Hence, a profound cellular and molecular characterization of GBM preclinical models, along with their correlation with human GBM subtypes is required to effectively translate the preclinical results to the most similar human GBM subtype. Otherwise, the general application of results from a single preclinical model to human GBMs will probably lead to very variable results. The GL261 model is the most widely used syngeneic model of GBM due to a series of advantages, such as the possibility to study the immune system. It has been used in studies of gene therapy, immune cell transfer, monoclonal antibodies, cytokine therapies, checkpoint inhibitors, and dendritic vaccines12,13,14,15,16. Intracranially implanted GL261 cells grow rapidly and form tumors with 100% penetrance and, although classified as GBM because of its aggressivity, GL261 cells show moderate immunogenicity, and this model does not exhibit all the histopathological GBM features17. GBMs contain populations of cells with stem cell properties, known as GBM stem cells (GSCs), which are highly tumorigenic, possess tumor-propagating potential, and exhibit resistance to chemotherapy and radiotherapy18,19. Several studies described a GBM model in which the tumor is initiated by the implantation of GSCs obtained from GL261 cells in immunocompetent mice (GL261-GSC GBM model). As GL261, GL261-GSCs show moderate immunogenicity, but the GL261-GSC GBM model recapitulates most of the typical features of human GBM20,21,22,23. Thus, although there is still some controversy in the field24, the comparison between the immunocompetent GL261-GSC and GL261 GBM models showed that GL261-GSC GBM models exhibit increased tumorigenicity, infiltration, chemoresistance and constitute a more reliable model for human GBM than GL261 GBM model20,21.
Here, we use snRNA-Seq to create a map of murine GBM cellular states from in vitro to in vivo using the GL261-GSC GBM model. We performed snRNA-Seq in a total of 14 samples from tumor-bearing mice and 5 samples from cultured GBM cells, using a microfluidic-droplet-based method. Additionally, we performed Visium spatial transcriptomics in the in vivo model to confirm the location of annotated cells. To explore the technical bias of this approach, we performed two scRNA-seq methods in GBM cells. We thoroughly characterized the TME in the GL261-GSC GBM model at early and late stages of GBM development and upon treatment with temozolomide (TMZ), the actual standard of care chemotherapeutic drug for GBM, and a novel experimental treatment, the cell penetrating peptide Tat-Cx43266-283, which exerts important anti-tumor effects and enhances the survival in GL261-GSC GBM model22,25,26,27. The present study identified prominent GBM targets that can be addressed using this preclinical model. Importantly, we unveiled crucial similarities of GL261-GSC GBM model with TMEMed GBM, the most frequent human GBM subtype, supporting its suitability for studying specifically this human GBM.
Results
Identification of tumor and non-tumor cells in the GL261-GSC GBM model
To characterize the biology of murine GBM cells growing in the brain TME, 5000 GL261-GSCs were intracranially injected into the brains of immunocompetent C57BL/6 mice. We have previously characterized GL261-GSCs22 and in agreement with the results from different groups20,21,23, this low number of cells develops very aggressive tumors with poor mice survival22, indicating the high tumorigenicity of GL261-GSCs20,21. We collected samples of four GBM-bearing mouse brains, including tumor core and surrounding tissue, at two stages of tumor growth, early (7 days post-implantation) and late (28 days post-implantation), and isolated nuclei for snRNA-Seq. This process is described in detail in the Methods section and graphically summarized in Fig. 1A.
A GL261-GSCs were grown as neurospheres in stem cell medium. They were obtained from GL261 cells grown in adherence in differentiation medium. 5000 GL261-GSCs were intracranially injected into the right hemisphere of brains of syngeneic C57BL/6 mice in saline solution or 100 µM Tat-Cx43266-283 (Tat-Cx43) or 250 µM temozolomide (TMZ). After 7 days, these treatments were intraperitoneally (IP) administered twice per week. Samples were collected from GBM-bearing mouse brains, including tumor core and surrounding tissue, at two stages of tumor growth, early (7 days post-implantation) and late (28 days post-implantation), and isolated nuclei for snRNA-Seq. snRNA-Seq was performed in all the in vitro and in vivo samples. Created with Bioicons: 96_well_plate and and microfluids_chip icons by Marcel Tisch licensed under CC0, Illumina_miseq icon and microtube-closed icons by DBCLS https://togotv.dbcls.jp/en/pics.html licensed under CC-BY 4.0. B Illustrative IBA1 immunofluorescence images of brains at 7 days post-implantation showing a small group of DAPI-stained tumor cells surrounded by IBA1+ TAMs at the injection site (dashed circle in upper panels), scale bar: 2 mm. Lower panels: zoomed-in images showing tumor cells intermingled and surrounded by IBA1+ TAMs (dashed line), scale bar: 50 µm. C Illustrative IBA1 immunofluorescence images of brains at 28 days post-implantation showing large tumors infiltrated and surrounded by IBA1+ TAMs, scale bar: 2 mm. Lower panels: zoomed-in images showing the border of the tumor together with a group of tumor cells infiltrating the brain parenchyma (dashed line), scale bar: 50 µm. Note the abundance of IBA1+ TAMs infiltrating and surrounding these tumors.
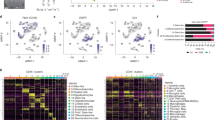
At 7 days post-implantation, tumors were small and contained few tumor cells (as judged by Sox6 expression, Suppl. Fig. 1A), which were surrounded by an active TME enriched in IBA1+ tumor associated microglia and macrophages (TAMs; Fig. 1B), as well as GFAP+ reactive astrocytes (Suppl. Fig. 1A). Therefore, we were able to take samples containing the whole tumor and its TME. In the case of 28-days tumors, we found large tumors with a high infiltration of IBA1+ TAMs (Fig. 1C) and GFAP+ reactive astrocytes at the tumor border (Suppl. Fig. 1B). Therefore, we obtained samples from the border of the tumor to capture tumor cells as well as the other cell types present in the brain TME (Fig. 1C and Suppl. Fig. 1B). Additionally, we analyzed 2 samples of GL261 cells growing in culture in differentiation conditions, and 3 samples of GL261-GSCs growing as neurospheres in stem cell medium (derived from GL261), which were the cells implanted into the mouse brain. In total, 28,833 cells passed our quality controls and were further analyzed. We used uniform manifold approximation and projection (UMAP) for dimensionality reduction28, followed by Leiden clustering29, which resulted in the identification of 22 separate clusters (Fig. 2A). We used hierarchical clustering and a Wilcoxon rank sum test to obtain differentially expressed genes (DEGs) in each cluster (Fig. 2B and Table S1). Clusters were classified by high expression of gene markers from published mouse CNS cell type databases30 (Fig. 2C). All clusters showed high expression of characteristic gene sets except for those corresponding to cultured GL261 cells (clusters 4 and 5), GL261-GSCs (clusters 1, 2, 6, and 11), and cluster 0, which we preliminarily classified as malignant (implanted GL261-GSCs).
A UMAP visualization of all cells collected, colored by Leiden clustering. k = 28,833 individual cells. B Heatmap showing relative expression of each of the 10 top expressed genes in each Leiden cluster. Columns correspond to cells, ordered by Leiden cluster. Dendrogram displays hierarchical clustering. See Table S1 for the full list. C UMAP visualization colored by cell type. D Inference of chromosomal CNVs on the basis of average expression in windows of 250 analyzed genes. Rows corresponds to cells, ordered by cell type. E UMAP visualization colored by CNV score. F Dotplot showing the expression of cell markers by each cell type. The size of the dot represents the percentage of cells expressing the gene and the color intensity represents the mean expression in that group. G Barplot showing the distribution of cell types obtained in tumors of 7 or 28 days. H Visual summary of the cell types from tumor and brain TME identified and for which transcriptomics is provided in this study, at two different stages of development and under two different treatments, as illustrated in Fig. 1A. Created in BioRender. Tabernero, A. (2025) https://BioRender.com/n16r525.
To confirm the identity of malignant cells, we inferred copy number variation (CNV) events based on the average expression of 250 genes in each chromosomal region4,31. We observed large-scale amplifications and deletions in most tumor cells (Fig. 2D). We calculated a CNV score for each cell and overlayed it with the UMAP, which revealed eight clusters of cells with high CNV score, including cultured GL261 and GL261-GSCs and cluster 0 (Fig. 2E). Figure 2F shows the expression of some of the main marker genes representing each cell type. We also analyzed the spatial topography of gene expression with Visium platform (Table S2) as described in methods. Interestingly, spatial gene expression results confirmed that tumor cell markers shown in Fig. 2F, i.e, Sox2, Olig1, Nes and Pdgfr, were significantly elevated in the tumor region identified by Hematoxylin-Eosin staining compared to healthy brain parenchyma. Conversely, astrocyte markers (GFAP, Aqp4 and Aldh1l1), neuron markers (Calb1, Slc17a7 and Gabra1) and oligodendrocyte markers (Mbp and Mag) were significantly elevated in the healthy brain parenchyma compared to the tumor region (Table S2).
Figure 2G shows the distribution of cell types based on time of tumor progression. Unsurprisingly, most of the cells in 28-day samples had a tumor origin, whereas we almost did not capture tumor cells in the samples from 7-day tumors. However, although 7-day samples contained few tumor cells, they were enriched in an active TME, which may provide important clues about the role of the TME in GBM development (Fig. 1B and Suppl. Fig. 1A). The high number of tumor cells at 28 days resulted in a relative detriment in the percentages of other cell populations, except for immune cells, whose proportion was increased, indicating high immune infiltration of the tumors, which is consistent with the high number of IBA1+ TAMs found by immunofluorescence in these tumors (Fig. 1C). Figure 2H shows a visual summary of the cell types from tumor and brain TME analyzed in this study, at two different stages of development and under two different treatments, as illustrated in Fig. 1A. The cell-type identity of each cell cluster, the number of cells originating from each brain or sample and QC metrics can be found in Table 1 and Suppl. Fig. 1C–E.
Analysis of the transcriptomes of GL261, GL261-GSCs and implanted GL261-GSCs
Given that GBM cells clustered separately depending on their origin (cultured in differentiation conditions, cultured in stem cell conditions, or implanted into the brain parenchyma), we analyzed the differences between them. Principal component analysis (PCA) revealed that the type of culture and the interaction with the brain microenvironment (brain implantation) are important sources of variation (PC1, Fig. 3A). We have previously shown that GL261-GSCs express higher levels of stem cell markers, Sox2, and reduced levels of GFAP when compared to GL261 cells22. Indeed, the intracranial injection of only 5,000 GL261-GSCs is sufficient to generate very aggressive tumors (see refs. 22,32 and Figs. 1C, 3H, 4D, 6E and Suppl. Fig. 1B), indicating their high tumorigenicity, one of the main features of GSCs. In this study, we performed Wilcoxon rank sum test to look for potential markers for these populations, with emphasis in the identification of additional GSC markers in this model, an essential subpopulation for GBM biology and therapy (Fig. 3B, Suppl. Fig. 2A, and Table S3). Among overexpressed genes in GL261-GSCs, we found Nkain2, Sema6a, Cdh19 or Cd81, all of them proposed as important targets in GBM33. Importantly, the transcription factors Olig2 and Olig1, which showed very high expression in most GL261-GSCs and, to a lesser extent, in implanted GL261-GSCs, were not expressed by GL261 cells (Fig. 3B, Suppl. Fig. 2A), suggesting that they are good markers for GSCs in this model. Protein tyrosine phosphatase receptor type Z1 (Ptprz1) or phosphacan, which has been reported to promote tumor invasion by a subset of human GSCs34,35, was also upregulated in GL261-GSCs and, even more, in implanted GSCs.
A PCA visualization of malignant cells, colored by cell type. k = 16,757 individual cells. B Heatmap showing relative expression of each of the 50 top expressed genes in each culture type. Columns correspond to cultured GL261 and GL261-GSCs and implanted GL261-GSCs. Selected genes are indicated (see Table S4 for the full list). C Top 10 enriched terms from gene ontology (GO) gene sets in cultured GL261 and GL261-GSCs and implanted GL261-GSCs compared to the others (see Tables S5–S7 for full lists). D Heatmap showing the meta-module scores described by Neftel et al.6 E Two-dimensional representation of the cellular states described by Neftel et al.6 for each group. Each quadrant corresponds to one cellular state and each dot represents a cell. F Barplot showing the percentage of cycling cells analyzed, calculated as the sum of percentages of S and G2/M phases, in GL261, GL261-GSCs and implanted GL261-GSCs (Suppl. Fig. 3B). G Venn diagram showing the overlap between differentially expressed genes across different types. Plotted data are provided in tabular form in Table S7. Differential gene expression was analyzed using Wilcoxon test with a p-value cutoff=0.05. H Illustrative Sox6 immunofluorescence images of brains at 28 days post-implantation showing that transplanted tumor cells express Sox6, scale bar: 2 mm. Lower panels: zoomed-in images showing tumor cells at the border of the tumor containing different levels of the transcription factor Sox6, scale bar: 100 µm. Quantification of Sox6 protein levels in the tumor area and contralateral hemisphere from the immunofluorescence images. Results are expressed in arbitrary units (a. u.) and are the mean±SEM from three independent experiments from which 5 fields were quantified. ***P < 0.001 (student’s t test). I Sox6 protein levels in the contralateral region and tumor area detected by western blotting (representative blots). GAPDH is shown as loading control. Full-length Western blots are shown in Suppl. Fig. 7A.
A UMAPs showing the expression of GBM-neuron synapsis genes. B UMAPs showing the expression of GBM network genes. Violin plots are included in Suppl. Fig. 2. C UMAPs showing the expression of immune-evasive genes. D Illustrative Gap43 and PD-L1 (Cd274) immunofluorescence images of brains at 28 days post-implantation, scale bar: 2 mm. Zoomed-in images showing the expression of Gap43 and PD-L1 (Cd274) in the border of the tumor, scale bar: 100 µm. Quantification of Gap43 and PD-L1 protein levels in the tumor area and contralateral hemisphere from the immunofluorescence images. Results are expressed in arbitrary units (a. u.) and are the mean±SEM from four and three independent experiments, respectively, from which 5 fields were quantified. **P < 0.01, ***P < 0.001 (student’s t test). Higher resolution images in Suppl. Fig. 2B. E Irf8 protein levels in the contralateral region and tumor area detected by western blotting (representative blots). GAPDH is shown as loading control. Full-length Western blots are shown in Suppl. Fig. 7B. F Gene set enrichment analysis (GSEA) plot of indicated gene sets for genes differentially expressed between implanted and cultured GSCs. NES Normalized enrichment score, FDR false discovery rate.
We used gene set enrichment analysis (GSEA) to identify key pathways and processes (Fig. 3C and Tables S4–S6). Interestingly, cellular components overrepresented in GL261-GSCs included some neuronal attributes (neuron projection, GO:0043005; dendrite, GO:0030425; axon, GO:0030424), which supports the idea that GL261-GSCs present the potential to differentiate towards a neuron-like lineage. GL261-GSCs upregulated cholesterol biosynthesis, which is linked to cancer development, counting GBM36,37,38. Cultured and implanted GL261-GSCs were very active in transcription, GTPase activity, and protein modification. Indeed, we scored a set of cell cycle genes from Satija et al.39 and assigned each cell to a cell cycle phase according to the scores obtained. This revealed a similar cell cycle pattern for GL261-GSCs and implanted cells, whereas GL261 samples contained less cells in G1 phase (Fig. 3F and Suppl. Fig. 3B).
To add additional value to our study, we analyzed GL261 and GL261-GSCs using single-cell methods, a microfluidic-droplet-based method on single cells from 10x (10x cells) and a full-length scRNA-seq using Smart-Seq2 method, based on the separation of cells in microtiter plates by fluorescence activated cell sorting (FACS)40 (Suppl. Fig. 3). The number of counts and genes captured per cell were higher using Smart-Seq2 (around 104 genes and 106 total counts), at the cost of sequencing less cells (Suppl. Fig. 3C). We detected lower similarity between samples sequenced using Smart-Seq2 compared to that between 10x samples. (Suppl. Fig. 3D). 10x nuclei showed the highest dissimilarity with the other technologies used. In the case of GL261-GSCs, the differences between samples sequenced using the same technology were bigger than those observed between GL261 samples, reinforcing the idea that GL261-GSCs are more diverse. We compared differentially expressed genes obtained independently with each technology (Suppl. Fig. 4A). From those genes that passed a p-value cutoff of 0.05 in GL261, only 17.51% were shared by the three technologies and 43.69% by two of them, whereas the remaining 38.8% were only detected with one technology. In GL261-GSCs, 14.76% were shared by the three technologies and 26.53% by two of them. 58.71% of the genes were detected only with one technology, with the main contributor to this being Smart-Seq2. We combined information from all three technologies for GO analysis and observed that adding technologies helps identifying different enriched terms within the same gene set, making the analysis more robust, but it does not substantially affect the processes obtained (Suppl. Fig. 4B).
Next, we calculated signature scores for the four cellular states that drive the heterogeneity of GBM cells described by Neftel el al.6, i.e., (1) neural-progenitor-like (NPC-like), (2) oligodendrocyte-progenitor-like (OPC-like), (3) astrocyte-like (AC-like), and (4) mesenchymal-like (MES-like). We observed that, whereas GL261 cells span between MES-like and AC-like programs, GL261-GSCs and implanted GL261-GSCs shift towards progenitor states (OPC-like and NPC-like) (Fig. 3D). The ‘cell-state plot’’ designed by Neftel and colleagues6 summarizes the distribution of cells across these states and their intermediates (Fig. 3E). This is in line with recent reports showing a MES-like phenotype of GBM cells grown in two-dimensional culture, with low transcriptional diversity and plenty of cycling cells, and over-representation of developmental states (OPC-, AC-, and NPC-like) in cells cultured in ex vivo human cortical tissue35.
Effects of the TME on GL261-GSC transcriptional activity
DEG analysis found 2568 genes unique for GL261 cells, 491 genes unique for GL261-GSCs, and 3695 genes unique for implanted GL261-GSCs (Fig. 3G and Table S7), suggesting that the TME strongly promotes transcriptional heterogeneity, as described in human GBMs35. Implanted GL261-GSCs shared some DEGs with GL261-GSCs (352), indicating that some implanted GSCs retain stem cell properties, and also shared some DEGs with GL261 cultured in differentiation conditions (180), which suggests that GL261-GSCs once implanted in the brain diverge in a great variety of phenotypes, including differentiation programs. Despite all the differences between human and murine GSCs, these data support the idea that implantation of GL261-GSCs might represent a good model to simulate the development of intratumor heterogeneity found in human GBMs. As described above, our results suggest that brain microenvironment importantly affects gene expression in tumor cells, as judged by the differential gene expression found between implanted and non-implanted GL261-GSCs. Thus, in addition to a downregulation in Olig2 and Olig1 expression, we found increased expression of a great number of neuron-related genes, such as Hexb (beta-hexosaminidase A subunit beta), Tnr (tenascin R), Nav3 (neuron navigator 3), Reln (reelin), and Tafa2 (TAFA chemokine like family member 2) in implanted GL261-GSCs compared to them in culture (Fig. 3B, Suppl. Fig. 2A, and Table S3). As expected, spatial transcriptomics, carried out as described in methods, confirmed that the expression of genes found in implanted GSCs by snRNA-seq (Fig. 3B, Suppl. Fig. 2A, and Table S3), such as Olig2, Olig1, Tnr, Nav3, Reln, and Tafa2, were much more abundant in the tumor area, compared with healthy brain parenchyma (Table S2). The GSC marker Sox6, found in implanted GSCs by snRNA-seq (Fig. 3B, Suppl. Fig. 2A, and Table S3), was also much more abundant in the tumor area than in the healthy brain parenchyma, as confirmed by immunofluorescence, Western blot and spatial transcriptomics (Fig. 3H, I, Suppl. Fig. 7A and Table S2). In fact, Sox6 emerged as an excellent marker for tracking the incipient infiltration of tumor cells in the early stages of brain tumor development (Suppl. Fig. 1A). Among the pathways significantly enriched in intracranially implanted GL261-GSCs when compared to cultured GL261-GSCs are many related to synaptic activity and neuronal signaling, such as transmitter-gated ion channel activity (GO:0022824), dendrite membrane (GO:0032590), modulation of chemical synaptic transmission (GO:0050804), inhibitory synapse assembly (GO:1904862), neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential (GO:0099529), neuron projection (GO:0043005), regulation of neurotransmitter receptor activity (GO:0099601), transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential (GO:1904315), synaptic transmission, GABAergic (GO:0051932) or synapse pruning (GO:0098883) (Table S8), which suggests that the brain TME promotes changes in GL261-GSC transcription towards the development of synaptic activity.
Because of the relevance of the interaction between GBM cells and neurons41,42, we manually interrogated the expression levels of some of the proposed effectors of this process (Fig. 4A). Thus, our study identified the expression of glutamate receptors of the AMPA type (Gria1, Gria2 and Gria3), NMDA type (Grin1) and Kainate type (Grik2) in implanted GL261-GSCs. It should be noted that the expression of AMPA receptors is nearly absent in cultured cells, and it is upregulated in some GL261-GSCs upon implantation. Tumor cells also express other proteins associated with neuron-glioma synapsis, such as Dlg4 (postsynaptic density protein 95, PSD95), Homer1 (postsynaptic density scaffolding protein PSD-Zip45), and Nlgn3 (neuroligin-3), one of the key mediators in the pro-tumor effects of neurons on GBM progression43. Interestingly, Nlgn3 levels were upregulated in tumor cells by the interaction with the TME, which supports its role as mediator in neuron-glioma synapsis in GL261-GSC GBM model. Some of the GL261-GSCs express the potassium channel KCa3.1 (Fig. 4B and Suppl. Fig. 2A; Kcnn4, also known as IK1 or SK4), with a prominent role in this process44. Implanted GL261-GSCs also express components of tumor microtubes, such as Gja1 (connexin43), and high levels of Gap43 (neuronal growth-associated protein 43) (Fig. 4B and Suppl. Fig. 2A), a key player on the pro-tumor effect of the microenvironment in GBM progression45,46. While Gja1 is expressed modestly in all the types of tumor cells, Gap43 expression importantly increased in tumor cells upon intracranial transplantation (Fig. 4B and Suppl. Fig. 2A). Thus, immunofluorescence analysis of Gap43 in brain tumor sections confirmed the high expression of Gap43 in tumor cells (Fig. 4D and Suppl. Fig. 2B), as well as in neurons that showed the expected pattern of expression of Gap43 in the brain parenchyma (Fig. 4D).
Among the genes upregulated in tumor cells by the interaction with the brain microenvironment, we also found increased expression of immune-related genes, including Nlrc5 (NOD-like receptor family CARD domain containing 5), a transactivator of MHC class I genes, and H2-K1 (histocompability complex 2 k1) in implanted GL261-GSCs compared to GL261-GSCs (Fig. 3B, Suppl. Fig. 2A, and Table S3). Because GBM cells may acquire an immune evasive phenotype47, we analyzed the expression of the immune evasion regulators Nt5e (CD73), Cd274 (PD-L1), and Irf8 (interferon regulatory factor 8). We found that Nt5e (CD73) was upregulated in implanted and cultured GL261-GSCs as compared to GL261, while the expression of Cd274 (PD-L1) and Irf8 was very low in GL261 and GL261-GSCs in culture, but it was upregulated in GL261-GSCs when implanted in the brain for 28 days (Fig. 4C). Immunofluorescence analysis of brain sections confirmed a high expression of PD-L1 in the tumor area (Fig. 4D and Suppl. Fig. 2B). In addition, spatial transcriptomics showed that the expression of Nlrc5, Nt5e, Cd274, and Irf8 was higher in the tumor area, compared with healthy brain parenchyma (Table S2). The higher level of Irf8 in the tumor area compared with the contralateral hemisphere was also confirmed by Western blot (Fig. 4E and Suppl. Fig. 7B). As a myeloid-specific master transcription factor48, Irf8 was also expressed by the immune cell cluster (Fig. 4C). GSEA analysis showed an increase in gene sets related to “Regulation of interferon-gamma production”, which might be responsible for Irf8 transcription47, and “Cytokine receptor activity” in GL261-GSCs at 28 days after implantation into the brain when compared to GL261-GSCs in culture (Fig. 4F and Table S8).
The immune microenvironment in the GL261-GSC GBM model
Recomputing Leiden clustering on the 2427 cells annotated as immune gave 5 clusters (Fig. 5A). All 5 clusters expressed the immune cell marker Ptprc (CD45, Suppl. Fig. 5A). Cluster 3 exhibited higher expression of Ptprc, together with lymphocyte markers Cd4, Cd3e, and Cd3g (CD4 T-cell), and Foxp3 (Treg), being thus classified as lymphoid cells, and was almost exclusively composed of cells from the 28-days tumors (Fig. 5A, B and Suppl. Fig. 5A). We did not find expression of B-cell markers. Clusters 0, 1, 2, and 4 were classified as myeloid cells, i.e., tumor-associated microglia/macrophages (TAMs). These data fitted with the immunofluorescence analyses, as we found a high number of myeloid IBA1+ TAMs infiltrating these tumors (Fig. 1C), while the number of CD3+ lymphoid cells was low in the border and core of these tumors (Suppl. Fig. 5B). We observed an increase in the number of cells in G2/M and S phases of the cell cycle in TAMs at 28 days (Fig. 5C and Suppl. Fig. 5C), suggesting an increase in proliferation of myeloid cells throughout GBM development. Scoring gene sets for microglia activation, differentiation, and migration, showed higher activation in immune cells at 28 days, whereas differentiation and migration were reduced (Fig. 5D, E). Both stages of tumor development showed similar records of phagocytosis. At 28 days, we observed increased expression of genes associated to major histocompatibility complex (MHC-II; Cd74, H2-Aa, H2-Ab1, H2-Eb1), and macrophage markers (Tgfbi), concurring with the idea that macrophages are enriched in the myeloid compartment at late stages of tumor development (Fig. 5F and Suppl. Fig. 5D)49 presumably caused by the breakage of the blood-brain barrier, as observed in human GBMs. Although it is difficult to distinguish tissue resident microglia from blood-derived macrophages, the expression of some genes can help this purpose50,51,52 and allowed us to annotate these myeloid populations (Fig. 5G). Thus, microglia has been traditionally identified by the expression of Tmem119, P2ry12, and Cx3cr1, whereas blood-derived macrophages express Tgfbi, Mrc1 (mannose receptor C type 1, also called CD206), Spp1, Nt5e, S100a4 and Hmox1, but adapt their transcriptional profile over time (Fig. 5H and Suppl. Fig. 5D). Expression of markers of immunosuppression in GBM-associated macrophages, such as Tgfbi, Mrc1 or Arg1 (arginase 1)53,54, increased throughout tumor development (Fig. 5H, Suppl. Fig. 5D, E and Suppl. Table S9). Indeed, our immunofluorescence analysis confirmed the expression of Mrc1 in these tumors at 28 days (Suppl. Fig. 5F). Interestingly, GSEA revealed upregulation of genes related to transforming growth factor beta (TGFß) and Hedgehog signaling, as well as epithelial-mesenchymal transition at 7 days (Fig. 5I and Table S10). Immune cells at late stage upregulated Myc and E2F targets and G2-M checkpoint genes, all indicative of proliferation. IFN-γ, TNF-α signaling, IL-2/STAT5, mTORC1, IL-6/JAK/STAT3, IFN-α, and p53 signaling were also upregulated in 28-days immune cells (Fig. 5I and Table S11). These pathways contribute to immunosuppression, including mTORC1, whose activation leads to the secretion of anti-inflammatory cytokines and immunosuppression55.
A UMAP visualization of immune cells, colored by Leiden cluster, time of tumor development, and cell type annotations. k = 2427 individual cells. B Barplot showing the proportion of cells at each time point per cluster. C Barplot showing the distribution of cells in each cell cycle phase at 7d or 28d. D UMAP visualization of microglia activation, microglia differentiation, microglia migration, and microglia phagocytosis scores. E Violinplots showing microglia activation, microglia differentiation, microglia migration, and microglia phagocytosis scores at 7 and 28 days. F Heatmap showing relative expression of each of the 50 top expressed genes in immune cells at 7 or 28 days. Columns correspond to cells, ordered by time. Selected genes are indicated (see Table S9 for the full list). G UMAP visualization of immune cells, colored by cell type annotations. H Stacked violinplot showing the expression of microglia and macrophage gene markers in 7d and 28d myeloid cells. I Barplot showing all 8 significantly overexpressed terms within the MSigDB Hallmark 2020 geneset in immune cells at 7 days and top 20 overexpressed terms at 28 days (see Tables S10, S11 for the full lists).
Cytokine and immune checkpoint landscape in the GL261-GSC GBM model
To provide a tentative map of the factors that mediate the interaction between the developing tumor and the immune system, we analyzed the expression of a panel of cytokines, including chemokines (CCL, CXCL), interleukins (IL), and tumor necrosis factor (TNF) family cytokines and their canonical receptors in all cell clusters (Fig. 6A). We found expression of most of them in the immune compartment, remarkably the chemokines Cxcl10, Cxcl9, Cxcl16, and Ccl25, Tgfb1, Il1b, Tnfsf8, and Tnfsf13b. Malignant cells predominantly expressed the chemokines Ccl25 and Cxcl10, Colony stimulating factor 1 (CSF-1; Csf1), Il7, Il33, Il34, Il17d, Tnfsf10, and Tnfsf13b. Ccl25 encodes an important cytokine in tumor progression56 highly expressed in all clusters in our data, and especially in implanted GL261-GSCs. However, its canonical receptor, CCR9, was scarcely expressed only by endothelial cells. Cxcl10 was mainly expressed by tumor cells, and to a lesser extent by other TME cells, such as immune cells, astrocytes, endothelial cells and OPCs. Intriguingly, the expression of Cxcr3, the canonical receptor of CXCL10, was not found in our data. Csf1 was expressed by tumor cells and other TME cells, such as astrocytes and endothelial cells, while its receptor, Csf1r, was highly expressed by immune cells. IL-34 is another ligand for CSF-1R, which regulates microglia and macrophages, and it has the ability to interact with PTPRZ57, whose transcript we found within the most overexpressed in GL261-GSCs, especially in implanted GL261-GSCs (Fig. 3B and Suppl. Fig. 2). GL261-GSCs also expressed Il17d, which can suppress the activity of CD8+ T cells by regulating dendritic cells58 and Tnfsf10 and Tnfsf13b, which encode two important proteins in cancer biology, TNF-related apoptosis-inducing ligand (TRAIL59; and TNF- and APOL-related leukocyte expressed ligand 1 (TALL-1), also called B-cell activating factor (BAFF60;). Among the receptors for these proteins, in this study we found a modest expression of Tnfrsf10b and Tnfrsf13b and Tnfrsf13c in tumor cells and astrocytes, and Tnfrsf13b and Tnfrsf13c in immune cells. Endothelial cells showed high expression of Cxcl12, as previously described49, Il34, and Tnfsf10. Oligodendrocytes showed strikingly high expression of Il33, which is known to be released upon CNS injury as an alarmin that acts on local astrocytes and microglia to induce chemokines for monocyte recruitment61, while being implicated in oligodendrocyte maturation62. Tgfb1 transcript was found in all cell types, although immune and endothelial cells (Fig. 6A), expressed the higher levels, which is consistent with the location of this protein in the GL261-GSC GBM model (Fig. 6B). Indeed, immunofluorescence analyses confirmed the expression of TGFß1 in these tumors (Suppl. Fig. 6E), with high levels in tumor areas containing TAMs associated to blood vessels (Fig. 6B and Suppl. Fig. 6F). In addition, high expression of its receptors Tgfbr1 and Tgfbr2 in endothelial, tumor and immune cells, and Tgfbr3 in endothelial cells was found (Fig. 6A), suggesting that the important role played by TGFß signaling pathway in GBM63 is recapitulated in the GL261-GSC GBM model. Strangely, we did not find expression of the interferon gamma (Ifng) transcript, although expression of its receptors Ifngr1 and Ifngr2 was found in all cell types, especially immune cells.
A Dotplot showing expression of cytokines and receptors per cluster. Orange lines indicate interactions described in the text; gray dotted lines indicate other described interactions. B Illustrative TGFß1, IBA1 and CD31 triple immunofluorescence at the tumor border 28 days post-implantation. Merge image showing the expression of TGFß1 in tumor areas enriched in IBA1+ TAMs associated to blood vessels (CD31+), scale bar: 50 µm. Zoomed-in images in Suppl. Fig. 6F. C Dotplot showing expression of checkpoint ligands and receptors per cluster. Orange lines indicate interactions described in the text; gray dotted lines indicate other described interactions. D UMAPs showing the expression of selected genes. E Illustrative TIM-3 immunofluorescence images of brains at 28 days post-implantation suggesting the recruitment of TIM-3+ cells towards the tumor border (dashed line), scale bar: 2 mm. Zoomed-in images showing the enwrapping of some tumor cells by TIM-3+ cells, scale bar: 50 µm. Quantification of TIM-3 protein levels in the tumor area (circular dots) and contralateral hemisphere (square dots) from the immunofluorescence images. Results are expressed in arbitrary units (a. u.) and are the mean±SEM from three independent experiments from which 5 fields were quantified. ***P < 0.001 (student’s t test).
Given the relevance of checkpoints inhibitor in immunotherapy, we analyzed the expression of the main checkpoint ligands and receptors in all the cell clusters (Fig. 6C). Immune cells showed expression of checkpoint receptors, with strikingly high levels of Havcr2 (TIM-3) and Vsir (VISTA) transcripts in immune cells (Fig. 6C, D). Indeed, the expression of TIM-3, a protein directly related to GBM progression64, is high in these tumors (Fig. 6E). A closer examination suggests that TIM-3+ immune cells are recruited towards these tumors, in which some TIM-3+ immune cells appear to enwrap tumor cells (Fig. 6E). Checkpoint ligands were expressed by tumor cells and immune cells, accordingly with recent studies that show the expression of checkpoint molecules by myeloid cells49,65. Galectin-9 (Lgals9), the ligand of TIM-3 and VISTA, was expressed in both tumor and immune cells. We observed low levels of PD-1 transcripts (Pdcd1), restricted, as expected, mostly to the T cell cluster. PD-L1 transcript (Cd274) levels were more prominent and were mostly present in myeloid immune cells and in implanted tumor cells, as confirmed by immunofluorescence (Fig. 4D). PD-L2 (Pdcd1lg2) levels were very low, maybe due to the low abundance of dendritic cells in our data. Expression of the co-stimulatory molecule Cd276 (B7-H3) increased in GSCs, especially after transplantation. Ctla4 expression is low and restricted to T cells, whereas its B7 ligands (Cd80, and mainly Cd86) were also found in myeloid cells.
Of note, spatial transcriptomic results showed that expression of most of the immune-related genes found in this study was significantly higher in the tumor area compared to healthy brain parenchyma (Table S2). This is the case for markers of different immune cell types (Ptprc, Cd4, Cd3e, and Cd3g, Cd74, H2-Aa, H2-Ab1, H2-Eb1, Tmem119, Cx3cr1, Mrc1, Nt5e, S100a4, Hmox1, Arg1, Mrc1 or Tgfb1), cytokines and receptors (Ccl25, Cxcl10, IFN-γ, Csf1, Csf1r, Il17d, Tnfsf10, Tnfsf13b, Tnfrsf10b, Tnfrsf13b, Tnfrsf13c, Tgfb1, Tgfbr1, Ifng, Ifngr1 and Ifngr) and checkpoints and receptors (Havcr2, Vsir, Lgals9, Cd274, Pdcd1lg2, Cd276, Ctla4 and Cd86). Immunofluorescence and Western blot analyses confirmed the higher expression of some of these immune markers in the tumor area when compared to the contralateral region. This is the case of IBA1+ (Fig. 1C), PD-L1 (Fig. 4D), Irf8 (Fig. 4E and Suppl. Fig. 7B), TGFß1 (Fig. 6B and Suppl. Fig. 6E), TIM-3 (Fig. 6E), CD3 (Suppl. Fig. 5B) and Mrc1 (Suppl. Fig. 5F).
snRNA-Seq analysis of TMZ and Tat-Cx43266-283 treatments in GL261-GSC GBM model
To explore the effects of treatments by snRNA-seq in this preclinical model, we selected TMZ, the standard of care for GBM patients, and the cell penetrating peptide Tat-Cx43266-283, because it exerts important anti-tumor effects and enhances the survival in the GL261-GSC GBM model22,25,26,27, as well as in other preclinical GBM models66. We analyzed the brain tumors and their microenvironment and clusters were classified by treatments, annotations, time and samples (Fig. 7A, B, Suppl. Fig. 6A), as previously described. Although extensive information can be obtained from these data, we focused on cell clusters with a significant number of cells that were highlighted as more prominent in previous sections.
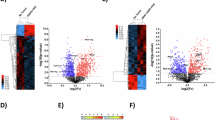
A UMAP visualization of all cells collected, colored by treatment and annotations. B Barplot showing the distribution of cell types obtained in untreated and treated animals. C Venn diagram showing the overlap between differentially expressed genes across treatments in the immune cell cluster at 7 dpi. Differential gene expression was analyzed using Wilcoxon test with a p-value cutoff=0.05. See Table S12 for full list. D Top 10 enriched terms from gene ontology (GO) gene sets in immune cell cluster from Tat-Cx43266-283-treated mice compared to the control (see Table S13 for full list). E Violinplots showing early (high levels in controls at 7 days) and late (high levels in controls at 28 days) immune scores in immune cells at 28 days post-implantation under different treatments. F Two-dimensional representation of the cellular states described by Neftel et al.6 in tumor cells of temozolomide-treated mice at 28 dpi. Each quadrant corresponds to one cellular state and each dot represents a cell. G Barplot showing the percentage of cycling cells analyzed, calculated as the sum of percentages of cells in S and G2/M phases, in control and TMZ-treated tumor cells at 28 dpi. H Heatmap showing the expression of selected cytokines and checkpoint molecules in control and TMZ-treated tumor cells at 28 dpi. I Illustrative TGFß1 immunofluorescence images of brains at 28 days post-implantation in control, Tat-Cx43266-283 and temozolomide (TMZ) treated animals, scale bar: 50 µm. J Quantification of TGFß1 protein levels in the immunofluorescence images. Results are expressed in arbitrary units (a. u.) and are the mean±SEM from three independent experiments from which 5 fields were quantified. *P < 0.05, **P < 0.01 (one-way ANOVA).
As shown in Fig. 7C, the treatment with Tat-Cx43266-283 peptide extensively affected the transcription in immune cells at 7 days post tumor implantation (see Table S12 for the full list of DEGs). Indeed, 1926 genes were differentially expressed by immune cells in Tat-Cx43266-283-treated animals, while 607 were modified in TMZ-treated animals. Only 67 genes were commonly affected by TMZ and Tat-Cx43266-283. The analysis of top 10 enrichment biological processes, cellular components, and molecular functions from gene ontology (GO) unveiled interesting GTPase, kinase, translation, transcriptional, and immune-related activity of immune cells in brain tumors treated with Tat-Cx43266-283 (Fig. 7D and Table S13). These analyses showed only molecular functions significantly modified by TMZ (Suppl. Fig. 6B and Table S14). By comparing the expression of genes in immune cells in untreated tumor-bearing mice, at 7 vs 28 days post-implantation, we obtained an early (7 days) and late (28 days) immune score. Interestingly, the treatment with Tat-Cx43266-283 and TMZ for 28 days, decreased the expression of those genes elevated in untreated animals at 28 days (late up) and showed an expression pattern more similar to that found at 7 days (early up) in untreated animals (Fig. 7E). Suppl. Fig. 6C shows the reduction in the expression of some checkpoint molecules and immune modulators, such as Hexb, Lars2, Rack1, H2-K1, Mrc1, Tgfb1, Cd74, Lglas9 or Nav3 promoted by Tat-Cx43266-283 and TMZ in immune cell cluster at 28 days post tumor implantation. Given the pivotal role played by TGFß1 in GBM63, we selected this protein to validate the snRNA-seq data obtained with these treatments. Our results confirmed that Tat-Cx43266-283 and TMZ reduced the levels of TGFß1 in GL261-GSC GBM model at 28 days post tumor implantation (Fig. 7I, J and Suppl. Fig. 6E).
Because of the low number of malignant cells obtained from peptide-treated 28-day samples, we could only study differences in malignant cells from control and TMZ-treated 28-day samples. Figure 7F shows that, upon treatment with TMZ for 28 days, tumor cells mostly fall into OPC and, to a lesser extent, NPC categories in the cellular-state map from Neftel et al.6. As expected, the analysis of cell cycle by snRNA-Seq showed that tumor cell proliferation was decreased by TMZ treatment (Fig. 7G). Interestingly, the expression of key genes involved in immune evasion, such as Irf8, Cd274, Lgals9, Havcr2, and Cd276 were downregulated by TMZ in tumor cells (Fig. 7H and Table S15). Furthermore, TMZ significantly decreased the expression of key genes related to neuron-glioma synapsis, such as Gap43, Gja1, Gria3, Grik2 and Homer1 (Table S15) in tumor cells. However, some genes related to bad prognosis were upregulated in tumor cells treated with TMZ, including Ahnak, Atrx, Cd44, Jarid2, Plcd4, Cdk8 and Myc (Suppl. Fig. 6D and Table S15).
Discussion
Preclinical models are essential to advance our understanding of GBM biology and to improve their treatment. However, a thorough characterization of each GBM preclinical model is essential to identify the cellular and molecular targets that can be studied within that specific model. In addition, the clinical application of this information requires the identification of the most similar type of human GBM, which would benefit from these results. In this study, we provide a comprehensive characterization of the preclinical GL261-GSC GBM model, in which tumors are originated by murine GSCs intracranially implanted in immunocompetent mice. We chose this model because it emulates the origin of human brain tumors from GSCs67,68 and allows the study of the GBM immune system and its evolution throughout tumor development. Our snRNA-Seq data showed that GL261-GSCs recapitulate some key human GSC attributes, cellular pathways, and processes. Although the genetic heterogeneity of mouse GBM cells is different from that of human GBM cells, once implanted into the brain, GL261-GSCs diverged and gave rise to heterogenous GBM cells, which shared the expression of some genes with differentiated GL261 cells and GL261-GSCs, but mainly expressed unique genes, indicating that these tumors are heterogenous and include stem-like and differentiated GBM cell subpopulations.
Interestingly, we found a bidirectional interdependence between tumor cells and brain microenvironment, which is very similar to that described in human GBM35. Although these results can be mined with a great variety of purposes, we focused on the transcriptional changes promoted by the brain microenvironment on tumor cells and those promoted by tumor cells on the immune system. These analyses unveiled and validated the expression of key GBM targets that can be studied in the GL261-GSC GBM model. Furthermore, we found similarities with a subtype of human GBM, paving the way to translate the preclinical results generated from the GL261-GSC GBM model into clinical trials in this specific human GBM subtype.
Our study shows the feasibility of the GL261-GSC GBM model to study GBM-neuron synapsis and GBM networks. Thus, Increasing evidence has demonstrated that GBM progression is robustly regulated by neuronal activity, which stimulates synaptic activity in GBM cells41,42,44,69. Our results show that among the pathways significantly enriched in intracranially implanted GL261-GSCs when compared to cultured GL261-GSCs are many related to synaptic activity and neuronal signaling, which suggests that the brain TME promotes changes in transcription towards the development of synaptic activity. Indeed, implanted GL261-GSCs expressed diverse ionotropic glutamate receptors, which might activate intercellular calcium signaling networks to orchestrate GBM cell growth, invasion, and drug resistance as it has been previously shown41,42,44,69.
Neuronal input facilitates the formation of tumor microtubes, which are ultra-long membrane tube protrusions that connect single tumor cells to create functional and communicating multicellular networks by intercellular Ca2+ waves. The neuronal growth-associated protein 43 (Gap43) is important for microtube formation and function, and drives microtube-dependent tumor cell invasion, proliferation, interconnection, radioresistance, contributes to mitochondria transfer from astrocytes to tumor cells and enhances glioblastoma tumorigenicity45,46. Microtube-associated gap junctions formed by connexin43 (Gja1) also contributes to communication in the multicellular network. The increased expression of Gap43 found in GL261-GSCs upon intracranial transplantation, together with the location of Gap43 protein in these tumors and the increased Gja1 (Cx43) expression, is compatible with a network of communicating tumor microtubes in the GL261-GSC GBM model. In agreement with this, we found that some of the implanted GL261-GSCs expressed channel KCa3.1 (Kcnn4), which is present in a small population of human GBM cells and is responsible for rhythmic Ca2+ oscillations within the connected network that sustain tumor growth44. Furthermore, the high levels of TGFß1 mRNA and protein found in in the GL261-GSC GBM model might contribute to the formation of microtubes70. Implanted GL261-GSCs were predominantly enriched with the OPC-like NPC-like cell states, which coincides with the most migratory GBM subpopulation described by Venkataramani et al.69. Therefore, the GL261-GSC GBM model might be useful to study the mechanism by which GBM-neuron synapsis are developed and to carry out preclinical studies of drug designed to target key candidates, such as GAP43, ionotropic glutamate receptors, neuroligin-343 or KCa3.1, specifically in GBM cells.
Among the changes found in tumor cells promoted by the TME, we found that intracranially-injected GL261-GSCs develop an immune evasive phenotype. Previous studies showed that GBM cells may acquire an immune evasive phenotype via epigenetic immunoediting47, a process in which immune attack promotes transcriptional changes in tumor cells that are stabilized and selected in those cells with increased immune evasive qualities, resulting in highly immune evasive and transcriptionally altered descendants. We found that key genes whose methylation is erased and are immune evasion regulators, such as Nt5e (CD73), Cd274 (PD-L1), and Irf8 are expressed by GL261-GSCs upon brain implantation. In fact, most of them are barely expressed in GL261-GSCs but are upregulated when implanted in the brain for 28 days, indicating that they are expressed in response to TME. CD73 is a critical component in the formation of an immunosuppressive microenvironment in cancer and its expression correlates with bad prognosis in distinct types of tumors71. CD73 is secreted in GBM cell exosomes72 and has been identified as a relevant target to improve immune checkpoint therapy in GBM73. Cd274, also known as programmed death-ligand 1 (PD-L1) is one of the best immune scape mediators and the development of anti-PD1/PD-L1 antibodies is a hot topic in cancer immunotherapy71. Upon binding to PD1, it activates downstream signaling pathways and inhibits T-cell activation. Irf8 is the key master transcription factor for immune evasion, and it is upregulated in tumor cells following immune attack via IFN- γ -mediated activation47. Our study unveiled “Regulation of interferon-gamma production” among the gene sets upregulated in GL261-GSCs after implantation into the brain when compared to GL261-GSCs in culture. Altogether, these results indicate that the in vivo immune attack triggers significant transcriptional changes in GL261-GSCs to promote an immune evasive phenotype.
In line with the immune evasion developed by tumor cells in response to TME, the expression of cytokine and cytokine receptors found in this study suggests that the development of these tumors reshapes the TME, with a shift towards a supportive immune system where TAMs are the main allies. In fact, the number of TAMs surrounding and infiltrating the entire tumor is notably high, whereas that of lymphocytes is low in the GL261-GSC GBM model. These data suggest a notable difference with the GL261 GBM model that contains an important number of CD4+ cells that maintained CD8+ cells active to respond to anti PD1 therapy74. Csf1r, Arg1, Mrc1 or Tgfb153,54 are among the markers of immunosuppressive and tumor supportive myeloid cells highly expressed by the immune cluster, mainly at late stages of tumor development. The expression of PD-L1, Irf8, Mrc1 and TGFß1 and their location in these tumors were validated by Western blot or immunofluorescence. Therefore, this is an interesting preclinical model to study the development of immune evasion in GBM cells promoted by the brain TME and to explore therapies against crucial targets for this process in tumor cells, such as Nt5e (CD73), PD-L1(Cd274) or Irf8, and in immune cells, such as Csfr1, Arg1, Mrc1 or Tgfb1. In this study, we confirmed that TMZ, the standard chemotherapy in GBM75, and Tat-Cx43266-283, a preclinical therapy that improves the survival of GL261-GSC GBM-bearing mice22, reduced TGFß levels in these tumors. Given the relevance of TGFß in GBM progression, these results suggest that the reduction of TGFß may be involved in the antitumor mechanism of these therapies.
A major finding of this study is that the GL261-GSC model resembles a specific subtype of human GBM with a heterogeneous immune population. Indeed, one of the hallmarks of human GBM is the infiltration of immune cells that may constitute up to a 30% of the cells that integrate these tumors76. Immunofluorescence analysis and spatial transcriptomics showed that IBA1+ cells and the expression of most of the immune-related genes were significantly higher in the tumor area compared to healthy brain parenchyma, indicating that immune cells, mainly TAMs, infiltrated brain tumors in the GL261-GSC GBM model, as described in human GBMs. Three novel human GBM subtypes with significantly different TME compositions have been recently proposed based on the analysis of more than 800 human GBM samples3. These subgroups were defined as TMELow (Immune-Low), TMEMed (heterogenous immune populations) and TMEHigh (Immune-High), comprising 24%, 46% and 30% of human GBMs, respectively. Only those patients classified as TMEHigh showed a significant improved survival after immunotherapy, which highlights the relevance of patient stratification to rationally select patients who might benefit most from treatments. By comparing these human GBM subtypes with the GL261-GSC GBM murine model, we found substantial similarities with TMEMed subtype, the most frequent human GBMs3. TMEMed GBMs are characterized by heterogeneous immune populations, enrichment in endothelial cell gene expression profiles and in pathways related to neuronal signaling. These and other TMEMed GBM features, such as the downregulation of the B lymphocyte chemoattractant and TLS marker, Cxcl13, are found in the GL261-GSC GBM murine model. More relevant for immunotherapy, Pdcd1 (PD1), Ctla4 and Lag3 are scarcely expressed in TMEMed GBM subtype, as well as in the GL261-GSC GBM model, which is in contrast with the high level of expression of these immune checkpoints in TMEHigh GBM subtype tumors. Retrospective studies showed that only TMEHigh GBMs have a positive response to anti-PD1 immunotherapy3, which agrees with the high levels of PD1 expression. Interestingly, GBM models derived from differentiated GL261 cells are highly responsive to anti-PD1 and anti-CTLA4 immunotherapy77, indicating high levels of PD1 and CTLA4 in the differentiated GL261 model. The low expression of PD1 and CTLA4 predicts a reduced response to anti-PD1 and anti-CTLA4 immunotherapies in GL261-GSC GBM model. However, other key targets for immunotherapy, such as PD-L1, TIM-378 or B7-H3 are highly expressed in the GL261-GSC GBM model, which coincides with TMEMed GBM subtype3, indicating the utility of GL261-GSC GBM model as a preclinical model to test therapies against these immunotherapy targets and support the translation of GL261-GSC GBM results to TMEMed GBM subtype.
Among the study limitations, we observed significant differences in the results obtained with three technologies for transcriptome analysis, i.e., snRNA-seq, a microfluidic-droplet-based method on single cells from 10x (10x cells) and a full-length scRNA-seq using Smart-Seq2 method. Our results highlight that, despite the growing interest and considerable utility of these methods, they also have limitations and further validation, that in our case was performed by immunohistochemistry and/or Western blot, is necessary to reinforce the results. In addition, although most data analyzed through the manuscript is robust, the low number of nuclei in some clusters under specific conditions, for example GBM cells after 7 days post-implantation, is limited and these results should be interpreted with caution. However, these samples were enriched in an interesting TME that can provide information to understand GBM development. As all preclinical models, GL261-GSC GBM model has inherent limitations, such as the moderate immunogenicity of GL261 cells or their different genetic heterogeneity compared to human GBM cells. It is noteworthy that not all human GBMs have low immunogenicity, in fact different subtypes of human GBMs have been identified according to their immune TME3. Importantly, our data indicate that the GL261-GSC GBM model recapitulates some features attributed to the TMEMed GBM subtype.
In conclusion, this study offers crucial information for future preclinical research with the potential to improve outcomes in clinical trials, particularly for patients with TMEMed GBM subtype. Furthermore, this preclinical model could be integrated into ongoing clinical trial frameworks to understand how the drug affects tumor cells and TME, including the immune system, which could be particularly relevant for immunotherapy. This knowledge could help in the design of amendments for an ongoing clinical trial, as well as in proposing the most promising therapy combinations. We provide extensive information, at two stages of tumor development, that can be an important resource for future research on GBM development. In addition, the mechanism of action of TMZ and Tat-Cx43266-283, their possible side effects or more effective drug combinations, can be further explored by exploiting the data included in this work. Overall, this study supports the utility of the GL261-GSC GBM model to study specific GBM targets and potential treatments with a strong rationale and better options for successful clinical translation.
Materials and methods
Animals
An equal number of 6-week-old male and female C57BL/6 mice were shipped from Charles River to the animal facility of the University of Salamanca at INCYL (SEA-INCYL). Animals were housed on a 12h/12h light/dark cycle and provided with food and water ad libitum for 1 week before the experimental procedure. Mice were maintained singly from the start of the experiments, and were monitored for signs of humane endpoints, including changes in behavior and weight. We have complied with all relevant ethical regulations for animal use. All animal procedures were approved by the ethics committee of the University of Salamanca and the Junta de Castilla y León (Spain) and were carried out in accordance with European Community Council directives (2010/63/UE), and Spanish law (RD 53/2013 BOE 34/11370–420, 2013) for the use and care of laboratory animals.
Cells
GL261 cells were obtained from DSMZ and grown adherently in differentiation medium containing DMEM supplemented with 10% fetal calf serum (FCS), dissociated using trypsin/EDTA, and split to convenience. Neurospheres (GL261-GSCs) were obtained from GL261 adherent cultures by successive passages with decreasing concentrations of FCS until cell detachment and the appearance of neurospheres21, which were cultured in stem cell medium22 containing Dulbecco’s modified Eagle’s medium (DMEM)/Nutrient Mixture F-12 Ham supplemented with 1% Minimum Essential Medium-Non-Essential Amino Acids (MEM-NEAA), 3.9 mM glucose, 1 mM L-glutamine, 0.07% β-mercaptoethanol, 121.8 μg/mL bovine serum albumin (BSA), 1% B-27 supplement, 0.5% N-2 supplement, 10 ng/ml epidermal growth factor (EGF), and 10 ng/ml basic fibroblast growth factor (b-FGF). Neurospheres were dissociated using Accutase and subcultured at a density of 104 cells/ml every 8-10 days. GL261-GSCs were stably transfected with pcDNA3.1-mCherry plasmid (a kind gift from C. Naus) using Lipofectamine 2000 Transfection Reagent according to manufacturer’s instructions. Cells were plated at low density and mCherry+ colonies were selected and amplified. All culture media were supplemented with 50 U/ml penicillin G, 37.5 U/ml Streptomycin and 0.23 µg/ml Amphotericin B to avoid bacterial and fungal contamination. Cells were maintained at 37 °C in an atmosphere of 95% air/5% CO2 and with 90-95% humidity.
Intracranial implantation of glioma cells
mCherry-GL261-GSCs were intracranially injected into 8-week-old C57BL/6 mice22. Mice were anesthetized by isoflurane inhalation, placed on a stereotaxic frame, and window-trephined in the parietal bone. A unilateral intracerebral injection to the right cortex was performed with a Hamilton microsyringe. 1 μl of physiological saline containing 5000 cells was injected at the following coordinates: 1 mm rostral to lambda, 1 mm lateral, and 2 mm deep. To minimize the inflammatory response from damaged brain tissue due to the needle injection, tumoral cells were slowly injected into the brain and the needle was held in place for an additional 2 min before removal. Cellular suspensions were kept on ice while the surgery was being performed and allowed to temperate for 5 min once loaded into the microsyringe. At the indicated times, mice were anesthetized with pentobarbital (120 mg/kg, 0.2 ml) and transcardially perfused with 15 ml of physiological saline. Brains were removed, fresh-frozen in liquid nitrogen, and kept at -80 °C until used.
Treatments
Mice were randomly marked with ear notching before the surgery. Then, they were allocated in experimental control and treatment groups (TMZ and Tat-Cx43266-283). To do so, 250 μM TMZ or 100 μM Tat-Cx43266-283 were intracranially injected in 1 μl of physiological saline together with the cells22. For 28-day experiments, 10 nmol/g TMZ or 4 nmol/g Tat-Cx43266–283 were intraperitonially injected twice per week, starting on day 8 and until the end of the experiment. An equivalent amount of saline was injected to control mice.
Synthetic peptides (>95% pure) were obtained from GenScript. YGRKKRRQRRR was used as the Tat sequence, which enables the cell penetration of peptides. The Tat-Cx43266-283 sequence was Tat-AYFNGCSSPTAPLSPMSP (patent ID: WO2014191608A1).
Tissue dissociation and sample preparation
A tissue piece was collected from each brain including tumor core and peritumoral space and the rest of the procedure was performed blindly. Nuclei suspensions from frozen tissue were obtained using a density gradient medium (https://www.protocols.io/view/nuclei-isolation-prep-and-protocol-5qpvoneedl4o/v1). Tissue pieces were mechanically digested in ice-cold buffer containing protease and RNAse inhibitors using a Dounce homogenizer and incubated in 0.35% IGEPAL® CA-630. Homogenized solutions were filtered using a 40 µm-pore cell strainer and carefully added to ultracentrifuge tubes with layered 40% and 30% Iodixanol solutions. Tubes were centrifuged at 10,000 g for 18 min and nuclei were collected from the interface between 40% and 30% Iodixanol solutions. Nuclei suspensions from fresh cells were obtained following 10xGenomics protocol (https://www.10xgenomics.com/products/nuclei-isolation). Cells and neurospheres were dissociated using Accutase and lysed in lysis buffer containing 0.3% IGEPAL® CA-630. Nuclei were counted using a Countess Automated Cell Counter and nuclei quality was assessed in an inverted microscope. Cells and nuclei were loaded with a target output of 6000 nuclei per sample.
Microfluidic droplet single-cell analysis
Single cells or nuclei were captured in droplet emulsions using the Chromium instrument (10x Genomics) and scRNA-seq libraries were constructed as per the 10x Genomics protocol using GemCode Single-Cell 5’ Bead and Library kit. Single nuclei were processed using the 10x Multiome ATAC + Gene Expression kit. All reactions were performed in the Biorad C1000 Touch Thermal cycler with 96-Deep Well Reaction Module. The number of cycles used for cDNA amplification and sample index PCR was determined following 10x guidelines. Amplified cDNA and final libraries were evaluated on a TapeStation System using a High Sensitivity D5000 ScreenTape. The average fragment length of the libraries was quantitated on a TapeStation System, and by qPCR with the Kapa Library Quantification kit for Illumina. Each library was diluted to 2 nM and equal volumes of up to 8 libraries were pooled for each sequencing run. Pools were sequenced with the number of cycles indicated by 10x guidelines. A PhiX control library was spiked in at 1%. Libraries were sequenced on a NovaSeq 6000 or a NextSeq 2000 Sequencing System (Illumina).
Visium spatial transcriptomics
After intracranial tumor implantation, frozen brain sections were obtained with a cryostat and processed for methanol fixation, hematoxylin-eosin staining to visualize brain tumor region and healthy brain region for use with 10x Genomics Visium Spatial protocols (https://doi.org/10.1101/2022.10.06.510405) following manufacturer’s instructions (https://www.biorxiv.org/content/10.1101/2023.07.11.548639v1.full).
Sequencing data extraction and pre-processing
Sequences were de-multiplexed using bcl2fastq v2.19.0.316. Reads were aligned to the mm10plus genome using STAR v2.5.2b with parameters TK. Gene counts were produced using HTSEQ v0,6,1p1 with default parameters, except ‘stranded’ was set to ‘false’, and ‘mode’ was set to ‘intersection-nonempty’. Sequences from the microfluidic droplet platform were de-multiplexed and aligned using CellRanger v2.0.1, with default parameters. Gene count tables were combined with the metadata variables using the Scanpy Python package v.1.4.2. We removed genes that were not expressed in at least 3 cells and then cells that did not have at least 250 detected genes. For FACS we removed cells with fewer than 5000 counts, and for the droplet method we removed cells with fewer than 2500 unique molecular identifiers (UMIs). The data was then normalized using size factor normalization such that every cell has 10,000 counts and log transformed. We computed highly variable genes using default parameters and then scaled the data to a maximum value of 10. We then computed principal component analysis, neighborhood graph and clustered the data using the Leiden algorithm29. The data was visualized using UMAP projection28. When performing batch correction to remove the technical artifacts introduced by the technologies, we replaced the neighborhood graph computation with batch balanced KNN (bbknn)79.
FACS
All the details of the protocols used in this study, including preparation of lysis plates, FACS sorting, cDNA synthesis using the Smart-Seq2 protocol 5,6, library preparation using an in-house version of Tn5 7,8, library pooling and quality control, and sequencing are described here (https://doi.org/10.17504/protocols.io.2uwgexe). Single-cell suspensions were sorted by a SONY SH800 Cell Sorter. All events were gated with forward scatter-area (FCS-A)/side scatter-area (SSC-A) and FCS-height (FCS-H)/FCS-width (FCS-W). Single cells were sorted in 96-well plates containing 4 μL lysis buffer (4U Recombinant RNase Inhibitor, 0.05% Triton X-100, 2.5 mM dNTP mix, 2.5 μM Oligo-dT30VN (5′-AAGCAGTGGTATCAACGCAGAGTACT30VN-3′), spun down for 2 min at 1,000 × g, and snap frozen. Plates containing sorted cells were stored at −80 °C until processing. Reverse transcription and PCR amplification were performed according to the Smart-seq2 protocol40. In brief, 96-well plates containing single-cell lysates were thawed on ice followed by incubation at 72 °C for 3 min and placed immediately on ice. Reverse transcription was carried out after adding 6 μL of reverse transcription-mix (100 U SMARTScribe Reverse Transcriptase, 10 U Recombinant RNase Inhibitor, 1× First-Strand Buffer, 8.5 mM DTT, 0.4 mM betaine, 10 mM MgCl2, and 1.6 μM TSO (5′-AAGCAGTGGTATCAACGCAGAGTACATrGrG+G-3′) for 90 min at 42 °C, followed by 5 min at 70 °C. Reverse transcription was followed by PCR amplification. PCR was performed with 15 μL PCR mix (1× KAPA HiFi HotStart ReadyMix), 0.16 μM ISPCR oligo (5′-AAGCAGTGGTATCAACGCAGAGT-3′), and 0.56 U Lambda exonuclease according to the following thermal-cycling protocol: (1) 37 °C for 30 min; (2) 95 °C for 3 min; (3) 21 cycles of 98 °C for 20 s, 67 °C for 15 s, and 72 °C for 4 min; and (4) 72 °C for 5 min. PCR was followed by bead purification using 0.7× AMPure beads, capillary electrophoresis, and smear analysis using a Fragment Analyzer. Calculated smear concentrations within the size range of 500 and 5000 bp for each single cell were used to dilute samples for Nextera library preparation.
Immunofluorescence
20-µm-thick coronal sections were processed for immunostaining27. After fixing with 4% PFA for 25 min, brain sections were blocked for 2h in PBS containing 10% NGS, 5% BSA, 0.1% Tween, and 0.02% azide and incubated overnight at 4 °C with rabbit monoclonal antibodies against IBA1 (1:500; WAKO #019-19741), Gap43 (1:250; Ab5220), Sox6 (1:200; Proteintech 14010-1-AP), CD3 (1:500; Ab16669) or mouse monoclonal antibody against GFAP (1:500; Sigma G3893); PD-L1 (1:200; Proteintech 66248-1-Ig); CD206 (Mrc1) (1:200; SC-58986), TIM-3 (1:300; Proteintech 60355-1-Ig) or TGFß1 (1:200; SC-52893), prepared in the same solution (with 0.1% Tween). After repeating washing in PBS with 0.1% Tween, sections were incubated at room temperature for 2 h with goat anti-rabbit IgG Alexa Fluor A647-conjugated secondary antibody in case of IBA1 (1:500, Invitrogen, ref: #A-21244); goat anti-mouse IgG Alexa Fluor A594-conjugated secondary antibody in case of GFAP, PD-L1, CD206, TIM-3 and TGFß1 (1:500, Invitrogen, ref: #A-11032); and goat anti-rabbit IgG Alexa Fluor A488-conjugated secondary antibody in case of Gap43, Sox6 and CD3 (1:500, Invitrogen, ref: #A-11034), prepared in all cases in PBS containing 10% NGS, 5% BSA, 0.1% Tween, and 0.02% azide. For CD31 immunofluorescence, after fixation with 4% PFA for 25 min, brain sections were blocked overnight at 4 °C in PBS containing 10% donkey serum and 1% Triton X-100. Afterwards, sections were incubated overnight at 4 °C with rat monoclonal antibody against CD31 (1:100; BD Pharmigen 550274) prepared in the same solution (with 1% Triton X-100). After repeating washing In PBS with 0.1% Triton X-100, sections were incubated at room temperature as long as possible with goat anti-rat IgG Alexa Fluor A488-conjugated secondary antibody (1:500, Invitrogen, ref: #A-11006) prepared in PBS containing 10% donkey serum and 0.5% Triton X-100. Nuclear DNA was stained with DAPI for 5 min and sections were mounted using SlowFade Light antifade (Life Technologies). Mosaic images of the sections were acquired using a Leica Stellaris 8 confocal microscope, using a 10X objective (whole section images) or a 63X objective (zoomed-in images).
Western blotting
Equal amounts of proteins across conditions were separated on NuPAGE Novex Bis-Tris 4-12% Midi gels (Life Technologies) at room temperature and constant voltage. Proteins were transferred to a nitrocellulose membrane (iBlot Gel Transfer Stacks Nitrocellulose) using an iBlot dry blotting system (Life Technologies). Some membranes were cut to be immunoblotted with distinct antibodies, thus allowing for comparative analysis of the amount of each protein in the same sample. After blocking, the membranes were incubated overnight at 4 °C with primary anti- bodies: mouse monoclonal antibody against Irf8 (1:200; Thermo Fisher Scientific GW4CML3) and rabbit polyclonal antibody against Sox6 (1:500; Proteintech 14010-1-AP). After washing, the membranes were incubated with peroxidase-conjugated anti-rabbit IgG or anti-mouse IgG antibodies (1:5,000; Jackson ImmunoResearch) and developed with a chemiluminescent substrate (Western Blotting Luminol Reagent; Santa Cruz Biotechnology) in a MicroChemi imaging system (Bioimaging Systems). GAPDH protein levels in the same lanes were used as loading controls.
Statistics and reproducibility
All data analysis was performed using Python. Principal component analysis, differential gene expression and enrichment analysis were performed using Scanpy. Wilcoxon rank sum test was performed with a p-value cutoff=0.01. CNV were inferred using the infercnvpy tool. Cells were assigned to cell cycle phases by scoring a set of cell cycle genes39.
Two-dimensional representation of malignant cellular states described by Neftel et al.6 was followed. Cells were first separated into OPC/NPC versus AC/MES by thesign of D = max (SCopc,SCnpc) - max(SCac,SCmes), and D defined the y axis of all cells. Next, for OPC/NPC cells (i.e., D>0), the x axis value was defined as log2(|SCopc–SCnpc|+1) and for AC/MES cells (i.e., D<0), the x axis was defined as log2(|SCac-SCmes|). Please for further details refer to the code available at github: https://github.com/laugarvi/GL261-GSC-GBM.
Differences in protein levels between groups were analyzed using GraphPad Prism software. Two-tailed Student’s t-test and one-way ANOVA, followed by a post hoc Tukey test, were used for comparisons between 2 groups or more than 2 groups, respectively.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The transcriptomics data are available in the public functional genomics data repository Gene expression Omnibus (GEO) identifier: GSE246154 - Single-nucleus RNA-sequencing (snRNA-Seq) of brain cells from tumor-bearing C57BL/6 mice- https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE246154. GSE245263 - Spatial transcriptomics of brains from tumor-bearing C57BL/6 mice- https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE245263. GSE244301 - Single-cell RNA-sequencing (scRNA-Seq) of GL261 and GL261-GSCs. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE244301. GSE246262 - Single-cell RNA-sequencing (scRNA-Seq) of GL261 and GL261-GSCs (SmartSeq) https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE246262. Numerical source data for Figs. 3H, 4D, 6E and 7J are available in Figshare: https://doi.org/10.6084/m9.figshare.28742636.v1. Supplementary tables (Tables S1–S15) contain complete analyses performed in the present study and are available in Figshare: Table S1. https://doi.org/10.6084/m9.figshare.28722128.v1. Table S2. https://doi.org/10.6084/m9.figshare.28722137.v1. Table S3. https://doi.org/10.6084/m9.figshare.28722122.v1. Table S4. https://doi.org/10.6084/m9.figshare.28722113.v1. Table S5. https://doi.org/10.6084/m9.figshare.28722131.v1. Table S6. https://doi.org/10.6084/m9.figshare.28722125.v1. Table S7. https://doi.org/10.6084/m9.figshare.28722116.v1. Table S8. https://doi.org/10.6084/m9.figshare.28722134.v1. Table S9. https://doi.org/10.6084/m9.figshare.28722119.v1. Table S10. https://doi.org/10.6084/m9.figshare.28722146.v1. Table S11. https://doi.org/10.6084/m9.figshare.28722143.v1. Table S12. https://doi.org/10.6084/m9.figshare.28722152.v1. Table S13. https://doi.org/10.6084/m9.figshare.28722140.v1. Table S14. https://doi.org/10.6084/m9.figshare.28722149.v1. Table S15. https://doi.org/10.6084/m9.figshare.28722155.v1.
Code availability
The code is available at github: https://github.com/laugarvi/GL261-GSC-GBM. All other data are available from the corresponding author on reasonable request.
References
Louis, D. N. et al. The 2021 WHO Classification of Tumors of the Central Nervous System: a summary. Neuro Oncol. 23, 1231–1251 (2021).
Mellinghoff, I. K. et al. Vorasidenib in IDH1- or IDH2-Mutant Low-Grade Glioma. N. Engl. J. Med. https://doi.org/10.1056/NEJMoa2304194 (2023).
White, K. et al. Identification, validation and biological characterisation of novel glioblastoma tumour microenvironment subtypes: implications for precision immunotherapy. Ann. Oncol. 34, 300–314 (2023).
Patel, A. P. et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science 344, 1396–1401 (2014).
Wang, Q. et al. Tumor Evolution of Glioma-Intrinsic Gene Expression Subtypes Associates with Immunological Changes in the Microenvironment. Cancer Cell 32, 42–56.e46 (2017).
Neftel, C. et al. An Integrative Model of Cellular States, Plasticity, and Genetics for Glioblastoma. Cell 178, 835–849.e821 (2019).
Suvà, M. L. & Tirosh, I. Single-Cell RNA Sequencing in Cancer: Lessons Learned and Emerging Challenges. Mol. Cell 75, 7–12 (2019).
Slyper, M. et al. A single-cell and single-nucleus RNA-Seq toolbox for fresh and frozen human tumors. Nat. Med. 26, 792–802 (2020).
Habib, N. et al. Massively parallel single-nucleus RNA-seq with DroNc-seq. Nat. Methods 14, 955–958 (2017).
Fischer, J. & Ayers, T. Single nucleus RNA-sequencing: how it's done, applications and limitations. Emerg. Top. Life Sci. 5, 687–690 (2021).
Johnson, J. I. et al. Relationships between drug activity in NCI preclinical in vitro and in vivo models and early clinical trials. Br. J. Cancer 84, 1424–1431 (2001).
Ciesielski, M. J. et al. Antitumor effects of a xenogeneic survivin bone marrow derived dendritic cell vaccine against murine GL261 gliomas. Cancer Immunol. Immunother. 55, 1491–1503 (2006).
Pellegatta, S. et al. Dendritic cells pulsed with glioma lysates induce immunity against syngeneic intracranial gliomas and increase survival of tumor-bearing mice. Neurol. Res. 28, 527–531 (2006).
Maes, W. & Van Gool, S. W. Experimental immunotherapy for malignant glioma: lessons from two decades of research in the GL261 model. Cancer Immunol. Immunother. 60, 153–160 (2011).
Xu, M. et al. Mouse glioma immunotherapy mediated by A2B5+ GL261 cell lysate-pulsed dendritic cells. J. Neurooncol 116, 497–504 (2014).
Wainwright, D. A. et al. Durable therapeutic efficacy utilizing combinatorial blockade against IDO, CTLA-4, and PD-L1 in mice with brain tumors. Clin. Cancer Res. 20, 5290–5301 (2014).
Szatmári, T. et al. Detailed characterization of the mouse glioma 261 tumor model for experimental glioblastoma therapy. Cancer Sci. 97, 546–553 (2006).
Eyler, C. E. & Rich, J. N. Survival of the fittest: cancer stem cells in therapeutic resistance and angiogenesis. J. Clin. Oncol. 26, 2839–2845 (2008).
Sherriff, J. et al. Patterns of relapse in glioblastoma multiforme following concomitant chemoradiotherapy with temozolomide. Br. J. Radio. 86, 20120414 (2013).
Pellegatta, S. et al. Neurospheres enriched in cancer stem-like cells are highly effective in eliciting a dendritic cell-mediated immune response against malignant gliomas. Cancer Res. 66, 10247–10252 (2006).
Yi, L. et al. Implantation of GL261 neurospheres into C57/BL6 mice: a more reliable syngeneic graft model for research on glioma-initiating cells. Int J. Oncol. 43, 477–484 (2013).
Jaraíz-Rodríguez, M. et al. Connexin43 Peptide, TAT-Cx43266-283, Selectively Targets Glioma Cells, Impairs Malignant Growth, and Enhances Survival in Mouse Models in Vivo. Neuro-Oncol. 22, 493–504 (2020).
Pan, T. et al. (2024). Glioma-Stem-Cell-Derived Exosomes Remodeled Glioma-Associated Macrophage via NEAT1/miR-125a/STAT3 Pathway. Cancers 16, https://doi.org/10.3390/cancers16142500.
Ahmad, M. et al. How stemlike are sphere cultures from long-term cancer cell lines? Lessons from mouse glioma models. J. Neuropathol. Exp. Neurol. 73, 1062–1077 (2014).
Jaraíz-Rodríguez, M. et al. A Short Region of Connexin43 Reduces Human Glioma Stem Cell Migration, Invasion, and Survival through Src, PTEN, and FAK. Stem Cell Rep. 9, 451–463 (2017).
Gangoso, E. et al. A c-Src Inhibitor Peptide Based on Connexin43 Exerts Neuroprotective Effects through the Inhibition of Glial Hemichannel Activity. Front Mol. Neurosci. 10, 418 (2017).
Pelaz, S. G. et al. Targeting metabolic plasticity in glioma stem cells in vitro and in vivo through specific inhibition of c-Src by TAT-Cx43. EBioMedicine 62, 103134 (2020).
Becht, E. et al. Dimensionality reduction for visualizing single-cell data using UMAP. Nat. Biotechnol. https://doi.org/10.1038/nbt.4314 (2018).
Traag, V. A., Waltman, L. & van Eck, N. J. From Louvain to Leiden: guaranteeing well-connected communities. Sci. Rep. 9, 5233 (2019).
Zeisel, A. et al. Molecular Architecture of the Mouse Nervous System. Cell 174, 999–1014.e1022 (2018).
Tirosh, I. et al. Single-cell RNA-seq supports a developmental hierarchy in human oligodendroglioma. Nature 539, 309–313 (2016).
Pelaz, S. G. et al. A proteomic approach supports the clinical relevance of TAT-Cx43. Transl. Res. 272, 95–1 (2024).
Zhao, S. C., Zhou, B. W., Luo, F., Mao, X. & Lu, Y. J. The structure and function of NKAIN2-a candidate tumor suppressor. Int J. Clin. Exp. Med. 8, 17072–17079 (2015).
Bhaduri, A. et al. Outer Radial Glia-like Cancer Stem Cells Contribute to Heterogeneity of Glioblastoma. Cell Stem Cell 26, 48–63.e46 (2020).
Ravi, V. M. et al. Spatially resolved multi-omics deciphers bidirectional tumor-host interdependence in glioblastoma. Cancer Cell 40, 639–655.e613 (2022).
Bovenga, F., Sabbà, C. & Moschetta, A. Uncoupling nuclear receptor LXR and cholesterol metabolism in cancer. Cell Metab. 21, 517–526 (2015).
Kuzu, O. F., Noory, M. A. & Robertson, G. P. The Role of Cholesterol in Cancer. Cancer Res. 76, 2063–2070 (2016).
Cheng, C., Geng, F., Cheng, X. & Guo, D. Lipid metabolism reprogramming and its potential targets in cancer. Cancer Commun. (Lond.) 38, 27 (2018).
Satija, R., Farrell, J. A., Gennert, D., Schier, A. F. & Regev, A. Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 33, 495–502 (2015).
Picelli, S. Full-Length Single-Cell RNA Sequencing with Smart-seq2. Methods Mol. Biol. 1979, 25–44 (2019).
Venkatesh, H. S. et al. Electrical and synaptic integration of glioma into neural circuits. Nature 573, 539–545 (2019).
Venkataramani, V. et al. Glutamatergic synaptic input to glioma cells drives brain tumour progression. Nature 573, 532–538 (2019).
Venkatesh, H. S. et al. Neuronal Activity Promotes Glioma Growth through Neuroligin-3 Secretion. Cell 161, 803–816 (2015).
Hausmann, D. et al. Autonomous rhythmic activity in glioma networks drives brain tumour growth. Nature 613, 179–186 (2023).
Osswald, M. et al. Brain tumour cells interconnect to a functional and resistant network. Nature 528, 93–98 (2015).
Watson, D. C. et al. GAP43-dependent mitochondria transfer from astrocytes enhances glioblastoma tumorigenicity. Nat. Cancer 4, 648–664 (2023).
Gangoso, E. et al. Glioblastomas acquire myeloid-affiliated transcriptional programs via epigenetic immunoediting to elicit immune evasion. Cell 184, 2454–2470.e2426 (2021).
Driggers, P. H. et al. An interferon gamma-regulated protein that binds the interferon-inducible enhancer element of major histocompatibility complex class I genes. Proc. Natl Acad. Sci. USA 87, 3743–3747 (1990).
Yeo, A. T. et al. Single-cell RNA sequencing reveals evolution of immune landscape during glioblastoma progression. Nat. Immunol. 23, 971–984 (2022).
Müller, S. et al. Single-cell profiling of human gliomas reveals macrophage ontogeny as a basis for regional differences in macrophage activation in the tumor microenvironment. Genome Biol. 18, 234 (2017).
Van Hove, H. et al. A single-cell atlas of mouse brain macrophages reveals unique transcriptional identities shaped by ontogeny and tissue environment. Nat. Neurosci. 22, 1021–1035 (2019).
Hammond, T. R. et al. Single-Cell RNA Sequencing of Microglia throughout the Mouse Lifespan and in the Injured Brain Reveals Complex Cell-State Changes. Immunity 50, 253–271.e256 (2019).
Pyonteck, S. M. et al. CSF-1R inhibition alters macrophage polarization and blocks glioma progression. Nat. Med. 19, 1264–1272 (2013).
Kaminska, B. & Cyranowski, S. Recent Advances in Understanding Mechanisms of TGF Beta Signaling and Its Role in Glioma Pathogenesis. Adv. Exp. Med Biol. 1202, 179–201 (2020).
Dumas, A. A. et al. Microglia promote glioblastoma via mTOR-mediated immunosuppression of the tumour microenvironment. EMBO J. 39, e103790 (2020).
Xu, B. et al. CCR9 and CCL25: A review of their roles in tumor promotion. J. Cell Physiol. 235, 9121–9132 (2020).
Nandi, S. et al. Receptor-type protein-tyrosine phosphatase ζ is a functional receptor for interleukin-34. J. Biol. Chem. 288, 21972–21986 (2013).
Lee, Y., Clinton, J., Yao, C. & Chang, S. H. Interleukin-17D Promotes Pathogenicity During Infection by Suppressing CD8 T Cell Activity. Front Immunol. 10, 1172 (2019).
Snajdauf, M. et al. The TRAIL in the Treatment of Human Cancer: An Update on Clinical Trials. Front Mol. Biosci. 8, 628332 (2021).
Ullah, M. A., and Mackay, F. (2023). The BAFF-APRIL System in Cancer. Cancers (Basel) 15, https://doi.org/10.3390/cancers15061791.
Gadani, S. P., Walsh, J. T., Smirnov, I., Zheng, J. & Kipnis, J. The glia-derived alarmin IL-33 orchestrates the immune response and promotes recovery following CNS injury. Neuron 85, 703–709 (2015).
Sung, H. Y. et al. Down-regulation of interleukin-33 expression in oligodendrocyte precursor cells impairs oligodendrocyte lineage progression. J. Neurochem 150, 691–708 (2019).
Han, J., Alvarez-Breckenridge, C. A., Wang, Q. E. & Yu, J. TGF-β signaling and its targeting for glioma treatment. Am. J. Cancer Res 5, 945–955 (2015).
Guo, Q. et al. Cancer cell intrinsic TIM-3 induces glioblastoma progression. iScience 25, 105329 (2022).
Strauss, L. et al. Targeted deletion of PD-1 in myeloid cells induces antitumor immunity. Sci. Immunol. 5, https://doi.org/10.1126/sciimmunol.aay1863 (2020).
Álvarez-Vázquez, A. et al. EGFR amplification and EGFRvIII predict and participate in TAT-Cx43266-283 antitumor response in preclinical glioblastoma models. Neuro Oncol. 26, 1230–1246 (2024).
Lee, J. H. et al. Human glioblastoma arises from subventricular zone cells with low-level driver mutations. Nature 560, 243–247 (2018).
Dirks, P. B. Brain tumor stem cells: the cancer stem cell hypothesis writ large. Mol. Oncol. 4, 420–430 (2010).
Venkataramani, V. et al. Glioblastoma hijacks neuronal mechanisms for brain invasion. Cell. https://doi.org/10.1016/j.cell.2022.06.054 (2022).
Joseph, J. V. et al. TGF-β promotes microtube formation in glioblastoma through thrombospondin 1. Neuro Oncol. 24, 541–553 (2022).
Jiang, T. et al. Comprehensive evaluation of NT5E/CD73 expression and its prognostic significance in distinct types of cancers. BMC Cancer 18, 267 (2018).
Wang, M., Jia, J., Cui, Y., Peng, Y. & Jiang, Y. CD73-positive extracellular vesicles promote glioblastoma immunosuppression by inhibiting T-cell clonal expansion. Cell Death Dis. 12, 1065 (2021).
Goswami, S. et al. Immune profiling of human tumors identifies CD73 as a combinatorial target in glioblastoma. Nat. Med 26, 39–46 (2020).
Khan, S. M. et al. Impact of CD4 T cells on intratumoral CD8 T-cell exhaustion and responsiveness to PD-1 blockade therapy in mouse brain tumors. J. Immunother Cancer. 10, https://doi.org/10.1136/jitc-2022-005293 (2022).
Stupp, R. et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 10, 459–466 (2009).
Landry, A. P., Balas, M., Alli, S., Spears, J. & Zador, Z. Distinct regional ontogeny and activation of tumor associated macrophages in human glioblastoma. Sci. Rep. 10, 19542 (2020).
Genoud, V. et al. Responsiveness to anti-PD-1 and anti-CTLA-4 immune checkpoint blockade in SB28 and GL261 mouse glioma models. Oncoimmunology 7, e1501137 (2018).
Ausejo-Mauleon, I. et al. TIM-3 blockade in diffuse intrinsic pontine glioma models promotes tumor regression and antitumor immune memory. Cancer Cell. https://doi.org/10.1016/j.ccell.2023.09.001 (2023).
Polański, K. et al. BBKNN: fast batch alignment of single cell transcriptomes. Bioinformatics 36, 964–965 (2020).
Acknowledgements
This work was funded by the grants FEDER PID2021-128549OB-I00 (Arantxa Tabernero) funded by MCIN/AEI/ 10.13039/501100011033 and “ERDF A way of making Europe,” PDC2022-133652-I00 (Arantxa Tabernero) funded by MCIN/AEI/ 10.13039/501100011033 and “European Union NextGenerationEU/PRTR”, SA125P20 and SA212P24 (Arantxa Tabernero), CLU-2023-1-01(Arantxa Tabernero) funded by FEDER and Junta de Castilla y León, and grants from Chan Zuckerberg Biohub SF (Norma Neff). L. García-Vicente was supported by the Spanish Ministerio de Universidades. M. Martínez-Fernández, A. Álvarez-Vázquez and R. Flores-Hernández were fellowship recipients from the Junta de Castilla y León and the European Social Fund. Y. Ding was a “Programa Investigo” fellowship recipient and R. González-Sánchez was a technician from the Junta de Castilla y León, Servicio Público de Empleo Estatal and the European Social Fund, NextGenerationEU. We are grateful to Spyros Darmanis and Ashley Byrne for initial support and guidance to the project. We thank T. del Rey for technical assistance.
Author information
Authors and Affiliations
Contributions
L.G.V. contributed to the experimental design and development, data acquisition, analysis and interpretation of the data. L.G.V. was aware of the group allocation at the different stages of the experiment. L.G.V., A.A.V., and R.F.H. performed cell culture and in vivo procedures. L.G.V., M.B., and V.T. performed snRNA-Seq experimental procedures. A.G., E.M., Y-J.K., and A.P. provided computational pipelines and helped with data analysis. A.D., H.M., and S.P. performed sequencing. N.N. helped to design snRNA-Seq experiments and supervised them. M.M.-F., R.F.-H., Y.D and R.G.S. performed the validation of the snRNA-seq results. A.T. conceived the study, supervised the experimental development, and interpreted the data. L.G.V. and A.T. drafted the article. All the co-authors revised the article for important intellectual content and approved the final version for publication.
Corresponding author
Ethics declarations
Competing interests
A.T. is an inventor on the Tat-Cx43266-283 patent (ID: ES2526109B1) from the University of Salamanca and all other authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks the anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editors: Kaliya Georgieva. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
García-Vicente, L., Martínez-Fernández, M., Borja, M. et al. Single-nucleus RNA sequencing reveals a preclinical model for the most common subtype of glioblastoma. Commun Biol 8, 671 (2025). https://doi.org/10.1038/s42003-025-08092-x
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s42003-025-08092-x
This article is cited by
-
Dissecting neuroblastoma heterogeneity through single-cell multi-omics: insights into development, immunity, and therapeutic resistance
Oncogene (2026)
-
Single-Cell Transcriptomic Profiling of GL261 Glioblastoma Cells Reveals Gene Expression Signatures Underlying Tumorigenicity
Cellular and Molecular Neurobiology (2025)