Abstract
Identifying causal relations or causal networks among molecules/genes, rather than just their correlations, is of great importance but challenging in biology and medical field, which is essential for unraveling molecular mechanisms of disease progression and developing effective therapies for disease treatment. However, there is still a lack of high-quality causal inference algorithms for any observed data in contrast to time-series data. In this study, we developed a causal concept for any observed data based on cross-validated predictability (CVP). The CVP can quantify the causal effects among observed variables in a system. The causality was extensively validated by combining a large variety of statistical simulation experiments and available benchmark data (simulated data and various real data). Combining the predicted causal network and the real benchmark network, the CVP algorithm demonstrates high accuracy and strong robustness in comparison with the mainstream algorithms.
Similar content being viewed by others
Introduction
Causal inference from observed data is a core problem in various research disciplines of natural science and engineering, such as biology, earth science, economics, medicine, neuroscience and machine learning. Molecular networks are the essential issue of biological systems in view of causal inference1,2,3,4 and building high-quality molecular networks based on the observed/measured data has always been a vital problem of computational biology5. Effective identification of causal relations in a complex biological system can better reveal regulatory mechanisms and explain biological functions, such as gene regulations, signaling processes, metabolic pathways and disease progression. In particular, deriving a specific disease causal/regulatory network can reveal the basic mechanism of molecular effects to provide quantitative studies for further precise treatment.
Causal effects between molecules can often be represented by causal diagrams, with nodes representing different molecules and directed edges characterizing the direct causality between molecules6. Existing mainstream algorithms for monitoring causality include the well-known Granger causality (GC)7,8, the convergent cross-mapping algorithm (CCM)9, transfer entropy (TE)10, Bayesian theory11 and some biological systems inference methods12,13,14. Granger causality (GC) inference as a representative method, that is based on time-series data to infer the potential causality, was proposed in 19697, and since then GC-based methods have been widely used in many fields. Specifically, GC is based on the information of time lags or time-series data, and explains the current state to be affected by past information. TE as a nonlinear version of GC method considers the asymmetry of information in time-series to determine causality and is commonly used in biological systems such as neuroscience15 and physiology16. Both GC and TE are measured at the original state space. On the other hand, CCM9 as a complement to GC theory is measured in the delay embedding space, which is also based on time-series data and reflects causal relationship between two variables. All of the above algorithms measure causality with a requirement of time-dependent data or time-series data, but most of the biological data are not based on time-series data, such as phenotypes, stages or phases, thus unsuitable for applying such methods. In contrast, Bayesian network and structural causal model (SCM)17,18 are able to handle time independent data based on statistical independence and intervention (e.g. do-calculus operation), and can identify the directed causal relations between molecules. However, these methods depend on the structure of directed acyclic graph (without loop structure) to infer causality, which limits to the application to biomolecular networks commonly with feedback loops or ring-like interactions19. Therefore, a high-quality inference method of causality in real biological/molecular systems without time or structural limitation, rather than relying on time-dependent data, remains an open question.
In this study, we proposed a causal concept and the method termed cross-validation predictability (CVP), which is a data-driven model-free algorithm for any an observed data. The CVP method quantifies causal effect by cross validation and statistical test on any observed data. The CVP method is statistically tested with a large number of causal simulation experiments as well as fully performing validation on simulated data from existing benchmarks. Moreover, the method is also shown the superior performance and significant advance in a variety of real systems, including gene regulatory networks and other causal networks with feedback loops, by extensive comparisons with various existing methods. In particular, CRISPR-Cas9 knockdown experiments in the liver cancer have validated that the functional driver genes identified by the CVP algorithm effectively inhibit the growth and colony formation of liver cancer cells. By knockdown experiments, we demonstrated the accuracy and significance of the causality predicted by CVP and identified the regulatory targets of functional driver genes SNRNP200 and RALGAPB in the liver cancer. In summary, CVP is a general-purpose method for causal inference based on the cross-validation testing on predictability from any observed/measured data, and able to infer causality among the variables in accurate and robust manner.
Results
Causality detection based on cross-validation predictability
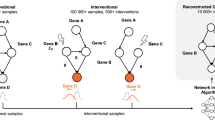
The causality from CVP method is a statistical concept based on cross-validation prediction of observed data. Assuming two variables \(X\) and \(Y\) are observed in m samples, we consider that \(X\) causes \(Y\) if the prediction of the values of \(Y\) is improved by including values of \(X\) in the sense of cross-validation (Fig. 1). Specifically, we assume that a variable set \(\left\{X,Y,{Z}_{1},{Z}_{2},\cdots ,{Z}_{n-2}\right\}\) includes n variables in the m samples, where \(\hat{Z}=\left\{{Z}_{1},{Z}_{2},\cdots ,{Z}_{n-2}\right\}\). In other words, \(X\) and \(Y\) are any two variables among all n observed variables. All m samples are randomly divided into a training group and a testing group, for the purpose of cross-validation, e.g., k-fold cross-validation. To test causal relation from X to Y, we formally define CVP causality framework, i.e. construct two contradictory models H0 (null hypothesis without causality) and H1 (alternative hypothesis with causality) by the same k-fold cross-validation and further define causal strength by the difference between H1 and H0 for quantifying CVP causality as follows.
The flowchart To infer/judge the causality from variable X to Y, all samples are divided into train set and test set based on cross-validation, and then modeling the causality equation for variable Y with and without X by train set. The test set was used to predict variable y and calculate the error ε. f means the linear model. By comparing the errors obtained under H1 and H0 assumptions, the causal strength of X to Y is finally determineds.
Train the regression \(\hat{f}\) by the training group samples, and then test \(\hat{f}\) by the testing group samples in a k-fold cross-validation manner. We have the error \({\hat{e}}_{i}\) of Eq. (1) in the i-th cross-validation test by the testing group samples. The total squared testing error is \(\hat{e}={\sum }_{i=1}^{m}{\hat{e}}_{i}^{2}\) for all k-fold cross-validation tests.
Train the regression \(f\) by the training group samples, and then test \(f\) by the testing group samples in a k-fold cross-validation manner. We have the error \({e}_{i}\) of Eq. (2) in the i-th cross-validation test by the testing group samples. The total squared testing error is \(e={\sum }_{i=1}^{m}{e}_{i}^{2}\) for all k-fold cross-validation tests. If \(e \, < \, \hat{e}\), then H1 holds, i.e. causal relation from \(X\) to \(Y\). And if \(\hat{e}\le e\), then H0 holds, i.e. no causal relation from X to Y.
Causal strength is the difference/distance measured between H0 and H1, i.e. \({{{\mathrm{ln}}}}\,\hat{{{\rm{e}}}}-\,{{\mathrm{ln}}}\,{{{\rm{e}}}}\). Of course, other statistical test (e.g., a paired Student’s t-test) can be also used to test the difference between \(e\) and \(\hat{e},\) i.e. significance test. In particularly, the e and ε are different in the manuscript for Eqs. (1) and (2). The ε is residual error from the training set, and the e is the error from testing set. So, the CVP use the e from testing set to infer the causal strength.
Here, \(f\) is the regression equation for \(Y\) fitting \(X\) and\(\hat{Z}\), and \(\hat{f}\) is the regression equation for \(Y\) fitting \(\hat{Z}\) without \(X\). In this work, we use the linear regression for both \(f\) and \(\hat{f}\). The \(\varepsilon \,\) and \(\hat{\varepsilon }\) are the error terms of Eqs. (2) and (1), respectively. The error is defined as the difference between the predicted value and the true value in testing group.
The Eqs. (1) and (2) are first fitted from the training group samples, and then are tested by the testing group samples for determining the testing/predicting errors, which are summarized as \(\hat{e}\) and \(e\) including the error of every sample in the testing group by Eqs. (1) and (2). We also ensure those errors are statistically independent of \(\hat{Z}\) and \(X\) by applying appropriate the regression algorithm, e.g., least-squares regression algorithm. Thus, causal criterion of CVP is essentially based on model predictability and statistical independence.
Clearly, if H1 or \(e\) is significantly less than \(\hat{e}\) in the testing group, it means that Eq. (2) is a better regression equation than Eq. (1), i.e. X causes Y (Fig. 1 and Supplementary Note 1). Otherwise, H0 holds, i.e. \(X\) does not cause \(Y\) (Fig. 1 and Supplementary Note 1). In this paper, we also introduce causal strength (Eq. (3)) to test the difference between \(e\) and \(\hat{e}\). Here, since the variable set \(\hat{Z}\) can be considered as other factors to affect variable \(Y\) except \(X\), hence this method considers or infers the direct causality from \(X\) to \(Y\) under all other factors fixed.
Benchmarking dataset
The superiority of the CVP algorithm is validated by different types of data, including the DREAM (Dialogue on Reverse Engineering Assessments and Methods, https://dreamchallenges.org) challenges (especially DREAM3 and DREAM4) and biosynthesis network (IRMA data) from Saccharomyces cerevisiae20, and further a variety of real data, such as the SOS DNA repair network in Escherichia coli21, yeast data (DDGni Yeast)22, the human HeLa data23, the TCGA cancer data (https://portal.gdc.cancer.gov/) and the E. coli data from GEO database24. The DREAM challenges and IRMA data demonstrate the adaptability of external disturbances to the CVP algorithm. For each dataset, the performance of the CVP algorithm is evaluated against eight of commonly used algorithms, i.e. Nonlinear ODEs25, GENIMS26, PLSNET27, GENIE3_ET_all, GENIE3_ET_sqrt, GENIE3_RF_all and GENIE3_RF_sqrt28, and TIGRESS29. In this study, we use the default parameters of the algorithm in order to make a fair comparison with those methods (Supplementary Note 2). Moreover, the DREAM challenges dataset is used for extensive benchmarking, and the algorithms that performed best in the DREAM challenges are included in the comparison results.
Causal effect estimation for simulated data
Two simulated causal networks are used to generate simulation data and test the effectiveness of the CVP algorithm. One network includes three nodes with different causal links (Figure S1 and Figure S2a-S2e), which produces corresponding simulation data (Supplementary Note 3 and Table S1). The CVP algorithm is used to quantify the causality from the simulated data (Figure S2f-S2j). The results show that the CVP algorithm can accurately measure the causal network structure among the three nodes from the simulated data with few false links or interactions (Supplementary Note 3 and Figure S2f-S2j). The CVP algorithm not only can identify the true causal relations but also eliminate the false causal links, i.e. indirect causal links caused by cascade or confounding factors (Fan-out) or collision factors (Fan-in) (Supplementary Note 3 and Figure S2).
Other nine causal networks are constructed from a basic regulatory network including 11 nodes with one center node and ten neighbor nodes and the center node is regulated by all the neighbor nodes, and then the neighbor nodes are removed from the initial regulatory network one by one to form nine structures or cases (Figure S3a). Each structure or case is used to produce the corresponding simulation data of 11 variables (Table S2). The CVP algorithm is also used to infer the causality between the center node and ten neighbors based on the simulation data of 11 variables (Supplementary Note 4 and Figure S3b-S3c). The results show that the CVP algorithm can accurately infer the network structure and causality among the 11 nodes from the simulated data with an accuracy rate to almost 100% for both existent and non-existent edges in the 9 cases (Supplementary Note 4 and Figure S3b-c).
Causal effect estimation for data in DREAM challenges
The DREAM challenges30 have been widely used as a benchmark dataset for causal inference. We obtain 4 datasets from 4 networks with 10 genes/nodes separately from DREAM3 and DREAM4, and the 4 networks are marked Network1~Network4 (Fig. 2a-d) and Supplementary Note 5).
a–d are the real networks from Network 1, Network 2, Network 3, and Network 4 from the DREAM challenges. e–h are the performance of the CVP and other eight methods for inferring Network 1, 2, 3 and 4 respectively. The horizontal coordinates are the AUPR values and the vertical coordinates are the AUROC values. The shape of the dots indicates the different algorithms, with dots closer to the top right corner indicating better algorithm performance. i, g, k, l show the network for IRMA data. i shows the ground truth network, and g and k, and l are the inferred networks of the CVP, TIGRESS, and PLSNET algorithms, respectively. The solid lines represent the accurate edges (true positive edges) to be inferred by the methods, and the dashed lines represent the wrong edges (false positive edges) to be inferred by the methods for g, k and l.
We compare the performance of the CVP algorithm with other eight algorithms, such as Nonlinear ODEs, GENIMS, PLSNET, GENIE3_ET_all, GENIE3_ET_sqrt, GENIE3_RF_all, GENIE3_RF_sqrt and TIGRESS based on two datasets of two networks in DREAM3 challenge (Fig. 2a, b), that one is a non-time series data (InSilico_Size10-Yeast3-heterozygous) for Network1 (Tab. S3 and Supplementary Note 5) and another is a time-series data (InSilico_Size10-Yeast2-trajectories) for Network2 (Table S3 and Supplementary Note 5). Clearly, the results demonstrate that the CVP algorithm performs better than other algorithms for the two networks (Fig. 2e, f). The closer the position locates on the top right of the coordinate system, the better performance of the algorithm is (Fig. 2e–h). For the Network1 (Fig. 2a), we can clearly find that CVP algorithm is much higher than other algorithms in AUROC (area under the receiver operating characteristic curve) and AUPR (area under the precision-recall curve)) values, with AUROC value reaching 0.77 and AUPR value 0.59 (Fig. 2e, Figure S4a and S4e). TIGRESS rank second among all the algorithms for Network1, with an AUROC of 0.59 and an AUPR of 0.31, which are much lower than the CVP algorithm (Fig. 2e, Figure S5a and S5e). For the Network2 (Fig. 2b), the CVP and PLSNET algorithms are close in performance, whose AUROC values are separately 0.7 and 0.67, and the AUPR values are separately 0.42 and 0.41, respectively (Fig. 2f, Figure S4b and S4f). The CVP algorithm slightly outperforms PLSNET algorithm and performs best (Fig. 2f).
Similar to the DREAM3 networks, DREAM4 also includes simulation data and corresponding real networks. We chose two datasets of two networks from DREAM4 challenge (Fig. 2e, d), and one is a non-time series data (insilico_size10_2_knockdowns) for Network3 (Table S3 and Supplementary Note 5) and another is a time-series data (insilico_size10_4_timeseries) for Network4 (Table S3 and Supplementary Note 5). The CVP algorithm outperforms the other algorithms for the two datasets by a significant margin, with AUROC values of 0.75 and 0.8 in Networks3 and Network4 (Figs. 2g, h and Figure S4c, S5d), respectively. In Network 3, all AUROC values of other algorithms are lower than 0.6, and AUPR values vary around 0.25 in similar performance. It may be because the complexity of the network leads to the unsatisfactory inference of the algorithm, while the AUPR value of CVP is 0.48, which still maintains the best performance (Fig. 2g). The GENIE3_ET_all, GENIE3_ET_sqrt, GENIE3_RF_sqrt, Nonlinear ODEs and TIGRESS algorithms performed well, with AUROC value of approximately 0.7 for Network4 (Fig. 2h and Figure S4d), but the CVP algorithm performs as high as 0.8 (Fig. 2h and Figure S4d), which still maintain the best performance.
For the DREAM challenges dataset, we totally test 40 datasets at different scales (Supplementary Note 5 and Figure S4-S7), and the CVP method performs better than other algorithms (Supplementary Note 5 and Figure S4-S7). For the network with size 10, the CVP algorithm always has far better AUROC and AUPR values than the other algorithms (Figs. 2e–h, Figure S4 and Figure S5). We also compare the accuracy of the algorithm for networks with size 50 and 100, and the CVP algorithm outperforms the other algorithms by a wide margin in 30 datasets, reflecting the accuracy and stability of CVP in synthetic datasets (Supplementary Note 5 and Figure S4-S7).
Moreover, we also compare the CVP method to other existing causal inference methods, e.g., PC31, FCI32, GES32, GeneNet33 and PORTIA34. From the causal results for different observed dataset (Figure S8-S10), the CVP not only infer the causality from observed data, but also perform better than other causal inference methods (Figure S8-S10), especially for real datasets (Figure S8 and Figure S10).
Causality detection in synthetic datasets
The IRMA network is a synthetic network20 embedded in Saccharomyces cerevisiae, cultured in vivo to obtain ground biological data, and is widely used as benchmark data in inferential modeling. This network contains five genes and eight regulatory edges, whose structure is known (Fig. 2i).
The IRMA network is relatively small, and the values of AUROC and AUPR are relatively sensitive to small changes of the threshold in the inferred results. Most existing methods, such as GENIE3, need delimit a threshold to judge whether there is a causal relationship, but the CVP judges the causal relationship by the validity of H0 and H1 hypothesis. So, the CVP algorithm is not subject to this limitation, and the AUROC and AUPR values are the highest compared with the other algorithms, which are 0.79 and 0.75 (Figure S11), respectively. For the eight edges in the ground truth network (Fig. 2i), the CVP algorithm accurately infers five edges, with only one false positive edge (Fig. 2g). Relatively good algorithms, such as TIGRESS infers four edges and two false positive edges (Fig. 2k and Supplementary Note 6), or PLSNET infers five edges and five false positive edges (Fig. 2l). However, these algorithms only provide the intensity of regulation without a specific threshold, which makes it difficult to determine the optimal subnet. The CVP algorithm not only yields the optimal directed network, but also quantifies the causal strength (Figure S11d). In the latest study, the best algorithm of this network inference is the BINGO algorithm6 with five inferred edges and one false positive, in agreement with our accuracy, but the method is restricted to the data based on time-series data. Therefore, the CVP algorithm has a high performance in in vivo synthesis networks.
Reconstruction of real gene regulatory networks
A real network is usually a complex nonlinear system, whose causal structure is generally difficult to be inferred. We focus on gene regulatory networks in biological systems, and apply the CVP algorithm to five real datasets of gene regulatory networks, i.e. the SOS DNA repair network data, Saccharomyces cerevisiae cell cycle data, human HeLa cell cycle data from literatures and BLCA dataset from TCGA. The performance of different algorithms is estimated using the AUROC or AUPR for each gene regulatory network.
The SOS DNA repair network
The SOS DNA repair network is the most commonly benchmark data set, which is the real non-time series data verified by experiments in E. coli21. It is a non-linear complex network consisting of 9 genes and 24 edges (Fig. 3i and Supplementary Note 7). By comparing the inference results based on the real data, the CVP algorithm outperformed the other algorithms for the SOS network (Fig. 3a, e, j). The AUROC value of the second highest ranking algorithm is 0.51, and the AUROC value of the CVP algorithm is 0.75, which is at least 24% higher than other algorithms (Fig. 3a and Figure S12a). The AUPR of the CVP is 0.63, and the second AUPR in the other algorithms is Nonlinear_ODEs and PLSNET with 0.35, that is 28% lower than the CVP (Fig. 3e and Figure S12b). At the same time, high false positives are always the main problem facing inferred GRNs (gene regulatory networks), while the CVP algorithm infers 16 edges in the SOS network with only 3 false positives (Fig. 3j), while other methods infers the optimal network with at least half of the false positive edges (Fig. 3k, l). It indicates that our algorithm introduces fewer redundant edges and is able to accurately infer the true network (the accuracy of CVP is 81%) (Fig. 3j, Supplementary Note 7 and Table S5).
a–h show the performance of the nine algorithms (Nonlinear ODEs, GENIMS, PLSNET, GENIE3_ET_all, GENIE3_ET_sqrt, GENIE3_RF_all, GENIE3_RF_sqrt, TIGRESS and CVP) on the SOS, DDGni, HeLa, and BLCA datasets respectively. a,b,c,d show the ROC curves for the SOS, DDGni, HeLa, and BLCA datasets, and e,f,g and h show the PR curves for the SOS, DDGni, HeLa, and BLCA datasets, where the different colored lines indicate different algorithms. i shows the real network for SOS dataset. j, k and l denote the inferred networks from CVP, PLSNET GENIE3-RF-sqrt algorithms for the SOS data, respectively. The solid lines represent the accurate edges (true positive edges) to be inferred by the methods, and the dashed lines represent the wrong edges (false positive edges) to be inferred by the methods for j, k and l. The PR (Precision Recall) curve is drawn with Precision and Recall. The ROC (Receiver Operating Characteristic) curve is drawn with False Positive Rate and True Positive Rate. SOS, DDGni, HeLa, and BLCA are the names of different datasets.
The yeast cell cycle data
This data is obtained by the authors of DDGni35 from the accession number GSE8799 on the Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/) database. The DDGni data is with 8 genes, 6 edges, and contains 30 time points of gene expression data from case of yeast cell cycle dataset. The time-series data and golden standard regulatory network is reported in literature35, and this data is also widely used as a benchmark for assessing the inference of GRNs25. We plot the ROC (Fig. 3b) and PR (Fig. 3f) curves for CVP and the other eight algorithms. Overall, each algorithm performs well for this network, but the CVP algorithm is still the highest in AUROC and AUPR values (Fig. 3b and f). The AUROC value of the CVP algorithm is 0.83 (Figure S12a), that for the other algorithms are basically between 0.7 and 0.8, and only PLSNET has a value of 0.53 (Fig. 3b and Figure S12a), probably due to a low AUROC due to high false positives. Similarly, the CVP algorithm has an AUPR of 0.47 (Figure S12b), while the other eight algorithms have an AUPR of no more than 0.4, with the highest being the GENIE3-RF-sqrt algorithm at 0.38(Fig.3b and Fig. S12b). Therefore, our algorithm is more robust and accurate to infer the real network of yeast cell cycle data.
The human HeLa cell cycle dataset
To compare the performance of the CVP algorithm with other algorithms, we use a biological experiment data, namely the HeLa cell cycle gene expression data23. Sambo et al.36 reports a subnetwork with corresponding time-series data, which is then often used as a benchmark37 for evaluating algorithms to construct GRNs. The expression data of the ground truth network with nine regulatory relationships among nine genes is used to evaluate the performance of the CVP and other algorithms. The ROC and PR curves of all algorithms are shown in Fig. 3c, g, and it is clearly that the curve of the CVP algorithm is always much higher than others (Fig. 3c, g). The AUROC value of the CVP algorithm is 0.86 (Fig. S12a), while other algorithms are close to 0.5, which is not higher than random enumeration (Fig. 3c and Fig. S12a). At the same time, the AUPR values of the all nine algorithms (CVP, Nonliner_ODEs, GENIMS, PLSNET, GENIE3_ET_all, GENIE3_ET_sqrt, GENIE3_RF_all, GENIE3_RF_sqrt and TIGRESS) are 0.56, 0.15, 0.10, 0.14, 0.13, 0.15, 0.14, 0.12, and 0.21 (Fig. 3g and Fig. S12b), respectively. And we can see that the CVP is also better than other eight algorithms in AUPR values. When inferring the network, the CVP algorithm identifies the largest number of true edges while ensuring the least number of false positives (Figure S13). Thus, the results show that the CVP algorithm outperforms other algorithms for different indexes and the infers results of the CVP algorithm are closer to the true network than other algorithms for the human HeLa cell cycle dataset (Fig. 3c, g, Figure S12 and S13).
The BLCA dataset in TCGA
The occurrence and development of cancer involve complex molecular regulation, monitoring regulation mechanism among genes can be beneficial to prevention and cure of different cancer efficiently. Here, we take BLCA (bladder urothelial carcinoma) cancer as an example to analyse the GRNs of oncogenes. The BLCA data is the RNA-Seq data from TCGA (the cancer genome atlas) database, but there is no ground truth network for this data. We extract the corresponding bladder cancer pathway (hsa05219) from KEGG (Kyoto Encyclopedia of Genes and Genomes) database (https://www.genome.jp/kegg/) and obtain a local network as the golden standard regulatory network for BLCA data. And the CVP algorithm infers a network with an accuracy of 87%, indicating that our algorithm constructed a network very close to the golden standard regulatory network. The CVP algorithm is also performed best among other algorithms (Fig. 3d, h). The highest AUROC value for the other algorithms is 0.66, and the CVP reached an AUROC value of 0.73 (Fig. 3d and Fig. S12a). The highest AUPR value for the other algorithms is 0.20, and the CVP reached an AUPR value of 0.24 (Fig. 3h and Fig. S12b). We also compare the results of other four cancer datasets, UCEC, LIHC, PAAD and STAD (Supplementary Note 8 and Figure S14), and the CVP algorithm still performs best in the AUROC index. Although the AUROC value is slightly lower relative to the other real data sets, the main reason is stemmed from the “false positives”. Due to the incomplete regulation relationship in KEGG pathways, the “false positives” in here are not necessarily real false positives by these algorithms. It is possible that some “false positives” edges are real regulations but not in the KEGG pathways, so they may be regulatory edges in tumors13.
A large-scale network
For validating the application scope of the CVP algorithm, we compare the performance of the CVP algorithm and other five algorithms on the data of a large-scale network, and the GSE2030524 dataset is used to provide a real time-series data for E. coli under cold stress conditions from GEO database. We obtain the standard network by integrating DREAM5 with the E. coli data and RegulonDB database (version 9.0)38 to form the final benchmark network containing 1484 genes and 3080 edges. We compare the accuracy of the CVP algorithm and other five algorithms (Nonlinear ODEs, GENIMS, PLSNET, GENIE3_RF_sqrt and TIGRESS) to infer the GRN for this network and the CVP algorithm still maintains the highest accuracy (Figure S15). For the large-scale network, the accuracy of the CVP algorithm is 0.89 (Figure S15), and it is better at least 20% than other five algorithms (Figure S15). It means that the inferred network from the CVP algorithm is closer to the true network than the networks from other algorithms (Figure S15) even if to a large-scale network.
All in all, the CVP algorithm recovers different causal networks from the real data (benchmark), and has consistently outperformed other advanced algorithms significantly.
Detecting causal networks in multiple contexts
The CVP algorithm is also considered to infer causal networks for other four systems, a planktonic food chain network, a causal network for the number of people treated for air pollution and respiratory diseases in Hong Kong, and two networks for the spread of new coronavirus infections in two regions of Japan.
The plankton food chain
The first example is from a time-series data on species abundance of plankton isolated from the Baltic Sea, where the food web is sampled and tested twice a week for over 2300 days39. The network is constructed by the five planktonic species cyclopoids, protozoa, rotifers, picocyanobacteria and nanophytoplankton with six regulations (Fig. 4a). The CVP algorithm inferred seven causal edges (Fig. 4b) that are basically consistent with the ground truth network, and these edges point in a way that is consistent with the biological laws of the food chain (Fig. 4a, b). It is worth noting that our algorithm infers two false positive edges, i.e., nanophytoplankton to cyclopoids and picocyanobacteria to cyclopoids, and the two edges do not exist in the ground truth network (Fig. 4a, b).
a A food chain network of plankton. b Causality of plankton detected by the CVP algorithm. c The detected the causal network for respiratory diseases by the CVP algorithm. The arrows represent the direction of causation, and the same colors represent the same dependent variables. d The inferred impact of environmental interactions on respiratory diseases by the CVP algorithm with darker heat map colors indicating stronger causal relationships. e and f show the reconstructed the network of the number of people spreading New Coronavirus by the CVP algorithm in Kanto and Tohoku of Japan, respectively.
The regulation edge from nanophytoplankton to cyclopoids is inferred by the CVP algorithm (Fig. 4b), and this regulation does not exist in the ground truth network (Fig. 4a). However, this predation or intake relationship for cyclopoids ingesting nanophytoplankton has been proved by a recent research work40, and it means that the causality from nanophytoplankton to cyclopoids is true by the CVP algorithm for the plankton food chain (Fig. 4b).
Another regulation edge from picocyanobacteria to cyclopoids is also inferred by the CVP algorithm (Fig. 4b), and this regulation is not marked in the ground truth network (Fig. 4a). A research work found that cyclopoids can take cyanobacteria as food41, and the picocyanobacteria is a kind of cyanobacteria with the smallest cell-size. So, the causality from picocyanobacteria to cyclopoids inferred by the CVP algorithm may be true for the food chain of the five planktonic species (Fig. 4b).
From the above results, we can see that the existent plankton food chain of the five planktonic species (Fig. 4a) is incomplete, and the CVP algorithm can be used to supplement the potential causality for the plankton food chain among the five planktonic species (Fig. 4b).
The causal network of respiratory inpatients and air pollution
The second example we considered is from Hong Kong air pollution data and the data of respiratory patients collected from major hospitals of Hong Kong from 1994 to 199742,43. The CVP algorithm is used to construct the causal network among NO2, temperature, humidity, O3, respirable suspended particulates, SO2 and respiratory system (Fig. 4c, d). The CVP algorithm identifies humidity, NO2 and respirable suspended particulates as the main causes of respiratory disease (Fig. 4c, d), which is consistent with prior research44,45,46. The CVP algorithm finds bidirectional causality for NO2 and respirable suspended particulates, and similar results are found in existing studies47. Moreover, this is also consistent with the causality inferred by the prior method, e.g. PCM algorithm48. The true causality between the pollutants SO2 and O3 is reported to be bidirectional49,50, which is the same as our results (Fig. 4c, d), but the PCM algorithm only monitors unidirectional causality from SO2 to O348. Specifically, the CVP algorithm reveals unidirectional causality for NO2 and O3, strongly related to the ability of NO2 in destroying O3, and similar discoveries are also reported in the currently available literature51,52. Here, we can find that the CVP algorithm is able to identify a causal network for the six pollutants on the respiratory system, including both unidirectional and bidirectional causation. Thus, the CVP algorithm screens for adverse factors for respiratory disease and then effectively assesses the relationship between air pollution and health.
New coronavirus transmission network in the region of Japan
COVID-19 is a highly infectious disease capable of causing mild or severe infection and even death in humans53. Therefore, it is crucial to forecast the transmission areas of new coronaviruses. Only by obtaining accurate transmission rules and then adjusting the prevention and control measures in time, the transmission area and scale can be effectively controlled. The CVP algorithm is a data-driven approach to build causal networks across regions of transmission based on the number of infected people in different regions, and indirectly infer the laws of infection.
Here, data of daily new COVID-19 cases are collected for 13 prefectures in the Kanto and Tohoku regions of Japan from January 15, 2020 to December 13, 2020 (Supplementary Note 11). We infers the spread network of the epidemic by the CVP algorithm in the Kanto (Fig. 4e) and Tohoku (Fig. 4f) regions of Japan54. As an international transportation hub, the Kanto region is also a key area for monitoring the situation of the novel coronavirus outbreak in Japan. Taking the economic hub of Tokyo as an example, it spread to Saitama and Ibaraki and cascaded to other regions (Fig. 4e). The Narita international airport, which locates in Chiba, is a major airport to handles international passengers for Tokyo. And many infected international passengers go to Tokyo from Narita international airport. Hence, it is reasonable that the transmission from Chiba to Tokyo in Kanto region (Fig. 4e). Thereafter, the number of infected people in various regions of Japan has an outbreak of the epidemic.
Compared to the Kanto region, the epidemic in the Tohoku region is relatively mild and the transmission network is relatively sparse, which is inextricably linked to the geographical location and mobility of the population in the Tohoku region. For example, Iwate, where the COVID-19 is last seen, has a low population density, few foreign visitors and a small transient population, and at the same time began early to study countermeasures to strictly prohibit the inflow of guests from outside the prefecture in order to prevent infection by the new coronavirus. This is in good agreement with our inference that Iwate shows a tendency towards more external transmission, mainly due to its strict control policy to avoid internal transmission as much as possible.
CVP reveals the gene regulations and functional genes during cancer progression
Presently, gastric and lung cancers are the major cancers that threaten human life55. Identifying genes and regulation in cancer progression will not only improve our understanding of the biology of the process, but also provide new targets for diagnosis and treatment. We get expression profiles for gastric cancer and lung cancer in TCGA database.
In this study, we use CVP to infer the regulatory network for early (Stage I and II) and late gastric cancer (Stage IV) and identify 15 out-degree hub genes involved in the regulation network of late gastric cancer as the functional genes in cancer progression (Table S17 and Supplementary Note 9). The 14 of 15 functional genes have been reported as association genes of gastric cancer and play a crucial role in progression of gastric cancer (Table S17 and Supplementary Note 9). By comparing the local regulatory network of the 15 functional genes in early and late gastric cancer (Fig. 5a, b), we find that there is a similar network structure for the 15 functional genes from early (Fig. 5a) with average out-degree 9 to late gastric cancer (Fig. 5b) with average out-degree 10. It is consistent with the reported genes, e.g., VEGFA is reported to take part in tumor growth and metastasis in gastric cancer56.
a, b are gene regulatory subnetworks for early and late stages of STAD, and the yellow nodes are central genes detected by the CVP algorithm in late cancer stages. c shows the comparison results of 15 functional genes predicted by CVP algorithm in literature verification. c, d are the gene regulatory subnetworks of early and late stages of LUAD, and f is the result of literature verification of 15 functional genes predicted by CVP algorithm.
Similarly, based on the CVP algorithm we construct the regulatory networks for two stages of lung adenocarcinoma (LUAD), the early (Stage I and II) and the late (Stage IV), identifying 15 out-degree hub genes as the functional genes associated with late cancer progression (Table S18 and Supplementary Note 9). Based on literature validation, 13 of the 15 central genes we found are strongly associated with the progression of lung cancer (Table S18 and Supplementary Note 9). In lung cancer, the late regulatory network of the 15 functional genes with average out-degree 10 is obviously larger than the early regulatory network with average out-degree 7 (Fig. 5c, d). Most of the functional genes gain more downstream regulation genes from early to late in lung cancer (Figs. 5c, d). It is consistent with some reported genes, e.g., IGF2 is an important oncogene and its activity is reported to take part in tumor growth and metastasis in lung cancer57.
Each type of cancer has its own unique pathogenesis. Using glioblastoma multiforme (GBM) as an example, we investigate the subnetwork of the oncogene tumor protein P53 (TP53) to reveal the specific regulatory pattern of TP53 in GBM. For GBM, we obtain 338 regulatory edges of TP53 in the cancer network, compared to 144 regulatory edges in normal samples (Figure S17). This reveal the regulatory pattern of TP53 in GBM, indicating that the regulatory molecules of TP53 are significantly increased in cancer. The TP53 may promote tumorigenesis mainly by inducing GBM related edges58, and this is consistent with the reported gain-of-function of TP53 in GBM59.
Identifying functional driver genes in a network level
The cancer is due to the accumulation of gene mutations, and now it is generally accepted that selective mutations of a small number of genes positively promote the occurrence of cancer, such genes that are called driver genes60. Identifying the genes that drive tumorigenesis and progression in cancer is key to cancer diagnosis and treatment. Despite the rapid development of a large number of algorithms for inferring driver genes, there is still a huge gap in the complete catalogue of driver genes in cancer61.
We identify hub genes from gene regulatory networks (GRNs) constructed using the CVP algorithm, aiming to predict driver genes in different types of cancer. The CVP algorithm is based on strict and consistent baseline analysis, when predicted the drivers. We apply the CVP to eight cancer datasets (GBM, CESC, PRAD, LUSC, OV, SARC, KIRP and LIHC) in TCGA (The Cancer Genome Atlas) to construct the GRNs and all the expression data are normalised using TPM (Transcripts Per Million). We consider that in directed networks, degree centrality alone does not provide a complete picture of the central gene, and often ignores nodes in the network that are critical but have few connected edges. Here, we use the PageRank centrality62, that an algorithm is originally used to rank web page popularity, and the genes with high PageRank centrality are defined as ‘functional driver genes’ involved in the regulation of multiple pathways. Eight GRNs are constructed by the CVP algorithm for the eight cancer datasets, and 100 genes with top PageRank centrality are identified from the GRN as the potential driver genes of each tumor (Table S7 and Figure S16). For validating the importance of these driver genes, we use the potential driver genes to do the enrichment analysis into the CGC(Cancer Gene Census)63 and InToGen databases64. The effectiveness of identifying cancer driver genes is further demonstrated by comparing the results with other 8 algorithms65,66,67,68,69,70,71,72 (Table 1).
Table 1 depicts the p-values of the nine methods (CVP, CoMDP65, Dendrix66, e-Driver67, ExInAtor72, MDPFinder68, MEMo69, OncoDriveCLUST70 and SCS71), which are used to predict the driver genes of cancer, for enrichment analysis of the predicted driver genes into CGC and InToGen. The lower p-values of the enrichment analysis, reflected the better effect to enrich to the existing cancer genes for the predicted driver genes from different algorithms. Specifically, the CVP based method has the best enrichment results in the 7 of 8 cancer datasets (Table 1), with the lowest p-values in all methods and orders of magnitude better than the other algorithms. The MEMo is relatively poor at predicting drivers, with a p-value of mostly 1, as no intersection of predicted genes and the tumor gene database (CGC or InToGen) or no drivers are predicted due to data limitations73. The fact that the network we have constructed uses only cancer expression data and does not require additional mutation information or methylation data, which means our algorithm is more broadly applicable and is sufficient to explain the importance of these genes to organisms even at the gene expression level. Overall, the network we have built provides a true measure of the complex information in biological systems and identifies potential driver genes and biological functions.
In the LIHC dataset only, our algorithm rank second (Table 1), which is still a positive result overall. Considering the overall excellent computational results described above, we have reason to believe that our predicted results are strongly likely to contain some drivers that have not yet been reported. Therefore, we use our inferred driver genes from LIHC dataset to do wet experiments to validate the driver genes and regulatory relationships.
Functional driver genes inhibited proliferation and colony formation in liver cancer cell
In order to investigate the influence of functional driver genes on tumor growth, we chose the top 10 functional driver genes from LIHC dataset. Then we knock out the 10 genes (SNRNP200, KHDRBS1, MTPN, MOB1A, XRN2, NCKAP, PPP1R12A, SP1, SRSF10 and RALGAPB) in liver cancer cell line Huh7 using CRISPR-Cas9 (Figure S18). For the exploration of the proliferative capacity of these cells, colony formation is used to detect the viability of liver cancer cells after knocking out these genes. Colony formation assay indicate that the formatting of cell colony-forming units is inhibited in KO (knock out) group, providing possible indicators for driver genes detection (Fig. 6a). Compared with control cells, these four factors (SNRNP200, XRN2, RALGAPB and SRSF10) induce about 50% growth inhibition in Huh7 cells (Fig. 6b), so these four gene KO cells are utilized in the following experiments.
a Colony formation of. Huh7 cells after knocked out the top ten functional driver genes. b The statistical analysis of colonyformation, n = 5 independent experiments, t-test. c, e were performed in Huh7 cels knocked out SNRNP200, XRN2. RALGAPB andSRSF10 genes. c BrdU assay of these cells by flow cytometer, d The statistical amalysis of BrdU assay. n = 3 independent experiments, t-test. e Cells were stained with anti-Ki67 antibodies (green) and DAPI to visualizeDNA (blue), Ki67 presents (green) and DAPI to visualize DNA (blue), Scale bars: 100 um. f The statistical analysis ofKi67 staining. n-5independent experiments, t-test.
Furthermore, BrdU assay is used to measure the proliferation in the above knocked out Huh7 cells. The flow cytometry assay results show that the BrdU-positive cells are markedly decreased in target-KO group (Fig. 6c). Especially, we find that there are very significant differences in SNRNP200-KO and MTPN-KO cells by statistical analysis (Fig. 6d). In addition, we assess the immunocytochemistry performance of Ki67, the widely used proliferation marker, to characterize the proliferation activity of tumor cells. The confocal microscopy results indicate that the Ki67 positive percent is reduced significantly in knocked out cells (Fig. 6e), and the results of this statistical analysis for the four groups (RALGAPB-KO, NCKAP-KO, SNRNP200-KO and MTPN-KO) show that they are a significantly lower than control group (Fig. 6f), which is liver cancer cells line Huh7 cells without any knockout. Taken together these results suggested that the top ten driver gene from LIHC can indeed suppress the proliferation and colony formation in liver cancer cells, and it can validate the power of the driver gene prediction from regulation network by the CVP algorithm.
The regulation validation in liver cancer cell
In order to validate the causal inference of the CVP algorithm, eight regulations from the regulation network of LIHC are used to verify the accuracy of the CVP algorithm by our biological experiment. To facilitate the biological experiment, we only chose the regulations from the two functional driver genes to their targets in liver cancer. So, four regulations from SNRNP200 to its target genes (MCM2, SMAD2, ORC2, and SMC1A), and other four regulations from RALGAPB to its target genes (REL, MAP2K1, NRAS and MAPK9) are chosen to validate the accuracy of the CVP algorithm by knockout experiment in liver cancer cell line Huh7 cells. When SNRNP200 is knocked out from the liver cancer cell, the gene expression of all the four target genes (MCM2, SMAD2, ORC2 and SMC1A) is strongly downregulated compared with the control cells that is liver cancer cell without SNRNP200 knock out (Figure S19a). It means that the SNRNP200 can regulate the expression of the four target genes, and it is consistent with the causality from SNRNP200 to the four genes by the CVP algorithm (Figure S19b).
Meanwhile, after RALGAPB is knocked out from the liver cancer cell, the expression of all four target genes (REL, MAP2K1, NRAS and MAPK9) is downregulated in RALGAPB knock out cells (Figure S19a). It also means that the RALGAPB can regulate the expression of the four target genes, and it is consistent with the causality from RALGAPB to the four genes by the CVP algorithm (Figure S19c). As far as we know, the eight regulations are not reported in liver cancer. Then, the eight regulations are predicted by the CVP algorithm in liver cancer and validated by knockout experiment (Figure S19). It can further reflect the power of the CVP algorithm.
Nonlinear simulation dataset
To further evaluate applicability to nonlinear relationships, we conduct simulation experiments analogous to Figure S2. These experiments involve Fan-in network structures with three variables (X, Y, and Z). The nonlinear relationships between variables are defined in Table S13. The results of the simulation demonstrate that CVP is capable of inferring causal relationships in nonlinear systems (Table S14). In the Fan-in network, CVP identifies causal strength values for the relationships X → Y and Z → Y (Table S14), which align with the true causal structure (Table S13). These findings highlight ability of CVP to capture nonlinear causal relationships. In summary, while current implementation of CVP is optimized for linear regression, so the sensitivity of CVP in nonlinear relationship is not as good as its performance in linear relationship. However, it is still applicable to nonlinear relationships, as shown in both real-world datasets and simulation experiments.
To further validate the method, we incorporate differential relationships into our simulations, as detailed in Table S15. We conduct 1000 simulation experiments and performed both t-tests and Wilcoxon tests on the results to determine whether the obtained causal strength values are significantly greater than zero (Table S16). The results demonstrate consistency with the actual causal relationships. Additionally, all parameter settings and the magnitude of the residual terms are provided in Table S15.
Discussion
The CVP causality concept with its algorithm is proposed for causal inference on any samples, and its effectiveness is validated by extensive studies of real datasets and also by biological experiments on cancer cell lines. The CVP algorithm quantifies causality between variables based on predictive ability of testing data in a cross-validation manner, which has two major differences from GC, i.e. data type and prediction process. GC is currently well accepted as being effective for inferring causality but on time-dependent or time-series data, whose prediction of testing data is processed in a time sequential manner. Interestingly, the CVP algorithm also performed better than GC-based method even for the time-series data (Table S6), which implies that on time-series data, our algorithm still has an advantage over the existing algorithms (Supplementary Note 10). A possible reason is the prediction process, i.e. cross-validation based on training and testing groups which are randomly divided, thus ensuring the robust inference of causality. On the other hand, comparing with the algorithms of time-independent data, CVP is able to infer a network with feedback loops common for many real systems, different from SCM mainly for directed acyclic graph without loops.
We show that the CVP algorithm performed better than the existing algorithms by extensive studies on various real cases, including gene regulatory and food chain networks. Our additional biological experiments also proved the predicted “functional driver genes” based on the inferred regulatory network. We highlight three noteworthy aspects. Firstly, a data-driven approach, the CVP algorithm explores the intrinsic characteristics of the data and does not require additional a priori information. Many of the existing algorithms may perform well in some cases but not in other cases, one possible reason is the requirement on some specific hypothesis or some pre-knowledge. Secondly, the CVP algorithm has few parameters to train, keeping consistent parameters under different data backgrounds, thus ensuring the universal operability of the algorithm (Figs. 3 and 4). Thirdly, high false positives have been a thorny problem for causal inference. Our CVP algorithm with condition on other variables (or partial correlation) can eliminate indirect causation by identifying individual causal variables one by one.
Causal inference is an important research topic in various fields including economics, physics, and biology. In biology, identifying causality between molecules benefits deep mechanism study on various diseases or biological processes at a network level. GC test is a classical method to infer the causality between variables, but requires time-series data, which limits the application to time independent data. Bayesian network or SCM can infer causality from time independent data, but it generally does not allow the cyclic or loop structure, which is common in the processes of biological regulations. In addition, the computational cost of Bayesian network is generally high, thus unsuitable for analyzing a large-scale biological network. To solve the problems, in this work we developed the CVP causality concept and its efficient algorithm. The CVP causality as well as its algorithm has significant implications for the interpretation of disease mechanisms and biological functions from the perspective of a causal network, and can be applied to analyze a wide range of network problems in various fields.
To further validate the performance of the t-test compared to causal strength, we conducted additional experiments using the SOS dataset as a benchmark. The results, summarized in Table S9, showed that while the t-test achieved reasonable performance with an AUC of 0.71 and an ACC of 0.70, these values are slightly lower than those obtained with causal strength (AUC of 0.75 and ACC of 0.81). Specifically, both methods outperformed other approaches, such as GENIMS, GENIE3, and TIGRESS, in capturing causal relationships (Table S9). These findings confirm that while the t-test can serve as a competitive method, causal strength remains the preferred approach due to its robustness and performance in this experiments.
In the comparison of results on simulated datasets, CVP is evaluated against five methods: Nonlinear ODEs, GENIMS, PLSNET, GENIE3, and TIGRESS, with GENIE3 being the most commonly used benchmark for gene regulatory network inference. Across both the DREAM and IRMA datasets, which include temporal and non-temporal data, CVP outperformed other methods in terms of AUC metrics (Fig. 2). On real control datasets, CVP demonstrated superior performance in TPR, FPR, precision, and recall. However, CVP is currently designed for linear problems, and its performance on nonlinear datasets may be suboptimal. To address this limitation, future work will focus on extending CVP to incorporate nonlinear causality.
To further explore driver genes from network inference, we employed the GENIE3 method to construct regulation network. We focused on the GBM dataset from TCGA and use GENIE3 to identify the potential functional driver genes. After constructing the network using GENIE3, the predicted functional driver genes are also identified based on degree hub and out-degree hub methods. We used the GBM driver genes from InToGen database (Table S10) as the gold standard, and the predicted functional driver genes from CVP and GENIE3 are used to do enrichment analysis with the gold standard gene in InToGen. And the p-value from enrichment analysis is used as the evaluation index (Table S10). The weak enrichment results obtained from GENIE3 highlight its limitations in effectively inferring cancer network structures, underscoring CVP’s superior performance in this domain (Table S10).
CVP is superior to the methods commonly used in their respective fields both in the task of gene regulatory network inference and network hub gene discovery. And CVP has shown some ability to solve some simple nonlinear problems. However, because the CVP method is limited by linear regression in its own method, more improvements will be made to the CVP method in the future, so that CVP can be more applicable to nonlinear problems.
Methods
The flow of the CVP method
The CVP method quantifies causal relations between variables based on any observed data by cross-validation predictability. Gene expression data with n genes and m samples is used as an example to explain the implement of the CVP method. Firstly, an initial correlation network is constructed by correlation coefficient (Pearson correlation coefficient or partial correlation coefficient) from the gene expression data, and we then eliminate the edges with low correlation to obtain the correlation network (without directions of edges) from the initial correlation network. Next, the correlation network and the gene expression data are used to infer the causal network (with directions of edges) by the CVP algorithm (Fig. 7a) as the following procedure.
-
1.
Initiation: Each node (or gene) \({g}_{j}\) and its first neighbors are considered as a unit (or subnetwork of \({g}_{j}\)) in the correlation network (Fig. 7b). We assume that all neighbors of node \({g}_{j}\) are the cause of \({g}_{j}\), i.e. the node \({g}_{j}\) is assumed to be causally regulated by all its neighbor nodes (Fig. 7b). All of the samples are randomly divided into two groups, i.e. training group and testing group, for cross validation (Fig. 7b).
-
2.
H1 model: The node \({g}_{j}\) is regressed on all its neighbors in the training samples to obtain regression Eq. (2) (Fig. 7b), and then the testing samples are used to calculate the error from the regression Eq. (2) (Fig. 7b). The \(e\) is the total squared error summation from the regression Eq. (2) in Fig. 7b for all of the testing samples, i.e. H1 model.
-
3.
H0 model: For judging the causality of a neighbor node \({g}_{i}\) to \({g}_{j}\), the node \({g}_{i}\) is removed from the subnetwork of \({g}_{j}\) and the gene expression of node \({g}_{i}\) is also removed from the subnetwork of node \({g}_{j}\) (Fig. 7b). We use the same two groups used in (2) for cross-validation of (1). In other words, for the perturbed subnetwork eliminating \({g}_{i}\), the node \({g}_{j}\,\) is also regressed on all its neighbors except node \({g}_{i}\) in the training samples to obtain regression Eq. (1) (Fig. 7b). Then, the new error is calculated by implementing the regression Eq. (1) in the testing samples without \({g}_{i}\) for predicting \({g}_{j}\) (Fig. 7b). The \(\hat{e}\) is the total squared error summation from the regression Eq. (1) in Fig. 7b for all of the testing samples, i.e. H0 model.
-
4.
Prediction testing: We used the sum of squared error \(e\) and \(\hat{e}\) to infer the potential causal regulation from \({g}_{i}\) to \({g}_{j}\). If \(e\) is less than \(\hat{e}\), it means that \({g}_{i}\) benefits the regression Eq. (1) to accurately predict the gene expression of \({g}_{j}\) in the testing group samples, and \({g}_{j}\) can be at least partly explained by \({g}_{i}\), thus the node \({g}_{i}\) is the cause of node \({g}_{j}\) in this case (Fig. 7b). If \(e\) is greater than \(\hat{e}\), it means that \({g}_{i}\) is unsuitable for predicting the gene expression of \({g}_{j}\) in the testing group samples, and \({g}_{j}\) cannot be explained by \({g}_{i}\), Hence, we consider that the node \({g}_{i}\) is not the cause of node \({g}_{j}\) in this case (Fig. 7b).
a The general idea of the algorithm, from a correlation network to a causal network. b The core steps of the CVP algorithm. For the correlation subnetwork of the variable \({g}_{j}\), candidate causal relations are obtained assuming that \({g}_{j}\) is modulated by multiple dependent variables. When judging whether there is causality between gi and gj, we first obtain sample data under H0 and H1 assumptions by removing gi or not. After that, the training set and test set are divided under each hypothesis, the regression equation is obtained in the training set, and the error e is obtained by bringing the regression equation into the test set. After that, whether the causal intensity is greater than 0 is calculated to determine whether there is causality between gi and gj. The causal subnetwork of gj is obtained by cross-validation testing to infer the causal relation of each candidate dependent variable on gj one by one. The red represents gj, and the slash in the matrix indicates that the gj representation is removed.
Here, we use causal strength \({\omega }_{i\to j}=\,{{{\mathrm{ln}}}}(\hat{{{\rm{e}}}}/{{{\rm{e}}}})\) of Eq. (3) to judge the causality from \({g}_{i}\) to \({g}_{j}\), i.e. if \({\omega }_{i\to j} > 0\), there is causality from \({g}_{i}\) to \({g}_{j}\), and if \({\omega }_{i\to j}\le 0\), there is no causality from \({g}_{i}\) to \({g}_{j}\) (Fig. 7b). After every neighbor node \({g}_{i}\) of \({g}_{j}\) is evaluated by the corresponding \({\omega }_{i\to j}\) in the subnetwork of \({g}_{j}\), we can obtain the causal subnetwork for \({g}_{j}\) (Fig. 7b). Based on the above procedure of CVP algorithm, the causal subnetwork for every node in the correlation network can be inferred one by one, i.e. j = 1, 2, …, n, and then the network combining all n causal subnetworks is the inferred causal network with n nodes (Fig. 7 and Supplementary Note 1).
Evaluation indicators
In this study, AUROC (the area under the receiver operating characteristic curve) and AUPR (the area under the precision-recall curve) are used to assess the effectiveness of the algorithm. AUROC is commonly used to evaluate classification models, and its value is the area under the ROC curve. The ROC (receiver operating characteristic) curve is used to weigh the TPR (true positive rate) and FPR (false positive rate) under different decision thresholds, where the horizontal coordinate is the FPR and the vertical coordinate is the TPR. AUROC has a high advantage over unbalanced data with high positive examples, but it does not capture the impact of a large number of negative examples on algorithm performance. Due to the sparsity of GRN, there may be relatively high negative examples, so we consider introducing AUPR. AUPR is the area under the PR (precision-recall) curve, and PR curve trade-offs precision and recall (TPR) at different decision thresholds, with the horizontal coordinate being recall and the vertical coordinate being precision. TPR, FPR, ACC and Precision are calculated as in Eq. (4).
Knock out
The targeted gRNA oligos were introduced into the lentiviral expression plasmids. The sequences of these oligos are listed in Table S7. In total 4.5 × 106 HEK293T cells were seeded on a 10 cm dish and transfected with lentiviral expression and packaging plasmids. After another 48 h, collect medium containing lentivirus particles. The culture medium was refreshed the following morning, and the supernatant containing lentiviral particles was collected after another 48 h. For experiments, Huh7 cells were transduced with the lentiviral vectors at a multiplicity of infection (MOI) of 0.5 and single colonies were selected with 2 mg/ml puromycin. Gene expression levels in each cell line were examined using Real-Time Quantitative PCR (QPCR).
Colony formation assay
Gene knocked out Huh7 cell lines were seeded and cultured into the six-well plate at a density of 2 × 103 cells/well in DMEM (Dulbecco’s Modified Eagle Medium) containing 10% FBS (Fetal bovine serum) for 3 weeks to allow colony formation. The plate was washed with cold PBS (Phosphate Buffered Saline). The colonies were fixed by 4% polyformaldehyde at room temperature, washed with water. Next, they were dyed with 1% crystal violet for 30 min at room temperature, washed with water several times and dried. Each experiment was done thrice in this study.
BrdU assay
BrdU staining was used to analyze the cancer cell proliferation according to manufacturer the protocol (BrdU APC flow Kit, 552598, BD). Briefly, knocked out cell lines were incubated with BrdU (10 mmol/L) for 2 h. The cells were fixed with 4% paraformaldehyde for 20 min at room temperature. After permeabilized and staining, cells were collected for flow cytometry analysis.
Ki67 staining
Huh7 cells were fixed in 1% formaldehyde. Incubate on ice for 5 min. Washed twice with PBS containing 0.05% Tween20 and permeabilized with 0.5% Triton X-100 in PBS. Cells stained with the anti-Ki67 antibody (abcam, ab15580, 1:1000) for 1 h at room temperature. Then the washed cells were stained with Donkey anti-rabbit IgG Alexa Fluor 488 (A32790, Invitrogen, 1:1000). DNA was stained with DAPI (D9542-1MG sigma).
Data availability
The data sets covered in this study are available from citations or links in the main text/supporting materials. All cancer datasets (BLCA, GBM, CESC, PRAD, LUSC, OV, SARC, KIRP and LIHC) are the transcriptome dataset from the TCGA database (https://portal.gdc.cancer.gov/), normalized by TPM (Transcripts Per Kilobase of exon model per Million mapped reads). E. coli dataset dataset24 (GSE20305) and yeast cell cycle dataset35 (GSE8799) are retrieved from the Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/) database.
Code availability
The codes are available at https://github.com/zyllluck/CVP.
References
Hill, S. M. et al. Inferring causal molecular networks: empirical assessment through a community-based effort. Nat. Methods 13, 310–318 (2016).
Liu, X., Wang, Y., Ji, H., Aihara, K. & Chen, L. Personalized characterization of diseases using sample-specific networks. Nucleic Acids Res. 44, e164 (2016).
Huang, Y., Chang, X., Zhang, Y., Chen, L. & Liu, X. Disease characterization using a partial correlation-based sample-specific network. Brief Bioinform. 22, https://doi.org/10.1093/bib/bbaa062 (2021).
Zhang, Y. et al. Identifying network biomarkers of cancer by sample-specific differential network. BMC Bioinforma. 23, 230 (2022).
De Smet, R. & Marchal, K. Advantages and limitations of current network inference methods. Nat. Rev. Microbiol 8, 717–729 (2010).
Aalto, A., Viitasaari, L., Ilmonen, P., Mombaerts, L. & Goncalves, J. Gene regulatory network inference from sparsely sampled noisy data. Nat. Commun. 11, 3493 (2020).
Granger, C. W. J. Investigating causal relations by econometric models and cross-spectral methods. Econometrica 37, 424–438 (1969).
Kamiński, M., Ding, M., Truccolo, W. A. & Bressler, S. L. Evaluating causal relations in neural systems: Granger causality, directed transfer function and statistical assessment of significance. Biol. Cybern. 85, 145–157 (2001).
Saberski, E. et al. Networks of causal linkage between eigenmodes characterize behavioral dynamics of Caenorhabditis elegans. PLoS Comput Biol. 17, e1009329 (2021).
Sun, J., Cafaro, C. & Bollt, E. Identifying the coupling structure in complex systems through the optimal causation entropy principle. Entropy 16, 3416–3433 (2014).
Choi, B. et al. Bayesian inference of distributed time delay in transcriptional and translational regulation. Bioinformatics 36, 586–593 (2020).
Mangan, N. M., Brunton, S. L., Proctor, J. L. & Kutz, J. N. Inferring Biological Networks by Sparse Identification of Nonlinear Dynamics. IEEE Trans. Mol., Biol. Multi-Scale Commun. 2, 52–63 (2016).
Zhang, Y., Chang, X. & Liu, X. Inference of gene regulatory networks using pseudo-time series data. Bioinformatics 37, 2423–2431 (2021).
Wang, J., Zhang, Y., Chen, L. & Liu, X. Reconstructing molecular networks by causal diffusion do-calculus analysis with deep learning. Adv. Sci. (Weinh.) 11, e2409170 (2024).
Vicente, R., Wibral, M., Lindner, M. & Pipa, G. Transfer entropy-a model-free measure of effective connectivity for the neurosciences. J. Comput. Neurosci. 30, 45–67 (2011).
Faes, L., Nollo, G. & Porta, A. Non-uniform multivariate embedding to assess the information transfer in cardiovascular and cardiorespiratory variability series. Comput Biol. Med. 42, 290–297 (2012).
Liang, Y. & Kelemen, A. Bayesian dynamic multivariate models for inferring gene interaction networks. Conf. Proc. IEEE Eng. Med. Biol. Soc. 2006, 2041–2044 (2006).
Pearl, J. Causal inference in statistics: An overview. Statistics Surveys 3, https://doi.org/10.1214/09-ss057 (2009).
Singh, N. & Vidyasagar, M. bLARS: An Algorithm to Infer Gene Regulatory Networks. IEEE/ACM Trans. Comput Biol. Bioinform. 13, 301–314 (2016).
Cantone, I. et al. A yeast synthetic network for in vivo assessment of reverse-engineering and modeling approaches. Cell 137, 172–181 (2009).
Shen-Orr, S. S., Milo, R., Mangan, S. & Alon, U. Network motifs in the transcriptional regulation network of Escherichia coli. Nat. Genet. 31, 64–68 (2002).
Park, S. et al. BTNET : boosted tree based gene regulatory network inference algorithm using time-course measurement data. BMC Syst. Biol. 12, 20 (2018).
Whitfield, M. L. et al. Identification of genes periodically expressed in the human cell cycle and their expression in tumors. Mol. Biol. Cell 13, 1977–2000 (2002).
Jozefczuk, S. et al. Metabolomic and transcriptomic stress response of Escherichia coli. Mol. Syst. Biol. 6, 364 (2010).
Ma, B., Fang, M. & Jiao, X. Inference of gene regulatory networks based on nonlinear ordinary differential equations. Bioinformatics 36, 4885–4893 (2020).
Wu, J., Zhao, X., Lin, Z. & Shao, Z. Large scale gene regulatory network inference with a multi-level strategy. Mol. Biosyst. 12, 588–597 (2016).
Guo, S., Jiang, Q., Chen, L. & Guo, D. Gene regulatory network inference using PLS-based methods. Bmc Bioinforma. 17, 545 (2016).
Huynh-Thu, V. A., Irrthum, A., Wehenkel, L. & Geurts, P. Inferring regulatory networks from expression data using tree-based methods. Plos One 5, https://doi.org/10.1371/journal.pone.0012776 (2010).
Haury, A.-C., Mordelet, F., Vera-Licona, P. & Vert, J.-P. TIGRESS: trustful inference of gene REgulation using stability selection. Bmc Syst. Biol. 6, 145 (2012).
Schaffter, T., Marbach, D. & Floreano, D. GeneNetWeaver: in silico benchmark generation and performance profiling of network inference methods. Bioinformatics 27, 2263–2270 (2011).
Maathuis, M. H., Colombo, D., Kalisch, M. & Buhlmann, P. Predicting causal effects in large-scale systems from observational data. Nat. Methods 7, 247–248 (2010).
Heinze-Deml, C., Maathuis, M. H. & Meinshausen, N. Causal Structure Learning. Annu. Rev. Stat. Its Application 5, 371–391 (2018).
Opgen-Rhein, R. & Strimmer, K. From correlation to causation networks: a simple approximate learning algorithm and its application to high-dimensional plant gene expression data. BMC Syst. Biol. 1, 37 (2007).
Passemiers, A., Moreau, Y. & Raimondi, D. Fast and accurate inference of gene regulatory networks through robust precision matrix estimation. Bioinformatics 38, 2802–2809 (2022).
Yalamanchili, H. K. et al. DDGni: dynamic delay gene-network inference from high-temporal data using gapped local alignment. Bioinformatics 30, 377–383 (2014).
Sambo, F., Camillo, B. D. & Toffolo, G. CNET: an algorithm for reverse engineering of causal gene networks. nettab varenna (2008).
Nguyen, P. & Braun, R. Time-lagged Ordered Lasso for network inference. BMC Bioinforma. 19, 545 (2018).
Gama-Castro, S. et al. RegulonDB version 9.0: high-level integration of gene regulation, coexpression, motif clustering and beyond. Nucleic Acids Res. 44, D133–D143 (2016).
Neutel, A. M., Heesterbeek, J. A. & De Ruiter, P. C. Stability in real food webs: weak links in long loops. Science 296, 1120–1123 (2002).
Jagadeesan, L., Jyothibabu, R., Arunpandi, N. & Parthasarathi, S. Copepod grazing and their impact on phytoplankton standing stock and production in a tropical coastal water during the different seasons. Environ. Monit. Assess. 189, 105 (2017).
Tõnno, I. et al. Algal diet of small-bodied crustacean zooplankton in a cyanobacteria-dominated eutrophic lake. PLoS ONE 11, e0154526 (2016).
Lee, B. J., Kim, B. & Lee, K. Air pollution exposure and cardiovascular disease. Toxicol. Res. 30, 71–75 (2014).
Wong, T. W. et al. Air pollution and hospital admissions for respiratory and cardiovascular diseases in Hong Kong. Occup. Environ. Med. 56, 679–683 (1999).
Davis, R. E., McGregor, G. R. & Enfield, K. B. Humidity: a review and primer on atmospheric moisture and human health. Environ. Res. 144, 106–116 (2016).
Hassoun, Y., James, C. & Bernstein, D. I. The effects of air pollution on the development of atopic disease. Clin. Rev. allergy Immunol. 57, 403–414 (2019).
Valavanidis, A., Fiotakis, K. & Vlachogianni, T. Airborne particulate matter and human health: toxicological assessment and importance of size and composition of particles for oxidative damage and carcinogenic mechanisms. J. Environ. Sci. health Part C., Environ. carcinogenesis Ecotoxicol. Rev. 26, 339–362 (2008).
Lam, G. C. K. et al. Street-level concentrations of nitrogen dioxide and suspended particulate matter in Hong Kong. Atmos. Environ. 33, 1–11 (1998).
Leng, S. et al. Partial cross mapping eliminates indirect causal influences. Nat. Commun. 11, 2632 (2020).
Gvozdić, V., Kovač-Andrić, E. & Brana, J. Influence of meteorological factors NO2, SO2, CO and PM10 on the concentration of O3 in the urban atmosphere of eastern croatia. Environ. Modeling Assess. 16, 491–501 (2011).
He, X., Pang, S., Ma, J. & Zhang, Y. Influence of relative humidity on heterogeneous reactions of O3 and O3/SO2 with soot particles: Potential for environmental and health effects. Atmos. Environ. 165, 198–206 (2017).
Zoran, M. A., Savastru, R. S., Savastru, D. M. & Tautan, M. N. Assessing the relationship between ground levels of ozone (O3) and nitrogen dioxide (NO2) with coronavirus (COVID-19) in Milan, Italy. Sci. Total Environ. 740, 140005 (2020).
Collivignarelli, M. C. et al. Lockdown for CoViD-2019 in Milan: What are the effects on air quality? Sci. total Environ. 732, 139280–139280 (2020).
Rai, P., Kumar, B. K., Deekshit, V. K., Karunasagar, I. & Karunasagar, I. Detection technologies and recent developments in the diagnosis of COVID-19 infection. Appl. Microbiol. Biotechnol. 105, 441–455 (2021).
Liu, R. et al. Predicting local COVID-19 outbreaks and infectious disease epidemics based on landscape network entropy. Sci. Bull. 66, 2265–2270 (2021).
Bray, F. et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: a cancer J. clinicians 68, 394–424 (2018).
Wang, C. Q. et al. MiR-377 suppresses cell proliferation and metastasis in gastric cancer via repressing the expression of VEGFA. Eur. Rev. Med Pharm. Sci. 21, 5101–5111 (2017).
Zheng, F. X., Wang, X. Q., Zheng, W. X. & Zhao, J. Long noncoding RNA HOXA-AS2 promotes cell migration and invasion via upregulating IGF-2 in non-small cell lung cancer as an oncogene. Eur. Rev. Med Pharm. Sci. 23, 4793–4799 (2019).
Liu, X., Wang, Y., Ji, H., Aihara, K. & Chen, L. Personalized characterization of diseases using sample-specific networks. Nucleic Acids Res. 44, e164–e164 (2016).
Wang, X. et al. Gain of function of mutant TP53 in glioblastoma: prognosis and response to temozolomide. Ann. Surg. Oncol. 21, 1337–1344 (2014).
Bashashati, A. et al. DriverNet: uncovering the impact of somatic driver mutations on transcriptional networks in cancer. Genome Biol. 13, R124 (2012).
Han, Y. et al. Corrigendum to article “DriverML: a machine learning algorithm for identifying driver genes in cancer sequencing studies”. Nucleic Acids Res. 49, 4196 (2021).
Gong, C. et al. PageRank tracker: from ranking to tracking. IEEE Trans. Cyber. 44, 882–893 (2014).
Futreal, P. A. et al. A census of human cancer genes. Nat. Rev. Cancer 4, 177–183 (2004).
Martinez-Jimenez, F. et al. A compendium of mutational cancer driver genes. Nat. Rev. Cancer 20, 555–572 (2020).
Zhang, J., Wu, L. Y., Zhang, X. S. & Zhang, S. Discovery of co-occurring driver pathways in cancer. Bmc Bioinforma. 15, 271 (2014).
Vandin, F., Upfal, E. & Raphael, B. J. De novo discovery of mutated driver pathways in cancer. Genome Res. 22, 375–385 (2012).
Porta-Pardo, E. & Godzik, A. e-Driver: a novel method to identify protein regions driving cancer. Bioinformatics 30, 3109–3114 (2014).
Zhao, J., Zhang, S., Wu, L. Y. & Zhang, X. S. Efficient methods for identifying mutated driver pathways in cancer. Bioinformatics 28, 2940–2947 (2012).
Ciriello, G., Cerami, E., Sander, C. & Schultz, N. Mutual exclusivity analysis identifies oncogenic network modules. Genome Res. 22, 398–406 (2012).
Tamborero, D., Gonzalez-Perez, A. & Lopez-Bigas, N. OncodriveCLUST: exploiting the positional clustering of somatic mutations to identify cancer genes. Bioinformatics 29, 2238–2244 (2013).
Guo, W. F. et al. Discovering personalized driver mutation profiles of single samples in cancer by network control strategy. Bioinformatics 34, 1893–1903 (2018).
Lanzos, A. et al. Discovery of cancer driver long noncoding RNAs across 1112 tumour genomes: new candidates and distinguishing features. Sci. Rep. 7, 41544 (2017).
Han, Y. et al. DriverML: a machine learning algorithm for identifying driver genes in cancer sequencing studies. Nucleic Acids Res. 47, e45 (2019).
Acknowledgements
This research was supported by the National Natural Science Foundation of China (NSFC) Grant No. T2341024, Zhejiang Provincial Natural Science Foundation of China under Grant No. LZ22C060001, the research funds of Hangzhou Institute for advanced study, UCAS (No. 2022ZZ01013 and 2024HIAS-Y016), the National Key Research and Development Program of China (2022YFA1004800).
Author information
Authors and Affiliations
Contributions
X.L., L.C., and X.C. designed the project. Y.Z., Q.L., and J.W. conducted the theoretical study, manuscript writing, experiment, and data analysis.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Physics thanks Jae Kyoung Kim and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zhang, Y., Li, Q., Wang, J. et al. Causal network inference based on cross-validation predictability. Commun Phys 8, 173 (2025). https://doi.org/10.1038/s42005-025-02091-4
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s42005-025-02091-4