Abstract
The facultative anaerobe and major human pathogen Staphylococcus aureus is able to sustain growth under a wide range of oxygen concentrations. Importantly, we have already demonstrated that under microaerobic conditions, sensed by the two-component system SrrAB, S. aureus efficiently induces the development of competence for natural transformation, one of the three main horizontal gene transfer mechanisms present in bacteria. Here, we show that when the oxygen concentration decreases even further (reaching almost anaerobic conditions) the development of competence for natural transformation is still allowed but with much less efficiency than under microaerobic conditions. This inhibition is controlled by a central competence regulator, named ComK2, that was not found involved under intermediate oxygen concentrations. This ComK2-dependent inhibitory pathway also involves the SA2107 protein, of unknown function, through a direct protein-protein interaction. Finally, we demonstrate that this inhibition of competence is controlled by this strong oxygen limitation, sensed by another two-component system named NreBC, probably involved in the same pathway as ComK2 and SA2107. All in all, our results show that the oxygen concentration, which varies drastically depending on the site in the human body but also during bacterial infections, is a key environmental factor that tightly modulates S. aureus genomic plasticity.
Similar content being viewed by others
Introduction
Staphylococcus aureus is an asymptomatic and permanent colonizer of the human population but is also considered as one of the foremost opportunistic bacterial pathogens of humans. S. aureus colonizes up to 30% of humans in the nose and frequently in other sites such as the skin, throat, axillae, groin and intestine1. Colonization is usually harmless but can occasionally lead to a wide range of infections including mild skin infections but also deadly invasive complications, such as osteomyelitis, septic arthritis, septicemia, pneumonia and endocarditis2. The diversity of colonization and infection sites, as well as the interplay with the host defenses and microbiota is only possible because of an important number of genes and pathways allowing the impressive adaptability of S. aureus3.
An important and fluctuating parameter that bacteria have to adapt to is the oxygen (O2) concentration within the host4. Indeed, the amount of dissolved O2 within the host tissues and cells depends on several factors including: barometric pressure, climatological conditions (temperature, relative humidity, latitude, altitude), as well as physiological, pathological, and physical-chemical processes within the organism itself5. In the presence of O2, aerobic bacterial species can conduct aerobic respiration during which O2 acts as the final electron acceptor for the electron transport chain. In the absence of O2, anaerobic bacteria use alternative metabolic pathways including anaerobic respiration and/or fermentation6.
As a facultative anaerobic organism, S. aureus is able to sustain growth under a wide range of O2 concentrations7,8. This ability is particularly important to promote an infection, on the onset of inflammation or in biofilm communities where microaerobic or anaerobic conditions are often encountered9. The switch from aerobic to anaerobic metabolisms in S. aureus is complex and highly regulated. The ability of S. aureus to adapt to extreme changes in external O2 concentration implies the presence of several O2-sensing systems (i.e. the SrrAB, NreBC and AirSR two-component systems, TCS10,11,12) that regulate the expression of genes involved during the transition from aerobic to anaerobic growth.
Importantly, we have recently shown that S. aureus is able to sense a decrease of the O2 concentration (leading to microaerobic conditions) and in response, optimally induces the development of a specific physiological state named competence and essential for natural transformation, one of the three main horizontal gene transfer mechanisms present in bacteria13. We hypothesized that similarly to other important model organisms, S. aureus induces competence for natural transformation in order to promote its genomic plasticity in stressing environments13.
Three main central competence regulators have been identified and characterized in S. aureus14,15: a secondary sigma factor, SigH (comparable to ComX in Streptococcus pneumoniae16), and two genes, comK1 and comK2, encoding putative homologs of the transcription factor ComK from Bacillus subtilis17. Among these three central competence regulators, we have demonstrated that only SigH and ComK1 are essential for the expression of the natural transformation genes under microaerobic conditions in CS2 medium13. We have then shown that the SrrAB TCS is important to activate SigH in response to low O2 concentrations13. Moreover, while ComK2 was not found as an activator of the natural transformation genes expression under microaerobic conditions, it was still considered as a central competence regulator13. Indeed, its regulon encompasses genes involved in competence-associated secondary functions such as the general stress response, toxin-antitoxin systems or amino and nucleic acids metabolisms13.
Moreover, the development of competence has not only been shown under microaerobic conditions but also in static and O2-deprived cultures (i.e. anaerobic conditions)18. Indeed, Morikawa and colleagues have developed a synthetic medium, called GS, in which S. aureus is also able to develop competence under strong O2 limitation18. Even though no correlation between the intensity of competence development and the O2 availability has been documented yet, this capacity to induce horizontal gene transfer under a wide variety of conditions clearly demonstrates S. aureus adaptability.
Interestingly, it has been proposed that the use of multiple regulators might have evolved to allow S. aureus to use a wider range of cues to decide whether or not to become competent for natural transformation13,15. Indeed, in this study we confirmed that S. aureus is able to induce competence under stronger O2 limitations even though much less efficiently than under microaerobic conditions. We particularly showed that while SigH and ComK1 are still activators of the natural transformation genes expression, the third central competence regulator, ComK2, represses the expression of some of these genes explaining the lower probability of inducing competence under strong O2 limitation. We also identified a protein of unknown function, named SA2107, interacting with ComK2 and potentializing this repressor function. Finally, we found that the natural transformation genes repression is controlled by the NreBC TCS in response to O2 rarefaction, acting in the same pathway as ComK2 and SA2107. Ultimately, we propose a model in which S. aureus modulates the development of competence, depending on the O2 concentration, the stress imposed to the bacteria and the energy necessary to adapt.
Results
Competence development is allowed, but restricted, under strong oxygen limitation
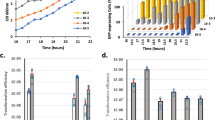
To monitor the development of competence under strong O2 limitation in GS medium, we optimized the protocol published by Morikawa and his colleagues in 201218, taking profit of our previous experience with CS2 medium13 (see material and methods for details). In order to compare our results, we used the same reporter strain as in13, expressing the gfp gene under the control of the comG late competence operon’s promoter (PcomG-gfp). Briefly, this reporter strain was first streaked on a BHI agarose plate. Isolated colonies are then used to inoculate a pre-culture in BHI medium. This pre-culture was then quickly stopped in exponential growth, centrifuged, washed and used to inoculate a fresh culture in GS medium incubated statically at 37 °C. Finally, cell density (OD600nm), the percentage of GFP-expressing cells as well as the O2 concentration were measured every 30 min.
Figure 1a shows how the O2 concentration first dropped from 21 to 0.03% during the first 15 h of growth and even lower in the second half of the experiment (compare to the 0.3% measured under microaerobic conditions, Supp. Fig. 1 and 13). As a consequence, we observed two distinct phases in the growth curve. Indeed, the growth rate of the culture was lower once the O2 concentration decreased below 15%, probably reflecting the metabolic adaptation of the cells to the less energetically efficient anaerobic metabolism. In addition, when the O2 concentration decreased below 8%, the expression of the comG operon was authorized, to finally reach around 5% of the total population after 26 h of growth in GS medium. This result is 10 times lower than what we obtained under microaerobic conditions13, suggesting that even though competence is allowed, an inhibitory mechanism must be present to limit its development under stronger O2 rarefaction.
a A wild-type strain (St29) expressing GFP under the control of PcomG was grown in a GS medium for 28 h. The oxygen concentration, the percentage of GFP-expressing cells, and OD600 nm were measured every 30 min. Results are presented as mean ± SD. Each experiment has been repeated three times (biological replicates). b Transformation efficiencies of wild type (N315ex w/o Phi), sigH (St45), comK1 (St37), comK2 (St38), sa2107 (St39), and nreC (St118) mutant strains using chromosomal DNA (gray bars) (see Material and methods for details). Results are presented as mean ± SD. For each strain, the experiment was repeated at least eight times (biological replicates). Individual experiments are shown as blue circles. Statistical significance was evaluated by two-way ANOVA with Tukey’s posthoc test as follows: ***P < 0.001 and *P < 0.05.
Accordingly, the transformation efficiency of a wild type strain grown in GS medium was evaluated around 2.1 × 10−7 (±3.7 × 10−7), a result 20 times lower than under microaerobic conditions (i.e. 4.2 × 10−6, 13) (Fig. 1b). Despite this concordance of results, it is important to mention that our natural transformation protocol presents an experimental limit. Indeed, to provide the exogenous DNA allowing the selection of transformants, we have to open the tubes and therefore disrupt the O2 limitation. Therefore, we cannot exclude the fact that some of the transformants appeared after the exogenous DNA was added under aerobic conditions. If that is the case, it means that we underestimate the observed repression of competence development occurring under anaerobic conditions.
SigH and ComK1 also activate the expression of the transformation genes under strong oxygen limitation
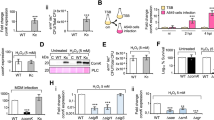
Next, we decided to test the effect of the deletion of the genes encoding the three central competence regulators, namely sigH, comK1 and comK2, on the expression detected from the promoter of two genes or operons representative of the two natural transformation genes classes: PcomG (a class I promoter controlled by both SigH and ComK1 under microaerobic conditions13), and Pssb (a class II promoter exclusively controlled by ComK113). Interestingly, the specific activation of class I and II natural transformation genes is still present under strong O2 limitation. Indeed, when sigH was deleted, only the expression from PcomG was lost (Fig. 2a, b) while the expression from both PcomG and Pssb was abolished when comK1 was inactivated (Fig. 2a, b). Consistently with these results, the absence of both sigH or comK1 abolished genetic transformation under strong O2 limitation (Fig. 1b).
The percentage of the population expressing GFP under control of PcomG (St29, St51, St40, and St 41) (a) or Pssb (St50, St61, St64, and St67) (b) in a wild-type background or in the absence of sigH, comK1, or comK2 was determined after 26 h of growth in GS medium. Results are presented as mean ± SD. Each experiment has been repeated at least four times (biological replicates). Individual experiments are shown as blue circles. Statistical significance was evaluated by two-way ANOVA with Tukey’s posthoc test as follows: ***P < 0.001 and *P < 0.05.
ComK2 inhibits the expression of the comG operon under strong oxygen limitation
Importantly, while no effect on the expression of the natural transformation genes could be associated to the deletion of comK2 under microaerobic conditions13, we found that the absence of ComK2 led to a 4-fold increase of the expression from the PcomG promoter under stronger O2 limitation (Fig. 2a). Accordingly, we showed that deletion of comK2 led to an increase of the natural transformation efficiency reaching 5.4 × 10−6 (±7.8 × 10−6) (Fig. 1b), consistent with the difference observed in the percentage of cells expressing the comG operon between comK2 and wild type strains (Fig. 2a). This result suggests that ComK2 might be a repressor of the comG operon expression, limiting the development of competence and hence natural transformation under strong O2 limitation. Furthermore, this ComK2 repression could not be confirmed with the Pssb promoter (Fig. 2b), potentially indicating a specific mode of action to regulate the comG operon expression.
Yeast two-hybrid reveals a potential interaction between ComK2 and SA2107
Because ComK2 is present in both microaerobic and stronger O2 limitation but the ComK2-associated inhibition of the comG operon is only detected in the latter, we first hypothesized that ComK2 needed to be potentialized by interacting with a partner. Interestingly, we identified an interaction between ComK2 and a protein of unknown function, SA2107, obtained in a yeast two-hybrid experiment performed as described previously (19, Fig. 3a). Briefly, a fusion between ComK2 and the GAL4 binding domain (BD) was used as bait to screen a S. aureus genomic library constructed in a GAL4 activation domain (AD) prey vector (see Material and methods). A fragment corresponding to the full length SA2107 protein was identified in this screen (Fig. 3a).
a Yeast two-hybrid experiment revealing a physical interaction between ComK2 and SA2107. A fusion between ComK2 and the GAL4 binding domain (BD) was used as a bait to screen a S. aureus genomic library constructed in a GAL4 activation domain (AD) prey vector. The full length SA2107 protein was identified in this screen. The interaction is revealed on a complete, synthetic and selective medium lacking leucine, uracile and adenine (to select for expression of the ADE2 interaction reporter). Negative controls harboring empty vectors (empty pBGDU, strain 121 and empty pGAD, strain 131). b Predicted structure of the ComK2-SA2107 complex using AlphaFold2. ComK2 is displayed in green and SA2107 in cyan. The amino acids at the interaction interface are highlighted in the “wire format” in yellow for SA2107 and in red for ComK2 (visualization of residues as sticks, see Supp. Fig. 2 for details). c Measured Hydrodynamic radius of SA2107 in complex with ComK2 using the Flow Induced Dispersion Analysis (FIDA26) method. A constant concentration of labelled SA2107 (20 nM) is exposed to increasing concentrations of ComK2 (15 nM-2 μM). SA2107 hydrodynamic radius is calculated based on three independent experiments (mean and SD). For each data point, individual hydrodynamic radius values are presented as blue circles. d Comparison of the calculated and estimated hydrodynamic radius of SA2107 alone and in the presence of ComK2 or ComK1. The dissociation constant (Kd) of the ComK2-SA2107 interaction is shown.
Structural modelling of the ComK2/SA2107 interaction
In addition, we used AlphaFold2 to further study the interaction between ComK2 and SA2107. First, the models proposed by AlphaFold2 for the individual proteins are robust with a pLDDT value superior to 70 for SA2107 and even 90 for ComK2 along the entire length of the proteins (Supp. Fig. 2). ComK2 is shown as a protein composed of 3 α-helices and 9 β-sheets while SA2107 is a short protein only displaying 4 α-helices (Supplementary Fig. 2). Then, Fig. 3b shows the best model generated by AlphaFold2 of the potential interaction between ComK2 and SA2107. Importantly, the predicted model for the complex formed by ComK2 and SA2107 seems very strong as the ipTM score (interface pTM) is superior to 0.8 (on a range from 0 to 1) (Supplementary Table 1). It is usually accepted that AlphaFold2 models of interactions displaying an ipTM superior to 0.7 can be considered as a strong starting point. Using the PyMol software, we next looked for the amino acids potentially involved in the ComK2-SA2107 interaction (Supplementary Table 2). We identified 8 hydrogen bonds as well as 1 salt bridge involving amino acids present in ComK2 and SA2107 that are less than 4 Å apart.
To further challenge our confidence in this model, as well as the specificity of this interaction, we then evaluated the ability of SA2107 to interact with ComK1. Indeed, in addition of displaying around 27% of sequence similarity, ComK1 and ComK2 predicted structures are very close (Supplementary Fig. 3). The two proteins are almost perfectly superposable apart from N-terminal extra amino acids and an additional α-helix present in the C-ter of ComK1 (Supplementary Fig. 3). However, despite this strong homology, AlphaFold2 revealed that the interaction between ComK1 and SA2107 was very unlikely as the ipTM and combined pTM score for these models were below 0.3 (Supplementary Table 3). This difference could be explained by the N- and C-terminal extremities of ComK1 that could prevent this interaction and ultimately underline the specificity of the ComK2-SA2107 interaction (Supplementary Fig. 3).
The interaction between ComK2 and SA2107 is binary and direct
In order to confirm that the interaction between ComK2 and SA2107 is binary and direct, we then purified the two recombinant His-tagged proteins (Supplementary Fig. 4). We then used Flow-Induced Dispersion Analysis (FIDA20) to investigate the ability of these proteins to interact. By measuring the fluorescence of a ligand in a laminar flow and analyzing its dispersion over time, FIDA allows the calculation of the ligand’s apparent hydrodynamic radius (Rh). Furthermore, when mixed with a potential interactant, this technique measures the change in size of the ligand (Rhi) as it selectively interacts with the target protein under native conditions. Interestingly, the dissociation constant (Kd) of the interaction may also be obtained in a titration experiment.
Here, we decided to fluorescently label SA2107, used as the indicator. When analyzed alone, FIDA measured SA2107 Rh around 1.97 ±0.02 nm (Fig. 3c, d). This result is very close from what we evaluated when using SA2107 AlphaFold structural models (i.e. Rha = 2,05 ±0.02 nm, Fig. 3d). We then mixed labelled SA2107 and unlabeled ComK2 in order to measure the change in size of the potential protein-protein complex. FIDA evaluated the SA2107-ComK2 Rhi around 2,75 ±0.09 nm, which is consistent with the AlphaFold structural model of the complex (Rha = 2,81 ±0.09 nm, Fig. 3c, d). Furthermore, a titration experiment, in which SA2107 concentration was fixed at 20 nM and mixed with ComK2 at concentrations ranging from 15 nM to 2 µM, we evaluated the Kd of the interaction at 420 ± 130 nM (Fig. 3c, d).
Finally, we decided to demonstrate that the SA2107-ComK2 interaction was specific and that as predicted by AlphaFold ComK1 could not interact with SA2107. This is why we reproduced the experiments presented above using the purified His-tagged ComK1 protein as a potential interactant (Supplementary Fig. 4). As expected, SA2107 Rh did not significantly change in the presence of ComK1, even at the higher concentrations (Supplementary Fig. 5), proving that despite the structural similarities observed between ComK1 and ComK2, SA2107 specifically interacts with the latter.
ComK2 and SA2107 inhibit comG operon expression
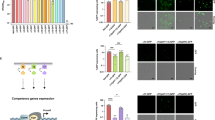
Finally, we wanted to verify if SA2107 could be involved, alongside ComK2, in the inhibition of the expression of natural transformation genes. This is why we investigated the impact of the sa2107 gene deletion on the expression from the PcomG promoter. Interestingly, the absence of SA2107 led to a 4-fold increase in the expression from the PcomG promoter (Fig. 4). Accordingly, the transformation efficiency of a sa2107 mutant strain was comparable to that of a comk2 mutant strain and evaluated around 3.3 × 10−6 ( ± 3.6 × 10−6) (Fig. 1b).
The percentage of the population expressing GFP under control of PcomG (St29, St41, St42 and St 161) in a wild type background or in the absence of comK2, sa2107, or both, was determined after 26 hours of growth in GS medium. Results are presented as mean ± SD. Each experiment has been repeated at least 4 times (biological replicates). Individual experiments are shown as blue circles. Statistical significance was evaluated by two-way ANOVA with Tukey’s posthoc test as follows: *P < 0.05.
Since ComK2 and SA2107 interact and that the deletion of both genes lead to a similar increase in the comG operon expression, we then asked if these two proteins could work together to inhibit the development of competence. Accordingly, the combination of the two genes deletion led to similar activation of the expression from the PcomG promoter as in the single mutant strains (Fig. 4). This last result strongly suggests that the two proteins act in the same regulatory pathway, probably through a direct protein-protein interaction, leading to the inhibition of the comG operon expression.
Strong oxygen limitation is sensed by the two-component system NreBC to inhibit comG operon expression
We previously showed that microaerobic conditions were sensed by SrrAB, one of the three TCS sensing O2 in S. aureus, to stimulate competence development13. Therefore, we wondered if O2-sensing TCS were involved in the regulation of competence development under stronger oxygen limitation. This is why we tested the impact of the deletion of the gene encoding each O2-sensing TCS’ response regulator on the percentage of comG-expressing cells. Remarkably, a srrA mutant did not affect the development of competence under strong O2 limitation (Fig. 5a). Similar results were obtained in a airR mutant strain (Fig. 5a). Strikingly, the percentage of comG-expressing cells was increased by a 4-fold factor in a nreC (encoding the TCS response regulator) mutant strain (Fig. 5a), a phenotype very close from what was obtained in comK2 and sa2107 mutant strains (Fig. 4b). This result was confirmed by the transformation efficiency of a nreC mutant strain (Fig. 1b), which we found higher than the wild type strain, reaching 2.5 × 10-6 ( ± 1.1 × 10-6), and similar to that of comK2 and sa2107 mutant strains.
a The percentage of the population expressing GFP under control of PcomG (St29, St145, St158 and St177) in a wild type background or in the absence of srrA, nreC, or airR was determined after 26 hours of growth in GS medium. The results are presented as mean ± SD. For each strain, the percentage of the population expressing GFP has been calculated from at least 4 independent experiments (biological replicates). Individual experiments are shown as blue circles. b The percentage of the population expressing GFP under control of PcomG (St29, St41, St158 and St273) in a wild type background or in the absence of comK2, nreC, or both, was determined after 26 hours of growth in GS medium. The results are presented as mean ± SD. For each strain, the percentage of the population expressing GFP has been calculated from at least 5 independent experiments (biological replicates). Individual experiments are shown as blue circles. Statistical significance was evaluated by two-way ANOVA with Tukey’s posthoc test as follows: *P < 0.05.
Next, we verified if the NreBC TCS could sense strong O2 rarefaction and in response activate the ComK2/SA2107 inhibitory pathway. To do so, we evaluated the percentage of comG-expressing cells in a mutant strain lacking both nreC and comK2. Interestingly, this double mutant strain displayed the same phenotype (i.e. a 4-fold increase in the percentage of comG-expressing cells) as the single mutant strains (Fig. 5b). This result probably reflects the fact that NreBC, ComK2 and SA2107 act in the same pathway to inhibit the development of competence under strong O2 limitation.
Discussion
Three central competence regulators control the expression of the NT genes in S. aureus
Three putative central competence regulators have been identified in the human pathogen S. aureus: the alternative sigma factor SigH and the two transcriptional regulators ComK1 and ComK215,18. We previously showed that only SigH and ComK1 were essential for the expression of the genes involved during natural transformation under microaerobic conditions (Fig. 6 and ref. 13). Meanwhile, ComK2 was found important for the general competence transcriptional program by controlling the expression of genes involved in secondary functions, such as the general response to stress, toxin-antitoxin systems or the nucleic and amino acid metabolism13. Interestingly, we show here that ComK2 also displays the ability to inhibit the expression of the natural transformation genes under stronger O2 limitation (Fig. 6).
a The development of competence for natural transformation is usually presented as a sequential process. First, a signal (stress) is sensed by the cells, which in turn activate signal transduction pathways. These pathways ultimately activate the central competence regulators responsible for the activation of the late competence genes, among which are found all the genes encoding for the proteins involved in natural transformation. b Under microaerobic conditions, we have shown that oxygen rarefaction is sensed by the SrrAB two-component system which in turn activate the alternative sigma factor SigH. Together with ComK1, SigH is required to activate the expression of the natural transformation genes13. c Under strong oxygen limitation, SigH and ComK1 still activate the expression of the natural transformation genes but much less efficiently than under microaerobic conditions. This inhibition is performed by the third competence regulator, ComK2, probably through a direct protein-protein interaction with SA2107, a small protein of so far unknown structure and function. Furthermore, we showed that the two-component system NreBC senses the strong O2 limitation and is involved in the ComK2/SA2107 inhibitory pathway, potentially through the activation of SA2107.
Ultimately, we demonstrate the existence of three true central competence regulators, modulating natural transformation, making the development of competence in S. aureus particularly complex and intricated. Each regulator can be induced or modulated by independent pathways allowing a fine tuning of the competence transcriptional program, of the percentage of cells inducing competence and as a consequence of the global transformation efficiency.
To our knowledge, S. aureus is the only model organism that has three central competence regulators. Indeed, the two historic model organisms only display one central competence regulator named ComX in S. pneumoniae16 and ComK in B. subtilis17. However, it has been shown that Vibrio cholerae, the causative agent of cholera epidemics, uses two central competence regulators (namely TfoX and HapR)21. It has been hypothesized that this multiplicity of central competence regulators could be used to finely tune the development of competence in response to changes in the environmental conditions. It seems that in S. aureus, the presence of three central competence regulators is dedicated, at least in part, to the modulation of the development of competence in response to the O2 concentration.
Protein-protein interaction modulates activity of ComK2
We have shown that ComK2 activates the expression of competence genes involved in secondary functions in CS2 under microaerobic conditions13. However, we could not detect any inhibition of the natural transformation genes by ComK2 in these conditions13. Since ComK2 is present under both microaerobic and stronger O2 limitation, but its inhibitory function is only present in the latter, we postulated that a co-repressor must be potentializing this new function. The identification of the interaction between ComK2 and SA2107 as well as the identical phenotypes of comK2 and sa2107 mutant strains led us to propose that SA2107 could be such co-repressor (Fig. 6). Following the same reasoning that SA2107 is the key to promote this new ComK2-associated inhibitory effect, we can propose that SA2107 expression or modification, could be controlled, directly or indirectly, by the NreBC TCS in response to strong O2 limitation (Fig. 6).
Interestingly, sa2107 is only found in S. aureus genomes and does not display any annotated domain. However, when searching the AlphaFold database (AFDB) proteome with FoldSeek, we found the BssR protein (also called YliH) from E. coli with a very similar tridimensional predicted structure. BssR’s name comes from the phrase “regulator of biofilm through signal secretion”22. Indeed, BssR is a small regulator involved in the inhibition of biofilm formation and the specific fold shared by BssR and SA2107 could represent a new family of small proteins, involved in the inhibition of bacterial environmental adaptations.
Finally, in order to explain how ComK2 and SA2107 could inhibit the expression of some natural transformation genes we can propose two hypotheses. First, the interaction between ComK2 and SA2107 could allow the complex to bind the promoter of natural transformation genes and prevent the access of SigH, ComK1 or the RNA-polymerase. However, preliminary results show that the ComK2-SA2107 complex cannot directly bind to the comG operon promoter (Supplementary Fig. 6). Alternatively, we can imagine that the complex formed by the two proteins is able to directly interact with SigH or ComK1, preventing the activation of the natural transformation genes expression through sequestration. Unfortunately, a strong unspecific binding of ComK1 to any DNA sequences prevented us to test the ability of the ComK2-SA2107 complex to inhibit ComK1 binding (Supplementary Fig. 6).
Oxygen concentration tightly modulates competence development in S. aureus
Facultative anaerobes, such as S. aureus, constitute a unique class of bacteria able to grow in the presence or absence of O2. On an evolutionary perspective, they can be considered as some of the most evolved and adapted bacteria for their ability to grow and disseminate within a wide range of microenvironments, especially during infection23. Therefore, it is not surprising to find in the list of priority and multi-resistant pathogens established by the WHO24 an overrepresentation of facultative anaerobes. Upon closer inspection of the WHO’s list, it has been established that a majority of species are known or suspected to be naturally transformable25. Therefore, it is striking to reveal that S. aureus, a facultative anaerobe, major human pathogen present in the WHO’s list is able to modulate the development of competence for natural transformation, and its need of genomic plasticity and antibiotic resistances, depending on the O2 concentration. Such correlation has already been proposed in S. pneumoniae26, without a clear regulatory pathway proposed.
Investigations have shown that S. aureus finds itself limited for O2 in vivo during the onset of infection or within biofilms. During bacterial infection, deprivation of O2 in a tissue may result from inflammation that prevents blood from reaching the infection site which cannot keep pace with O2 consumption from growing bacteria and recruited host immune cells27. Wound sites are example of tissues experiencing hypoxia and characterized by inflammation and bacterial infections28. Some diseases also create anoxic environments. This is the case of the sputum clogging the lungs of cystic fibrosis patients in which oxygen is depleted within the first few millimeters below the sputum-air interface29. As S. aureus is one of the main pathogens found in the lungs of young cystic fibrosis patients30, this important human pathogen is clearly exposed, over time, to strong O2 limitations. Finally, S. aureus is also known to grow microaerobically to anaerobically within biofilms which can lead to recurrent infections or septicemia31. Whether these biofilms are formed on medical devices or within the lungs of cystic fibrosis patients, they are all characterized by important gradients of O2 limitations stressing the bacterial cells that need to adapt their metabolism. All these types of infections are so inherently linked to S. aureus pathogenicity that this human pathogen has to constantly adapt its energetic metabolism but also its need of genomic plasticity.
Ultimately, to explain such behavior, we can propose that even though S. aureus is able to grow under strong O2 rarefaction, such environment remains stressing. Indeed, it has been shown that after a shift from aerobic to anaerobic growth (which occurs in our cultures), S. aureus growth rate was drastically reduced8. Therefore, it is plausible that stressed cells adapting to anaerobic conditions do not invest as much into competence for natural transformation, a well-known energy-consuming process. In addition, the low growth rate of S. aureus transformants under strong O2 limitation could impact the potential gain of fitness associated to the new sequences acquired. Contrastingly, under microaerobic conditions, S. aureus cells could still use a combination of respiration, a less stressing and more energetically efficient metabolism, and fermentation, allowing the cells to invest more into competence for natural transformation13.
Materials and methods
Bacterial strains and culture conditions
N31518 S. aureus strains used in this project are all listed in Supplementary Table 4. Staphylococcus aureus strains were grown in BHI medium (Becton, Dickinson and Company) or a complete synthetic medium, called GS (18, K2HPO4, 7 g/L; KH2PO4, 2 g/L; Na3citrate ∙ 2H2O, 0.4 g/L; (NH4)2SO4, 1 g/L; MgSO4, 0.05 g/L; Thiamine, 1 mg/L; Niacin, 1.2 mg/L; Biotin, 0.005 mg/L; D Pantothenate, 0.25 mg/L; Adenine, 5 mg/L, Guanine, 5 mg/L; Cytosine, 5 mg/L; Uracil, 5 mg/L; Thymine, 20 mg/L; L-alanine, 60 mg/L; L-arginine, 50 mg/L; L-aspartic acid, 90 mg/L; L-cystine, 20 mg/L; L-glutamic acid, 100 mg/L; L-histidine, 20 mg/L; L-isoleucine, 30 mg/L; L-leucine, 90 mg/L; L-lysine, 50 mg/L; L-methionine, 3 mg/L; L-phenylalanine, 40 mg/L; L-proline, 80 mg/L; L-threonine, 30 mg/L; L-tryptophan, 10 mg/L; L-tyrosine, 50 mg/L; L-valine, 80 mg/L) depending on the experiment. When necessary, antibiotics were used to select specific events (Kan, 200 µg/mL; Cm, 10 µg/mL).
Protocol to naturally induce competence in S. aureus in GS medium
Cells were isolated from −80 °C stock on BHI plate. Four clones were inoculated in 10 mL of BHI (Becton, Dickinson and Company) and incubated at 37 °C with shaking at 180 rpm until OD reached 2.5. This pre-culture was then centrifuged, washed in fresh GS medium18, and used to inoculate 2 mL of fresh GS medium (OD = 0.05) in closed 2 mL Eppendorf tubes. Several tubes of this culture were prepared in order to take individual samples along growth (i.e. each Eppendorf tube was only opened once for one sample). The cultures in GS medium were finally incubated overnight statically at 37 °C. Cells were collected throughout growth (GFP reporter strains, Fig. 1), after 16 h (transformation efficiencies, Fig. 2c) or 26 h (GFP reporter strains, Figs. 2, 4 and 5).
Construction of S. aureus deletion mutants
In order to investigate the role of sa2107 in the regulation of competence development in S. aureus, allelic replacement constructs were cloned into the temperature sensitive pIMAY plasmid32 and used as presented in ref. 13. All the primers (IDT) used for cloning in the present study are listed in Supplementary Table 5.
Flow cytometry to determine the percentage of the population expressing GFP
Following growth in GS, 4 mL of cells were harvested by centrifugation at 11,000 g for 4 min. Pellets were resuspended in 500 µL of cold 70% ethanol and incubated on ice for 30 min, in order to fix the cells. Then, S. aureus cells were resuspended in 500 µL of PBS (pH 7,4) after centrifugation at 11,000 g for 1 min. Finally, the percentage of the population expressing GFP was evaluated by Flow cytometry (Cytoflex top-bench cytometer, Beckman-Coulter). Following Forward- and Side-scatter detection to identify individual cells (20000 events), a 488 nm laser was used to distinguish GFP-expressing competent cells by comparison with the auto-fluorescence of a strain that did not express GFP (St12) (as already shown in ref. 13).
It is important to mention that GFP is a very stable protein. Therefore, once the maximum percentage of competent cells was reached, this number stayed constant for hours. This feature does not mean that competence stays ‘open’ for hours but rather that once the maximum is reached, no new competent cells appear.
Natural transformation of competent S. aureus cells
Wild type strain (St12) as well as sigH (St45), comK1 (St37), comK2 (St38), sa2107 (St39) and nreC (St118) mutant strains were first grown to competence in GS medium. Cells were naturally transformed following the protocol previously published13.
Transformation protocol
Briefly, at each time point (every half hour), 8 mL of cells were harvested by centrifugation at 10,000 g for 4 min at 4 °C, resuspended in 500 µL of fresh CS2 (Note that at this stage the medium used to resuspend the cells contains normal oxygen concentrations). Five µg of donor- chromosomal DNA were added to one of the tubes (the second tube is used as a “no DNA” control) and incubated at 37 °C for 2.5 hours with agitation at 180 rpm. 50 or 500 µL (tube with DNA) or 1 ml (“no DNA control”) from each tube where finally mixed with 25 mL of melted BHI (Becton, Dickinson and Company) agar pre-cooled to 55 °C together with antibiotic, and the mixture was poured into petri dishes. After solidification, the plates were incubated at 37 °C for 48 hours. At each time point, the viability was also evaluated by serial dilution on BHI (Becton, Dickinson and Company) agar plates. Transformation efficiencies were finally calculated by dividing the number of transformants detected in 1 mL of culture by the total number of cells in the same volume.
Numbers presented in Fig. 1C represent the mean of the highest transformation efficiencies detected along growth during each experiment. The experiments have been repeated for each strain at least 10 times to provide strong statistical relevance.
Donor DNA preparation
Chromosomal DNA: strain St294 was used to provide donor chromosomal DNA. In St294, the pIMAY-INT15 plasmid (Cm) was inserted in the chromosome at the INT chromosomal site15. The plasmid insertion was verified by PCR while no replicating plasmid could be detected. Briefly, 100 mL of culture were centrifuged and resuspended in 5 mL of TEG (Tris 5 mM, pH8; EDTA, 10 mM; Glucose, 1%) complemented with 500 µL of Proteinase K (10 mg/mL, Invitrogen), 2 mL of lysis buffer (NaOH, 0.2 N; SDS, 1%) and 20 g of glass beads (Stratech, #11079-105, 0.5 mm in diameter). The cells were then broken using 5 cycles of vortex (1 min each) with 1 min in ice between each cycle. To finish cell lysis, 3 mL of lysis buffer were added for 5 min at room temperature and neutralized with 6 mL of NaAc (3 M, pH 4.8). Finally, chromosomal DNA present in the supernatant was precipitated using 96% ethanol (1 mL of EtOH for 500 µL of supernatant) after 2 hours of incubation at -20 °C. After centrifugation, chromosomal DNA was washed using 300 µL of cold 70% ethanol. Precipitated chromosomal DNA was finally resuspended in 300 µL of Tris-HCl 5 mM, pH8.
Oxygen concentration measurements
Oxygen concentrations were measured using the SP-PSt3-SA23-D3-OIW oxygen sensor spots (PreSens GmbH, Regensburg, Germany). These self-adhesive sensor spots were attached to the inner wall of 2 mL Eppendorf tubes so that the spots would always be immerged during the experiments. The Eppendorf tubes were closed at T0 and remained closed for the entire experiment.
The sensor spots are covered with an oxygen-sensitive coating where molecular oxygen quenches the luminescence of an inert metal porphyrine complex immobilized in an oxygen-permeable matrix. This process guarantees a high temporal resolution and a measurement without drift or oxygen consumption.
The photoluminescence lifetime of the luminophore within the sensor spot was measured using a polymer optical fiber linked to an oxygen Meter (Fibox 4 trace; PreSens GmbH). Excitation light (505 nm) was supplied by a glass fiber, which also transported the emitted fluorescence signal (600 nm) back to the oxygen meter. Briefly, an oxygen measurement was realized, through the 2 mL Eppendorf tube plastic, by simply approaching the optical fiber from the sensor spot. At each time point, the oxygen concentration was measured three times and the results provided represent the mean of these three measurements. In our experiments, oxygen concentration was measured every 30 minutes.
Yeast two-hybrid in yeast
The comK2 gene cloned into a GAL4 BD (bait) vector was expressed in Saccharomyces cerevisiae to screen a S. aureus library essentially as described previously33. Briefly, the S. aureus library was constructed in a GAL4 AD (prey) vector in E. coli and transferred into yeast. Interactions were revealed by growth of diploid cells after 5 to 14 days at 30 °C on synthetic complete medium34 lacking leucine, uracil, and histidine (to select for expression of the HIS3 interaction reporter) and further tested on synthetic complete medium lacking leucine, uracil and adenine (to select for expression of the ADE2 interaction reporter). Controls with empty vector plasmids (i.e., carrying only the BD or AD domain) were systematically included.
Protein 3D structure prediction using AlphaFold2
AlphaFold2 is an artificial intelligence system developed by DeepMind capable of predicting the 3-dimensional structure of a protein only with its amino-acid sequence35,36,37. A user interface developed by the I2BC (host institute, https://bioi2.i2bc.paris-saclay.fr/tools/structural-biology/), facilitating the access to AlphaFold, has been used to predict the 3D structure of ComK1, ComK2 and SA2107, as well as their potential interaction. Additionally, the algorithm calculates different statistics quantifying the confidence in the predicted models. The pTM-score (represented by a number between 0 and 1) measures the accuracy of the entire structure, and it is accepted that a pTM-score above 0.5 means that the global predicted fold for the protein might be similar to the true structure. The ipTM-score (interface pTM-score) is a similar statistic, but only applied to the regions found at the interface between proteins in a complex. The “combined score” is a weighted sum of the pTM and ipTM and gives thus a better global estimation of the model’s accuracy. In the same way as previously, a value close to 1 corresponds to a high confidence predicted model. Finally, the plDDT (predicted local distance difference test) corresponds to a measure of confidence in the interpretation of the structure, by amino acids. The PyMol software (The PyMOL Molecular Graphics System, Version 3.0 Schrödinger, LLC.) was used to generate images of different protein structures and to highlight the residues involved in the interactions. The PISA tool38 (Proteins, Interfaces, Structures and Assemblies, EMBL-EBI) was then used to assign the amino-acids capable of forming hydrogen bonds, salt bridges, or hydrophobic interactions at the interface of proteins.
To compare the apparent hydrodynamic radius of SA2107 alone (Rh) or in a complex (Rhi) obtained through the FIDA method with estimated measurements, we used the AlphaFold2 predicted structures. The approximative diameter of 5 different models were obtained by tracing two lines covering the structures’ maximum length using the “Measurement” tool of PyMol. Mean of these diameters were calculated, and divided by two to obtain the estimated hydrodynamic radius of SA2107, SA2107+ComK1 and SA2107+ComK2 (referred to as Rha).
ComK1, ComK2 and SA2107 purification
After a PCR amplification of the comK1, comK2 and sa2107 genes from the St012 strain, using the NdeI-6His-comK1-F and XhoI-comK1-R, NdeI-comK2-F and Xho-comK2-His-R, and NdeI-His-SA2107-F and Xho-His-SA2107-R oligonucleotides respectively, followed by a cloning in pET21a (comK2) or pET29a (comK1 and sa2107) vectors, a His6 tagged version of ComK1 and ComK2 were expressed in Escherichia coli BL21-Gold (DE3) or Rosetta (DE3) pLysS (Novagen) for SA2107. Protein expression was induced in 800 mL of cultures by using 2xYT medium supplemented with IPTG (100 µg/mL). After an overnight incubation at 15 °C, cells were collected and the pellet resuspended in 80 mL of specific buffer (ComK1: 20 mM Phosphate buffer, 1 M NaCl, pH5.6; ComK2: 20 mM Phosphate buffer, 500 mM NaCl, pH5.6; SA2107: 50 mM Tris-HCl, 200 mM NaCl, pH7.5). After a night at -20 °C, to induce cell lysis, the cultures are sonicated (Branson Sonifier 250, 4 cycles of 50 sec, pulse at 90%, intensity at 40 arbitrary units). The cytoplasmic fraction was purified by centrifugation (18,000 × g, 30 min, 8 °C for ComK2 and SA2107, 20 °C for ComK1). The supernatant was then loaded into a Ni-NTA resin. Elutions obtained at 200 mM and/or 400 mM of Imidazole were then concentrated using ultrafiltration by centrifugation (Vivaspin protein concentrator spin columns, Cytiva). These samples were then further purified using an ÄKTA pure™ size-exclusion chromatography system (Superdex 200 column, Cytiva). Chosen fractions containing the purified ComK1 and ComK2 were then finally concentrated using ultrafitration (365 µM for ComK1, 536 µM for ComK2). To concentrate SA2107, chosen fractions were gathered, precipitated with 25% of Ammonium Sulfate, incubated overnight at 8 °C on a rotating wheel and centrifugated (18,000 x g, 1 h, 8 °C). The supernatant was then eliminated, and the pellet was resuspended in 1 mL of Tris Buffer (50 mM Tris-HCl, 200 mM NaCl, pH7.5), and dialyzed (in Phosphate Buffered Saline + Tween 0.1% solution to remove the Ammonium Sulfate) to obtain SA2107 at 50 µM. Finally, the protein samples were aliquoted in Eppendorf tubes, frozen in liquid nitrogen and stored at -80 °C.
Flow-induced dispersion analysis (FIDA20)
Labelled SA2107 (referred to as SA2107488) was prepared by conjugation with ATTO 488 NHS Ester (Sigma-Aldrich). 47 µL of SA2107 (50 µM) recovered in PBS (Bio-Rad) +Tween 0.1% (Sigma) was incubated with 2.5 µL of Sodium Bicarbonate pH9 0.2 M, and 0.5 µL of ATTO 488 NHS Ester 10 mM (2-fold molar ratio of dye-to-protein) for 2 hours at room temperature, protected from light. The labelled protein was then purified using PD SpinTrap G-25 column (Cytiva), pre-equilibrated with PBS (Bio-Rad) +Tween 0.1% (Sigma) buffer to remove the free dye excess. The concentration of SA2107488 was measured by UV-vis at 480 nm and 506 nm (absorbance of the fluorophore) using an absorption coefficient of 18 910 M-1 cm−1.
Binding experiments were performed with a Fida 1 instrument (Fida Biosystems ApS), using laser-induced fluorescence detection with an excitation wavelength of 480 nm. A permanently coated capillary (outer diameter 375 µm, inner diameter 75 µm, length to detector 84 cm, Fida Biosystems ApS) was used in the apparatus with a controlled temperature of 25 °C.
Binding curves of SA2107488 with ComK1 or ComK2 were obtained by using a PreMix method where the indicator sample (SA2107488) is preincubated with the analyte (Non-labelled ComK1 or ComK2). To form the complexes and let the reaction reach the equilibrium state, the indicator sample is prepared with a fixed concentration of 20 nM of SA2107488 preincubated for 10 minutes with ComK1 or ComK2 with a titration of 0-2 µM, in PBS (Bio-Rad) +Tween 0.1% (Sigma) buffer, in a final volume of 100 µL. The analyte sample is prepared by making a titration of 0-2 µM of ComK1 or ComK2 in PBS (Bio-Rad) +Tween 0.1% (Sigma), in a final volume of 100 µL. The capillary was first equilibrated with just the analyte sample at 3500 mbar for 30 seconds. The indicator sample is then injected at 50 mbar for 10 seconds. Finally, the indicator is mobilized and fluorescence is measured by injecting the analyte at 400 mbar for 180 seconds. Each binding experiments (SA2107488+ComK2 or SA2107488+ComK1) were performed thrice.
Taylorgrams obtained with this method were analyzed using Fida Software (version 3.0) with a multi-species fit to fix the ATTO 488 NHS Ester hydrodynamic radius (Rh) at 0.60 nm. This way, the apparatus is able to rigorously determine the apparent hydrodynamic radius of SA2107488 alone (Rh), or in a complex (Rhi).
Electromobility shift assay
To test the ability of our protein of interest (ComK1, ComK2, SA2107) to bind to the comG promoter, we constructed a 100 bp double-stranded probe corresponding to the comG operon upstream region labelled with a Cyanine5 (Cy5) fluorophore contained in one of the two primers. The promoter of bsaA (a gene not involved in NT) was used as a negative control. The different probes were then diluted at 1 nM in a buffer containing 20 mM of HEPES (pH 7.5, Sigma), 150 mM of NaCl and 0.01% of IGEPAL CA-630 (Sigma-Aldrich). In different Eppendorf tubes, the probes were then incubated for 15 minutes at room temperature with an increasing concentration of protein (0 to 2 µM). The samples were finally loaded on a 6% polyacrylamide gel (0.25% TBE, Bio-Rad, 5% glycerol), and migrated for 1h10min (100 V, 30 mA). The gels displaying Cy5-stained DNA were revealed in an Amersham Typhoon Biomolecular imager (Cytiva).
Statistics and reproducibility
Dynamic expression of the comG operon promoter (Fig. 1a) has been repeated three times to provide mean values and standard deviations at each time point. Transformation efficiencies (Fig. 1b) were calculated based on at least 8 independent experiments (biological replicates) to calculate mean values and standard deviation for each strain. Natural transformation genes expression (Figs. 2, 4 and 5) was evaluated for each strain (mean and standard deviation) based on at least 4 independent experiments (biological replicates). Finally, FIDA experiments were conducted three times independently for each protein couple to provide mean values and standard deviations.
Results were compared (multiple comparisons to a control condition) using a two-way ANOVA with Tukey’s correction (*p < 0.05, ***p < 0.001).
Data availability
All data supporting the findings of this study are available within the paper and its Supplementary Information. Numerical source data for graphs and charts are presented in the Supplementary Data 1 file. Other details are available from the corresponding author on reasonable request.
References
Lowy, F. D. Staphylococcus aureus Infections. N. Engl. J. Med. 339, 520–532 (1998).
Gordon, R. J. & Lowy, F. D. Pathogenesis of methicillin-resistant staphylococcus aureus infection. Clin. Infect. Dis. 46, S350–S359 (2008).
Howden, B. P. et al. Staphylococcus aureus host interactions and adaptation. Nat. Rev. Microbiol. 21, 380–395 (2023).
Dietz, I., Jerchel, S., Szaszák, M., Shima, K. & Rupp, J. When oxygen runs short: the microenvironment drives host–pathogen interactions. Microbes Infect. 14, 311–316 (2012).
Ortiz-Prado, E., Dunn, J. F., Vasconez, J., Castillo, D. & Viscor, G. Partial pressure of oxygen in the human body: a general review. Am. J. Blood Res. 9, 1–14 (2019).
Unden, G., Becker, S., Bongaerts, J., Schirawski, J. & Six, S. Oxygen regulated gene expression in facultatively anaerobic bacteria. Antonie Van. Leeuwenhoek 66, 3–22 (1994).
Masalha, M., Borovok, I., Schreiber, R., Aharonowitz, Y. & Cohen, G. Analysis of transcription of the Staphylococcus aureus Aerobic Class Ib and anaerobic class III ribonucleotide reductase genes in response to oxygen. J. Bacteriol. 183, 7260–7272 (2001).
Fuchs, S., Pané-Farré, J., Kohler, C., Hecker, M. & Engelmann, S. Anaerobic gene expression in Staphylococcus aureus. J. Bacteriol. 189, 4275–4289 (2007).
Archer, N. K. et al. Staphylococcus aureus biofilms. Virulence 2, 445–459 (2011).
Kinkel, T. L., Roux, C. M., Dunman, P. M. & Fang, F. C. The Staphylococcus aureus SrrAB two-component system promotes resistance to nitrosative stress and hypoxia. mBio 4, e00696-13 (2013).
Schlag, S. et al. Characterization of the oxygen-responsive NreABC regulon of Staphylococcus aureus. J. Bacteriol. 190, 7847–7858 (2008).
Sun, F. et al. AirSR, a [2Fe-2S] cluster-containing two-component system, mediates global oxygen sensing and redox signaling in Staphylococcus aureus. J. Am. Chem. Soc. 134, 305–314 (2012).
Feng, S. Y., Hauck, Y., Morgene, F., Mohammedi, R. & Mirouze, N. The complex regulation of competence in Staphylococcus aureus under microaerobic conditions. Commun. Biol. 6, 512 (2023).
Morikawa, K. et al. A new staphylococcal sigma factor in the conserved gene cassette: functional significance and implication for the evolutionary processes. Genes Cells 8, 699–712 (2003).
Fagerlund, A., Granum, P. E. & Havarstein, L. S. Staphylococcus aureus competence genes: mapping of the SigH, ComK1 and ComK2 regulons by transcriptome sequencing. Mol. Microbiol 94, 557–579 (2014).
Lee, M. S. & Morrison, D. A. Identification of a new regulator in Streptococcus pneumoniae linking quorum sensing to competence for genetic transformation. J. Bacteriol. 181, 5004–5016 (1999).
van Sinderen, D. et al. comK encodes the competence transcription factor, the key regulatory protein for competence development in Bacillus subtilis. Mol. Microbiol 15, 455–462 (1995).
Morikawa, K. et al. Expression of a cryptic secondary sigma factor gene unveils natural competence for DNA transformation in Staphylococcus aureus. PLoS Pathog. 8, e1003003 (2012).
Mortier-Barriere, I. et al. A key presynaptic role in transformation for a widespread bacterial protein: DprA conveys incoming ssDNA to RecA. Cell 130, 824–836 (2007).
Pedersen, M. E., Østergaard, J. & Jensen, H. Flow-Induced Dispersion Analysis (FIDA) for Protein Quantification and Characterization. Methods Mol. Biol. Clifton NJ 1972, 109–123 (2019).
Meibom, K. L., Blokesch, M., Dolganov, N. A., Wu, C. Y. & Schoolnik, G. K. Chitin Induces Natural Competence in Vibrio cholerae. Science 310, 1824–1827 (2005).
Domka, J., Lee, J. & Wood, T. K. YliH (BssR) and YceP (BssS) regulate Escherichia coli K-12 biofilm formation by influencing cell signaling. Appl Environ. Microbiol 72, 2449–2459 (2006).
André, A. C., Debande, L. & Marteyn, B. S. The selective advantage of facultative anaerobes relies on their unique ability to cope with changing oxygen levels during infection. Cell. Microbiol. 23, e13338 (2021).
World Health Organization. (WHO). Global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics. http://www.who.int/mediacentre/news/releases/2017/ (2017).
Blokesch, M. In and out—contribution of natural transformation to the shuffling of large genomic regions. Curr. Opin. Microbiol. 38, 22–29 (2017).
Echenique, J. R., Chapuy-Regaud, S. & Trombe, M. Competence regulation by oxygen in Streptococcus pneumoniae: involvement of ciaRH and comCDE. Mol. Microbiol. 36, 688–696 (2000).
Murdoch, C., Muthana, M. & Lewis, C. E. Hypoxia regulates macrophage functions in inflammation. J. Immunol. 175, 6257–6263 (2005).
Sawyer, R. G., Spengler, M. D. & Pruett, T. L. The peritoneal environment during infection. The effect of monomicrobial and polymicrobial bacteria on pO2 and pH. Ann. Surg. 213, 253–260 (1991).
Chu, V. H. et al. Staphylococcus aureus bacteremia in patients with prosthetic devices: Costs and outcomes. Am. J. Med. 118, 1416–1416 (2005).
Cowley E. S. et al. Pediatric cystic fibrosis sputum can be chemically dynamic, anoxic, and extremely reduced due to hydrogen sulfide formation. mBio. 6, e00767 (2015).
Mahomed T. G., Kock M. M., Masekela R., Hoosien E. & Ehlers M. M. Genetic relatedness of Staphylococcus aureus isolates obtained from cystic fibrosis patients at a tertiary academic hospital in Pretoria, South Africa. Sci. Rep. 8, 12222 (2018).
Monk, I. R., Shah, I. M., Xu, M., Tan, M.-W. & Foster, T. J. Transforming the untransformable: application of direct transformation to manipulate genetically staphylococcus aureus and staphylococcus epidermidis. mBio. 3, e00277-11 (2012).
Noirot-Gros, M.-F. et al. An expanded view of bacterial DNA replication. Proc. Natl Acad. Sci. 99, 8342–8347 (2002).
Guthrie, C. and Fink, G. R. Guide to Yeast Genetics: Functional Genomics, Proteomics, and Other Systems Analysis. (1991).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Richard. E. et al. Protein complex prediction with AlphaFold-Multimer. bioRxiv (2022).
Mirdita, M. et al. ColabFold: making protein folding accessible to all. Nat. Methods 19, 679–682 (2022).
Krissinel, E. & Henrick, K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 (2007).
Acknowledgements
The present work has benefited from the facilities and expertise of the I2BC platform PIM supported by French Infrastructure for Integrated Structural Biology (FRISBI) ANR-10-INBS-05. We also want to thank the Imagerie-Gif flow cytometry and proteomics (SICaPS) facilities (Institute for the Integrative Biology of the Cell, I2BC, Gif sur Yvette, FRANCE) for their help and support. We thank the BIOI2 platform for making the ColabFold pipeline easily accessible at the I2BC. This work was supported by a “Young Researcher grant” from the French National Research Agency (ANR-18-CE35-0004, GenTranSa) and by the MICROBES interdisciplinary object (Paris-Saclay university) both awarded to Nicolas Mirouze. This work was also supported by PhD scholarships attributed to S. Y. Feng (Chinese Scholarship Council), Y. Arab (Paris-Saclay University scholarship) and Pierre Poirette (Fondation pour la Recherche Médicale).
Author information
Authors and Affiliations
Contributions
S.Y.F.: Conceptualization, Methodology, Investigation; Y.A.: Conceptualization, Methodology, Investigation; Y.H.: Conceptualization, Methodology, Investigation; P.P.: Conceptualization, Methodology, Investigation; M.N.: Conceptualization, Methodology, Investigation and Supervision; S.Q.-C.: Conceptualization, Methodology, Investigation, Editing, Funding acquisition and Supervision; S.M.: Conceptualization, Methodology, Investigation, Editing, Funding acquisition and Supervision; J.A.: Conceptualization, Methodology, Investigation, Editing, Funding acquisition and Supervision; N.M.: Conceptualization, Methodology, Investigation, Writing - Review & Editing, Funding acquisition and Supervision.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks the anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editor: Tobias Goris. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Feng, S.Y., Arab, Y., Hauck, Y. et al. ComK2 represses competence development for natural transformation in Staphylococcus aureus grown under strong oxygen limitation. Commun Biol 8, 1416 (2025). https://doi.org/10.1038/s42003-025-08816-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s42003-025-08816-z