Abstract
The aging of hematopoietic stem cells (HSCs) substantially alters their characteristics. Mitochondria, essential for cellular metabolism, play a crucial role, and their dysfunction is a hallmark of aging-induced changes. The impact of mitochondrial mass on aged HSCs remains incompletely understood. Here we demonstrate that HSCs with high mitochondrial mass during aging are not merely cells that have accumulated damaged mitochondria and become exhausted. In addition, these HSCs retain a high regenerative capacity and remain in the aging bone marrow. Furthermore, we identified GPR183 as a distinct marker characterizing aged HSCs through single-cell analysis. HSCs marked by GPR183 were also enriched in aged HSCs with high mitochondrial mass, possessing a high capacity of self-renewal. These insights deepen understanding of HSC aging and provide additional perspectives on the assessment of aged HSCs, underscoring the importance of mitochondrial dynamics in aging.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
de Haan, G. & Lazare, S. S. Aging of hematopoietic stem cells. Blood 131, 479–487 (2018).
Mejia-Ramirez, E. & Florian, M. C. Understanding intrinsic hematopoietic stem cell aging. Haematologica 105, 22–37 (2020).
Dykstra, B., Olthof, S., Schreuder, J., Ritsema, M. & de Haan, G. Clonal analysis reveals multiple functional defects of aged murine hematopoietic stem cells. J. Exp. Med. 208, 2691–2703 (2011).
Cho, R. H., Sieburg, H. B. & Muller-Sieburg, C. E. A new mechanism for the aging of hematopoietic stem cells: aging changes the clonal composition of the stem cell compartment but not individual stem cells. Blood 111, 5553–5561 (2008).
Bowman, R. L., Busque, L. & Levine, R. L. Clonal hematopoiesis and evolution to hematopoietic malignancies. Cell Stem Cell 22, 157–170 (2018).
Challen, G. A. & Goodell, M. A. Clonal hematopoiesis: mechanisms driving dominance of stem cell clones. Blood 136, 1590–1598 (2020).
Rossi, D. J. et al. Deficiencies in DNA damage repair limit the function of haematopoietic stem cells with age. Nature 447, 725–729 (2007).
Kirschner, K. et al. Proliferation drives aging-related functional decline in a subpopulation of the hematopoietic stem cell compartment. Cell Rep. 19, 1503–1511 (2017).
Chen, Z. et al. Cohesin-mediated NF-κB signaling limits hematopoietic stem cell self-renewal in aging and inflammation. J. Exp. Med. 216, 152–175 (2019).
Chen, C., Liu, Y., Liu, Y. & Zheng, P. mTOR regulation and therapeutic rejuvenation of aging hematopoietic stem cells. Sci. Signal. 2, ra75 (2009).
Sun, D. et al. Epigenomic profiling of young and aged HSCs reveals concerted changes during aging that reinforce self-renewal. Cell Stem Cell 14, 673–688 (2014).
Tang, B. et al. Aging-disturbed FUS phase transition impairs hematopoietic stem cells by altering chromatin structure. Blood 143, 124–138 (2024).
Florian, M. C. et al. Cdc42 activity regulates hematopoietic stem cell aging and rejuvenation. Cell Stem Cell 10, 520–530 (2012).
Suda, T., Arai, F. & Hirao, A. Hematopoietic stem cells and their niche. Trends Immunol. 26, 426–433 (2005).
Nakamura-Ishizu, A., Ito, K. & Suda, T. Hematopoietic stem cell metabolism during development and aging. Dev. Cell 54, 239–255 (2020).
Xu, Y., Chiang, Y. H., Ho, P. C. & Vannini, N. Mitochondria dictate function and fate of HSCs and T cells. Cancer Immunol. Res. 11, 1303–1313 (2023).
Morganti, C. & Ito, K. Mitochondrial contributions to hematopoietic stem cell aging. Int. J. Mol. Sci. 22, 11117 (2021).
Filippi, M. D. & Ghaffari, S. Mitochondria in the maintenance of hematopoietic stem cells: new perspectives and opportunities. Blood 133, 1943–1952 (2019).
Umemoto, T., Hashimoto, M., Matsumura, T., Nakamura-Ishizu, A. & Suda, T. Ca2+–mitochondria axis drives cell division in hematopoietic stem cells. J. Exp. Med. 215, 2097–2113 (2018).
Harada, K. et al. Mitochondrial electron transport chain complex II dysfunction causes premature aging of hematopoietic stem cells. Stem Cells 41, 39–49 (2023).
Mohrin, M. et al. Stem cell aging. A mitochondrial UPR-mediated metabolic checkpoint regulates hematopoietic stem cell aging. Science 347, 1374–1377 (2015).
Sun, X. et al. Nicotinamide riboside attenuates age-associated metabolic and functional changes in hematopoietic stem cells. Nat. Commun. 12, 2665 (2021).
Girotra, M. et al. Induction of mitochondrial recycling reverts age-associated decline of the hematopoietic and immune systems. Nat. Aging 3, 1057–1066 (2023).
de Almeida, M. J., Luchsinger, L. L., Corrigan, D. J., Williams, L. J. & Snoeck, H. W. Dye-independent methods reveal elevated mitochondrial mass in hematopoietic stem cells. Cell Stem Cell 21, 725–729 (2017).
Pham, A. H., McCaffery, J. M. & Chan, D. C. Mouse lines with photo-activatable mitochondria to study mitochondrial dynamics. Genesis 50, 833–843 (2012).
Takihara, Y. et al. High mitochondrial mass is associated with reconstitution capacity and quiescence of hematopoietic stem cells. Blood Adv. 3, 2323–2327 (2019).
Hinge, A. et al. Asymmetrically segregated mitochondria provide cellular memory of hematopoietic stem cell replicative history and drive HSC attrition. Cell Stem Cell 26, 420–430 (2020).
Ho, T. T. et al. Autophagy maintains the metabolism and function of young and old stem cells. Nature 543, 205–210 (2017).
Balazs, A. B., Fabian, A. J., Esmon, C. T. & Mulligan, R. C. Endothelial protein C receptor (CD201) explicitly identifies hematopoietic stem cells in murine bone marrow. Blood 107, 2317–2321 (2006).
Beerman, I. et al. Functionally distinct hematopoietic stem cells modulate hematopoietic lineage potential during aging by a mechanism of clonal expansion. Proc. Natl Acad. Sci. USA 107, 5465–5470 (2010).
Montecino-Rodriguez, E. et al. Lymphoid-biased hematopoietic stem cells are maintained with age and efficiently generate lymphoid progeny. Stem Cell Rep. 12, 584–596 (2019).
Wilkinson, A. C. et al. Long-term ex vivo haematopoietic-stem-cell expansion allows nonconditioned transplantation. Nature 571, 117–121 (2019).
Kowalczyk, M. S. et al. Single-cell RNA-seq reveals changes in cell cycle and differentiation programs upon aging of hematopoietic stem cells. Genome Res. 25, 1860–1872 (2015).
Li, J. et al. STAT1 is essential for HSC function and maintains MHCIIhi stem cells that resist myeloablation and neoplastic expansion. Blood 140, 1592–1606 (2022).
Lauridsen, F. K. B. et al. Differences in cell cycle status underlie transcriptional heterogeneity in the HSC compartment. Cell Rep. 24, 766–780 (2018).
Wilson, N. K. et al. Combined single-cell functional and gene expression analysis resolves heterogeneity within stem cell populations. Cell Stem Cell 16, 712–724 (2015).
Sommerkamp, P. et al. Mouse multipotent progenitor 5 cells are located at the interphase between hematopoietic stem and progenitor cells. Blood 137, 3218–3224 (2021).
Chitteti, B. R. et al. CD166 regulates human and murine hematopoietic stem cells and the hematopoietic niche. Blood 124, 519–529 (2014).
Hu, M. et al. CD63 acts as a functional marker in maintaining hematopoietic stem cell quiescence through supporting TGFβ signaling in mice. Cell Death Differ. 29, 178–191 (2022).
Sawai, C. M. et al. Hematopoietic stem cells are the major source of multilineage hematopoiesis in adult animals. Immunity 45, 597–609 (2016).
Liberzon, A. et al. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 1, 417–425 (2015).
Rath, S. et al. MitoCarta3.0: an updated mitochondrial proteome now with sub-organelle localization and pathway annotations. Nucleic Acids Res. 49, D1541–D1547 (2021).
Mishra, P. & Chan, D. C. Metabolic regulation of mitochondrial dynamics. J. Cell Biol. 212, 379–387 (2016).
Flohr Svendsen, A. et al. A comprehensive transcriptome signature of murine hematopoietic stem cell aging. Blood 138, 439–451 (2021).
Bartlett, S. et al. GPR183 regulates interferons, autophagy, and bacterial growth during Mycobacterium tuberculosis infection and is associated with TB disease severity. Front. Immunol. 11, 601534 (2020).
Lima, T., Li, T. Y., Mottis, A. & Auwerx, J. Pleiotropic effects of mitochondria in aging. Nat. Aging 2, 199–213 (2022).
Silva Ramos, E. et al. Mitochondrial fusion is required for regulation of mitochondrial DNA replication. PLoS Genet. 15, e1008085 (2019).
Yamamoto, R. et al. Large-scale clonal analysis resolves aging of the mouse hematopoietic stem cell compartment. Cell Stem Cell 22, 600–607 (2018).
Tanaka, T. et al. Identification of differentiation roots of hematopoietic stem cells by a paired-daughter assay combined with multiple barcoding. Exp. Hematol. 111, S146 (2022).
Yien, Y. Y. & Perfetto, M. Regulation of heme synthesis by mitochondrial homeostasis proteins. Front. Cell Dev. Biol. 10, 895521 (2022).
Liang, R. et al. Restraining lysosomal activity preserves hematopoietic stem cell quiescence and potency. Cell Stem Cell 26, 359–376 (2020).
Watanuki, S. et al. SDHAF1 confers metabolic resilience to aging hematopoietic stem cells by promoting mitochondrial ATP production. Cell Stem Cell 31, 1145–1161 (2024).
Bonora, M. et al. A mitochondrial NADPH-cholesterol axis regulates extracellular vesicle biogenesis to support hematopoietic stem cell fate. Cell Stem Cell 31, 359–377 (2024).
Arosio, P., Elia, L. & Poli, M. Ferritin, cellular iron storage and regulation. IUBMB Life 69, 414–422 (2017).
Yi, W. et al. Ferritin-mediated mitochondrial iron homeostasis is essential for the survival of hematopoietic stem cells and leukemic stem cells. Leukemia 5, 1003–1018 (2024).
Jiang, X., Stockwell, B. R. & Conrad, M. Ferroptosis: mechanisms, biology and role in disease. Nat. Rev. Mol. Cell Biol. 22, 266–282 (2021).
Freitas, F. P. et al. 7-Dehydrocholesterol is an endogenous suppressor of ferroptosis. Nature 626, 401–410 (2024).
Hannedouche, S. et al. Oxysterols direct immune cell migration via EBI2. Nature 475, 524–527 (2011).
Liu, C. et al. Oxysterols direct B-cell migration through EBI2. Nature 475, 519–523 (2011).
Zhang, P. et al. G protein-coupled receptor 183 facilitates endothelial-to-hematopoietic transition via Notch1 inhibition. Cell Res. 25, 1093–1107 (2015).
Nevius, E. et al. Oxysterols and EBI2 promote osteoclast precursor migration to bone surfaces and regulate bone mass homeostasis. J. Exp. Med. 212, 1931–1946 (2015).
Arcangeli, M. L. et al. JAM-B regulates maintenance of hematopoietic stem cells in the bone marrow. Blood 118, 4609–4619 (2011).
Oedekoven, C. A. et al. Hematopoietic stem cells retain functional potential and molecular identity in hibernation cultures. Stem Cell Rep. 16, 1614–1628 (2021).
Wang, T. et al. Loss of Nupr1 promotes engraftment by tuning the quiescence threshold of hematopoietic stem cell repository via regulating p53-checkpoint pathway. Haematologica 107, 154–166 (2022).
Liu, J. et al. NUPR1 is a critical repressor of ferroptosis. Nat. Commun. 12, 647 (2021).
Matsumura, T. et al. Hematopoietic stem cells acquire survival advantage by loss of RUNX1 methylation identified in familial leukemia. Blood 136, 1919–1932 (2020).
Matsumura, T. et al. A Myb enhancer-guided analysis of basophil and mast cell differentiation. Nat. Commun. 13, 7064 (2022).
Hao, Y. et al. Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587 (2021).
Stuart, T., Srivastava, A., Madad, S., Lareau, C. A. & Satija, R. Single-cell chromatin state analysis with Signac. Nat. Methods 18, 1333–1341 (2021).
Hänzelmann, S., Castelo, R. & Guinney, J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics 14, 7 (2013).
Kanehisa, M. & Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 28, 27–30 (2000).
Ritchie, M. E. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015).
Kim, D., Paggi, J. M., Park, C., Bennett, C. & Salzberg, S. L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 37, 907–915 (2019).
Liao, Y., Smyth, G. K. & Shi, W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA 102, 15545–15550 (2005).
Argüello, R. J. et al. SCENITH: a flow cytometry-based method to functionally profile energy metabolism with single-cell resolution. Cell Metab. 32, 1063–1075 (2020).
Watanabe, A. et al. Noncanonical TCA cycle fosters canonical TCA cycle and mitochondrial integrity in acute myeloid leukemia. Cancer Sci. 116, 152–163 (2025).
Acknowledgements
The authors thank the CSI FACS facility for technical assistance and the NUS MD2 Vivarium for mouse husbandry. This research is supported by the Singapore Translational Research Investigator Award from the National Medical Research Council in Singapore (NMRC/STaR 18 may-0004 to T. Suda); JSPS Grant-in-Aid for Scientific Research (S) (18H05284 and 26221309 to T. Suda); the Chinese Academy of Medical Sciences Innovation Fund for Medical Sciences (CIFMS) (2024-12M-3-017 to T. Suda); the National Natural Science Foundation of China (W2441024 to T. Suda); JSPS Grant-in-Aid for Challenging Research (Exploratory) (JP21K19514 to T.M.); JSPS Grant-in-Aid for Scientific Research (C) (24K11298 to T.M.); the Daiwa Securities Health Foundation (T.M.); the Novartis Foundation (Japan) for the Promotion of Science (T.M.); the SENSHIN Medical Research Foundation (T.M.); the Takeda Science Foundation (T.M.); the Naito Foundation (T.M.); and the Open Fund-Young Individual Research Grant (OF-YIRG) from the National Medical Research Council in Singapore (OFYIRG21nov-0020 to H.T.).
Author information
Authors and Affiliations
Contributions
H.T., T.M. and T. Suda conceived and designed the experiments. H.T., T.M., T.U., Y.T., C.Y., L.H.C., A.W. and T. Sanda performed the experiments. T.M. and R.Y. analyzed bioinformatics data. H.T. and T.M. wrote the manuscript. T.M. and T. Suda supervised the research. All authors reviewed and approved the manuscript. H.T. and T.M. contributed equally to this work.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no financial conflicts of interest.
Peer review
Peer review information
Nature Aging thanks Simón Méndez-Ferrer, Danica Chen and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Flow cytometry gating strategy for HSCs.
(a) Gating Strategy for HSCs in flow cytometry. FSC, forward scatter; FSC-A, FSC-aria; FSC-H, FSC-height; FSC-W, FSC-width; SSC, side scatter; SSC-A, SSC-aria; SSC-H, SSC-height; SSC-W, SSC-width; PI, propidium iodide; Live cells, PI− cells; Lin− cells, lineage− (CD4−CD8−B220−Gr1−CD11b−Ter119−NK1.1−CD127−) cells; LS−K, Lin−Sca1−c-Kit+; LSK, Lin−Sca1+c-Kit+; MPP, CD150−CD48+ LSK; ST-HSC, CD150−CD48− LSK; SLAM (LT-HSC), CD150+CD48− LSK; ESLAM, EPCR+ SLAM-HSC. (b) Gating Strategy for classical HSCs in flow cytometry. LT-HSC, CD34−CD135− LSK; ST-HSC, CD34+CD135− LSK; MPP, CD34+CD135+ LSK; CMP, CD34+CD16/32− LS−K; GMP, CD34+CD16/32+ LS−K; MEP, CD34−CD16/32− LS−K.
Extended Data Fig. 2 Expression of mito-Dendra2 in HSCs and analysis of their phenotype during bone marrow transplantation.
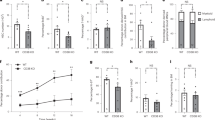
(a) Mitochondrial mass in young and aged HSPCs. Representative histogram of mito-Dendra2 for ST-HSC, MPP, LS−K, and Lin−. (b) Mitochondrial DNA copy number in mito-Dendra2 Low and High HSCs (P = 0.0118, n = 6). (c) EPCR expression in aged ESLAM-HSCs by mitochondrial mass. Relative MFI of EPCR (P = 0.0001, n = 6). (d) Complete blood count post-BMT (n = 6, 8 for Low and High groups). P-values by week: white blood cell - 4 (0.0015), 8 (0.4187), 12 ( < 0.0001), 16 (0.0529); red blood cell - 4 (0.8886), 8 (0.8816), 12 (0.0300), 16 (0.8269); hemoglobin - 4 (0.4995), 8 (0.3988), 12 (0.0037), 16 (0.8101); hematocrit - 4 (0.9067), 8 (0.4799), 12 (0.0092), 16 (0.4611); platelet - 4 (0.0672), 8 (0.8458), 12 (0.5581), 16 (0.6478). (e-g) Proportion of mito-Dendra2+ cell in LSK and LS−K fractions at 16 weeks post-BMT (Low: n = 6, High: n = 8). (e) HSPCs (Extended Data Fig. 1a). P-values: ST-HSC (0.1639), MPP (0.5754), LSK (0.1102), LS−K (0.5728), Lin− (0.5853). (f) HSPCs (Extended Data Fig. 1b). P-values: ST-HSC (CD34+CD135−) (0.2781), MPP (CD34+CD135+) (0.4255), CMP (0.6850), GMP (0.5686), MEP (0.5608). (g) MPP2 (CD150+CD48− ESLAM-HSCs). P = 0.1084. (h) mito-Dendra2 expression in donor-derived cells in the HSC at 16 weeks post-BMT. Due to the lower number of donor-positive cells in the HSC fraction originating from mito-Dendra2 Low cells, the sample size is smaller compared to that of mito-Dendra2 High cells (n = 3, 7 for mito-Dendra2 Low and High). P = 0.1510. (i) Proportion of donor-derived cells post-BMT in young mice (ref. 26; n = 5, 4 for Young-Low and Young-High). P-values: SLAM (0.0374), ST-HSC (0.1513), MPP (0.0975). Note: All charts show mean ± SD. Statistical analysis: two-tailed Student’s t-test (*P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001; ns = not significant).
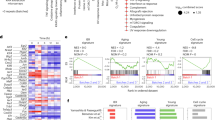
Extended Data Fig. 3 Single cell multiome analysis classified HSCs into four clusters.
(a) A heatmap of multiome analysis of young and aged mito-Dendra2 HSCs, showing the top differentially expressed genes in each cluster. The x-axis represents individual cells and clusters, and the y-axis represents differentially expressed genes. (b) Uniform manifold approximation and projection (UMAP) visualization of HSCs from young and aged mito-Dendra2 mice. scRNA-seq and scATAC-seq data were integrated with Weighted Nearest Neighbor (WNN) analysis. UMAP based on WNN analysis (top), scRNA-seq data only (middle), and scATAC-seq data only (bottom) were shown. In the right panels, young and aged mito-Dendra2 Low and High HSCs were depicted separately. The top leftmost panel is identical to Fig. 3b.
Extended Data Fig. 4 Single cell multiome analysis revealed that aged mito-Dendra2 High HSCs were characterized with HSC signature and oxidative phosphorylation genes.
(a) Violin plots showing differences in Surface Marker Overlap (SuMo) scores (upper) and HSC scores (lower) of young and aged mito-Dendra2 Low and High HSCs in each cluster. In the upper panel, P = 0, 2.99×10−209, 1.04×10−70, and 7.10×10−25 for Clusters 1, 2,3 and 4 (one-way Welch’s ANOVA). *P = 0, 0, 3.26×10−13, and 1.00×10−11 between aged mito-Dendra2 Low and High HSCs for Clusters 1, 2,3 and 4 (Games-Howell post hoc test). In the lower panel, P = 0, 1.90×10−172, 3.83×10−77, and 1.39×10−19 for Clusters 1, 2,3 and 4 (one-way Welch’s ANOVA). *P = 0, 3.77×10−14, 2.42×10−13, and 3.93×10−8 between aged mito-Dendra2 Low and High HSCs for Clusters 1, 2,3 and 4 (Games-Howell post hoc test). (b) Violin plot showing myeloid-biased (left) and lymphoid-biased (right) score. P = 0 and 0 (one-way Welch’s ANOVA). *P = 2.05×10−12 and 1.69×10−12 (Games-Howell post hoc test). (c) Violin plots showing differences in mRNA expression levels of ribosomal genes between aged mito-Dendra2 Low and High HSCs in clusters 1 (left) and 2 (right). *P = 1.13×10−261, 9.30×10−125, 1.93×10−264, 6.27×10−115, 9.12×10−227, 1.31×10−85, 1.62×10−273 and 2.65×10−150 from left to right, calculated by the Wilcoxon Rank Sum test, and adjusted based on Bonferroni correction with all genes using the Seurat package in R in a two-sided manner. (d) Gene set variation analysis (GSVA) using MitoCarta gene sets between aged mito-Dendra2 Low and High HSCs. Each dot represents one gene set. Two-sided adjusted P-values were calculated using the limma package. (e) Violin plots showing differences in mRNA expression levels of selected iron homeostasis genes between aged mito-Dendra2 Low and High HSCs. *P = 5.18×10−84 and 2.48×10−81 from left to right, calculated by the Wilcoxon Rank Sum test, and adjusted based on Bonferroni correction with all genes using the Seurat package in R in a two-sided manner.
Extended Data Fig. 5 scRNA-seq and bulk RNA-seq revealed genes specific to aged HSCs.
(a) Gene set enrichment analysis (GSEA) comparing young and aged LT-HSCs using 50 hallmark gene sets. The top 4 gene sets enriched in aged LT-HSCs are shown. Each solid bar represents one gene within the gene set. NES, normalized enrichment score; FDR, false discovery rate. (b) Violin plots showing mRNA expression levels of Kcnb2 and Clu in each cluster of young mito-Dendra2 Low (cyan), young mito-Dendra2 High (pink), aged mito-Dendra2 Low (blue), and aged mito-Dendra2 High HSCs (red). P = 0, 1.84×10−146, 9.94×10−67, and 2.00×10−38 (Kcnb2), and 5.76×10−100, 7.11×10−52, 8.86×10−25, and 5.37×10−11 (Clu) between aged mito-Dendra2 High HSCs and others from left to right, calculated by the Wilcoxon Rank Sum test and adjusted based on Bonferroni correction with all genes using the Seurat package in R. (c) Box plots of bulk RNA-seq showing mRNA expression levels of selected aged LT-HSC-specific genes in young (pink) and aged (cyan) LT-HSCs, ST-HSCs, and MPPs (n = 4). P = 4.30×10−3, 7.07×10−12, 1.78×10−100, 6.46×10−116, 3.07×10−86, and 0, calculated by the likelihood ratio test and adjusted by the Benjamini and Hochberg method using the DESeq2 package in R. The boxes show median values, 1st and 3rd quartiles. The whiskers extend to the most extreme data point which is no more than 1.5 times the interquartile range from the box.
Extended Data Fig. 6 The relationship between mitochondria and GPR183.
(a) Relative GPR183 expression following SR-18292 treatment, with normalized MFI value (P = 0.0029, n = 6). (b) Relative mito-Dendra2 expression following NIBR-189 and/or 7α,25-DHC treatment, with normalized MFI values (n = 3). Adjusted P-value: NIBR-189−/7α,25-DHC− vs NIBR-189−/7α,25-DHC+ (0.2551), NIBR-189+/7α,25-DHC− vs NIBR-189+/7α,25-DHC+ (0.1046), NIBR-189−/7α,25-DHC− vs NIBR-189+/7α,25-DHC− (0.2310), NIBR-189−/7α,25-DHC+ vs NIBR-189+/7α,25-DHC+ (0.7146). Note: All charts show mean ± SD. Statistical analysis: (a) two-tailed Student’s t-test and (b) one-way ANOVA (*P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001; ns = not significant).
Extended Data Fig. 7 BMT for GPR183neg and GPR183high HSCs.
(a) Complete blood count post-BMT (n = 6, 7 for GPR183neg and GPR183high groups). P-values by week: white blood cell - 4 (0.0817), 8 (0.0987), 12 (0.6245), 16 (0.0755); red blood cell - 4 (0.5566), 8 (0.0567), 12 (0.3505), 16 (0.3763); hemoglobin - 4 (0.4078), 8 (0.1669), 12 (0.8170), 16 (0.6456); hematocrit - 4 (0.4519), 8 (0.0957), 12 (0.4468), 16 (0.3169); platelet - 4 (0.0269), 8 (0.6668), 12 (0.9417), 16 (0.9568). (b) Proportion of CD45.1+ cells in PB. GPR183low (n = 6, 7 for GPR183neg and GPR183high groups). P-values by week: total cells - 4 (0.0195), 8 (0.0153), 12 (0.0219), 16 (0.0290); granulocytes - 4 (0.0049), 8 (0.0139), 12 (0.0056), 16 (0.0086); B-cells - 4 (0.0659), 8 (0.0829), 12 (0.0937), 16 (0.0933); T-cells - 4 (0.8150), 8 (0.1236), 12 (0.1091), 16 (0.2046). (c-d) Proportion of CD45.1+ cell in LSK and LS−K fractions at 16 weeks post-BMT (n = 6, 7 for GPR183neg and GPR183high groups). (c) HSPCs (Extended Data Fig. 1a). P-values: ST-HSC (0.0365), MPP (0.0268), LSK (0.0229), LS−K (0.0301), Lin− (0.0374). (d) HSPCs (Extended Data Fig. 1b). P-values: ST-HSC (CD34+CD135−) (0.0256), MPP (CD34+CD135+) (0.0370), CMP (0.0250), GMP (0.0205), MEP (0.0385). Note: All charts show mean ± SD. Statistical analysis: two-tailed Student’s t-test (*P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001; ns = not significant).
Supplementary information
Source data
Source Data Fig. 1
Statistical Source Data.
Source Data Fig. 2
Statistical Source Data.
Source Data Fig. 3
Statistical Source Data.
Source Data Fig. 4
Statistical Source Data.
Source Data Fig. 5
Statistical Source Data.
Source Data Fig. 6
Statistical Source Data.
Source Data Fig. 7
Statistical Source Data.
Source Data Extended Data Fig. 2
Statistical Source Data.
Source Data Extended Data Fig. 4
Statistical Source Data.
Source Data Extended Data Fig. 5
Statistical Source Data.
Source Data Extended Data Fig. 6
Statistical Source Data.
Source Data Extended Data Fig. 7
Statistical Source Data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Totani, H., Matsumura, T., Yokomori, R. et al. Mitochondria-enriched hematopoietic stem cells exhibit elevated self-renewal capabilities, thriving within the context of aged bone marrow. Nat Aging 5, 831–847 (2025). https://doi.org/10.1038/s43587-025-00828-y
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s43587-025-00828-y