Abstract
Cardiomyocyte maturation is crucial for generating adult cardiomyocytes and the application of human pluripotent stem cell-derived cardiomyocytes (hPSC-CMs). However, regulation at the cis-regulatory element level and its role in heart disease remain unclear. Alpha-actinin 2 (ACTN2) levels increase during CM maturation. In this study, we investigated a clinically relevant, conserved ACTN2 enhancer’s effects on CM maturation using hPSC and mouse models. Heterozygous ACTN2 enhancer deletion led to abnormal CM morphology, reduced function and mitochondrial respiration. Transcriptomic analyses in vitro and in vivo showed disrupted CM maturation and upregulated anabolic mammalian target for rapamycin (mTOR) signaling, promoting senescence and hindering maturation. As confirmation, ACTN2 enhancer deletion induced heat shock protein 90A expression, a chaperone mediating mTOR activation. Conversely, targeting the ACTN2 enhancer via enhancer CRISPR activation (enCRISPRa) promoted hPSC-CM maturation. Our studies reveal the transcriptional enhancer’s role in cardiac maturation and disease, offering insights into potentially fine-tuning gene expression to modulate cardiomyocyte physiology.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Murphy, S. A., Chen, E. Z., Tung, L., Boheler, K. R. & Kwon, C. Maturing heart muscle cells: mechanisms and transcriptomic insights. Semin. Cell Dev. Biol. 119, 49–60 (2021).
Guo, Y. & Pu, W. T. Cardiomyocyte maturation: new phase in development. Circ. Res. 126, 1086–1106 (2020).
Karbassi, E. et al. Cardiomyocyte maturation: advances in knowledge and implications for regenerative medicine. Nat. Rev. Cardiol. 17, 341–359 (2020).
Gomez-Garcia, M. J., Quesnel, E., Al-Attar, R., Laskary, A. R. & Laflamme, M. A. Maturation of human pluripotent stem cell derived cardiomyocytes in vitro and in vivo. Semin. Cell Dev. Biol. 118, 163–171 (2021).
Jiang, Y., Park, P., Hong, S. M. & Ban, K. Maturation of cardiomyocytes derived from human pluripotent stem cells: current strategies and limitations. Mol. Cells 41, 613–621 (2018).
Cho, G. S. et al. Neonatal transplantation confers maturation of PSC-derived cardiomyocytes conducive to modeling cardiomyopathy. Cell Rep. 18, 571–582 (2017).
Sjoblom, B., Salmazo, A. & Djinovic-Carugo, K. α-actinin structure and regulation. Cell. Mol. Life Sci. 65, 2688–2701 (2008).
Sequeira, V., Nijenkamp, L. L., Regan, J. A. & van der Velden, J. The physiological role of cardiac cytoskeleton and its alterations in heart failure. Biochim. Biophys. Acta 1838, 700–722 (2014).
Murphy, A. C. & Young, P. W. The actinin family of actin cross-linking proteins—a genetic perspective. Cell Biosci. 5, 49 (2015).
Frank, D. & Frey, N. Cardiac Z-disc signaling network. J. Biol. Chem. 286, 9897–9904 (2011).
Frank, D., Kuhn, C., Katus, H. A. & Frey, N. The sarcomeric Z-disc: a nodal point in signalling and disease. J. Mol. Med. (Berl.) 84, 446–468 (2006).
Cukovic, D., Lu, G. W., Wible, B., Steele, D. F. & Fedida, D. A discrete amino terminal domain of Kv1.5 and Kv1.4 potassium channels interacts with the spectrin repeats of α-actinin-2. FEBS Lett. 498, 87–92 (2001).
Ziane, R. et al. Cell membrane expression of cardiac sodium channel Nav1.5 is modulated by α-actinin-2 interaction. Biochemistry 49, 166–178 (2010).
Lu, L. et al. Molecular coupling of a Ca2+-activated K+ channel to L-type Ca2+ channels via α-actinin2. Circ. Res. 100, 112–120 (2007).
Chopra, A. et al. Force generation via β-cardiac myosin, titin, and α-actinin drives cardiac sarcomere assembly from cell-matrix adhesions. Dev. Cell 44, 87–96 (2018).
Arvanitis, M. et al. Genome-wide association and multi-omic analyses reveal ACTN2 as a gene linked to heart failure. Nat. Commun. 11, 1122 (2020).
Cai, W. et al. An unbiased proteomics method to assess the maturation of human pluripotent stem cell-derived cardiomyocytes. Circ. Res. 125, 936–953 (2019).
Guo, Y. et al. Sarcomeres regulate murine cardiomyocyte maturation through MRTF-SRF signaling. Proc. Natl Acad. Sci. USA 118, e2008861118 (2021).
Gao, T. et al. Identification and subcellular localization of the subunits of L-type calcium channels and adenylyl cyclase in cardiac myocytes. J. Biol. Chem. 272, 19401–19407 (1997).
Sadeghi, A., Doyle, A. D. & Johnson, B. D. Regulation of the cardiac L-type Ca2+ channel by the actin-binding proteins α-actinin and dystrophin. Am. J. Physiol. Cell Physiol. 282, C1502–C1511 (2002).
Sewanan, L. R. & Campbell, S. G. Modelling sarcomeric cardiomyopathies with human cardiomyocytes derived from induced pluripotent stem cells. J. Physiol. 598, 2909–2922 (2020).
Schwan, J. et al. Anisotropic engineered heart tissue made from laser-cut decellularized myocardium. Sci. Rep. 6, 32068 (2016).
Kannan, S., Farid, M., Lin, B. L., Miyamoto, M. & Kwon, C. Transcriptomic entropy benchmarks stem cell-derived cardiomyocyte maturation against endogenous tissue at single cell level. PLoS Comput. Biol. 17, e1009305 (2021).
Yang, X., Pabon, L. & Murry, C. E. Engineering adolescence: maturation of human pluripotent stem cell-derived cardiomyocytes. Circ. Res. 114, 511–523 (2014).
Murphy, S. A. et al. PGC1/PPAR drive cardiomyocyte maturation at single cell level via YAP1 and SF3B2. Nat. Commun. 12, 1648 (2021).
Castro-Mondragon, J. A. et al. JASPAR 2022: the 9th release of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 50, D165–D173 (2022).
ENCODE Project Consortiumet al. Expanded encyclopaedias of DNA elements in the human and mouse genomes. Nature 583, 699–710 (2020).
Akerberg, B. N. et al. A reference map of murine cardiac transcription factor chromatin occupancy identifies dynamic and conserved enhancers. Nat. Commun. 10, 4907 (2019).
He, A. et al. Dynamic GATA4 enhancers shape the chromatin landscape central to heart development and disease. Nat. Commun. 5, 4907 (2014).
Zhu, L. et al. Transcription factor GATA4 regulates cell type–specific splicing through direct interaction with RNA in human induced pluripotent stem cell–derived cardiac progenitors. Circulation 146, 770–787 (2022).
Funk, C. C. et al. Atlas of transcription factor binding sites from ENCODE DNase hypersensitivity data across 27 tissue types. Cell Rep. 32, 108029 (2020).
Akerberg, B. N. & Pu, W. T. Genetic and epigenetic control of heart development. Cold Spring Harb. Perspect. Biol. 12, a036756 (2020).
Zhou, P. et al. Dynamic changes in P300 enhancers and enhancer-promoter contacts control mouse cardiomyocyte maturation. Dev. Cell 58, 898–914 (2023).
Wahlstrom, G., Norokorpi, H. L. & Heino, T. I. Drosophila α-actinin in ovarian follicle cells is regulated by EGFR and Dpp signalling and required for cytoskeletal remodelling. Mech. Dev. 123, 801–818 (2006).
Brown, J. B. et al. Diversity and dynamics of the Drosophila transcriptome. Nature 512, 393–399 (2014).
Negre, N. et al. A cis-regulatory map of the Drosophila genome. Nature 471, 527–531 (2011).
Shokri, L. et al. A comprehensive Drosophila melanogaster transcription factor interactome. Cell Rep. 27, 955–970 (2019).
Luo, Y. et al. New developments on the Encyclopedia of DNA Elements (ENCODE) data portal. Nucleic Acids Res. 48, D882–D889 (2020).
Funakoshi, S. et al. Generation of mature compact ventricular cardiomyocytes from human pluripotent stem cells. Nat. Commun. 12, 3155 (2021).
Skorska, A. et al. Monitoring the maturation of the sarcomere network: a super-resolution microscopy-based approach. Cell. Mol. Life Sci. 79, 149 (2022).
Tzahor, E. & Poss, K. D. Cardiac regeneration strategies: staying young at heart. Science 356, 1035–1039 (2017).
Kannan, S. & Kwon, C. Regulation of cardiomyocyte maturation during critical perinatal window. J. Physiol. 598, 2941–2956 (2020).
Sciarretta, S., Forte, M., Frati, G. & Sadoshima, J. The complex network of mTOR signalling in the heart. Cardiovasc. Res. 118, 424–439 (2022).
Szwed, A., Kim, E. & Jacinto, E. Regulation and metabolic functions of mTORC1 and mTORC2. Physiol. Rev. 101, 1371–1426 (2021).
Garbern, J. C. et al. Inhibition of mTOR signaling enhances maturation of cardiomyocytes derived from human-induced pluripotent stem cells via p53-induced quiescence. Circulation 141, 285–300 (2020).
Liu, G. Y. & Sabatini, D. M. mTOR at the nexus of nutrition, growth, ageing and disease. Nat. Rev. Mol. Cell Biol. 21, 183–203 (2020).
Garbern, J. C., Escalante, G. O. & Lee, R. T. Pluripotent stem cell-derived cardiomyocytes for treatment of cardiomyopathic damage: current concepts and future directions. Trends Cardiovasc. Med. 31, 85–90 (2021).
Ladha, F. A. et al. Actinin BioID reveals sarcomere crosstalk with oxidative metabolism through interactions with IGF2BP2. Cell Rep. 36, 109512 (2021).
Roberts, R. J., Hallee, L. & Lam, C. K. The potential of Hsp90 in targeting pathological pathways in cardiac diseases. J. Pers. Med. 11, 1373 (2021).
Abeyrathna, P. & Su, Y. The critical role of Akt in cardiovascular function. Vascul. Pharmacol. 74, 38–48 (2015).
Hu, B. et al. Binding of the pathogen receptor HSP90AA1 to avibirnavirus VP2 induces autophagy by inactivating the AKT-MTOR pathway. Autophagy 11, 503–515 (2015).
Hutz, J. E., Manning, W. A., Province, M. A. & McLeod, H. L. Genomewide analysis of inherited variation associated with phosphorylation of PI3K/AKT/mTOR signaling proteins. PLoS ONE 6, e24873 (2011).
Zech, A. T. L. et al. ACTN2 mutant causes proteopathy in human iPSC-derived cardiomyocytes. Cells 11, 2745 (2022).
Yotti, R., Seidman, C. E. & Seidman, J. G. Advances in the genetic basis and pathogenesis of sarcomere cardiomyopathies. Annu. Rev. Genomics Hum. Genet. 20, 129–153 (2019).
Thompson, B. R. & Metzger, J. M. Cell biology of sarcomeric protein engineering: disease modeling and therapeutic potential. Anat. Rec. (Hoboken) 297, 1663–1669 (2014).
Li, K. et al. Interrogation of enhancer function by enhancer-targeting CRISPR epigenetic editing. Nat. Commun. 11, 485 (2020).
Hilton, I. B. et al. Epigenome editing by a CRISPR–Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat. Biotechnol. 33, 510–517 (2015).
Heintzman, N. D. et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature 459, 108–112 (2009).
VanDusen, N. J. et al. Massively parallel in vivo CRISPR screening identifies RNF20/40 as epigenetic regulators of cardiomyocyte maturation. Nat. Commun. 12, 4442 (2021).
Prondzynski, M. et al. Disease modeling of a mutation in α-actinin 2 guides clinical therapy in hypertrophic cardiomyopathy. EMBO Mol. Med. 11, e11115 (2019).
Lindholm, M. E. et al. Mono- and biallelic protein-truncating variants in α-actinin 2 cause cardiomyopathy through distinct mechanisms. Circ. Genom. Precis. Med. 14, e003419 (2021).
Ahmed, R. E., Tokuyama, T., Anzai, T., Chanthra, N. & Uosaki, H. Sarcomere maturation: function acquisition, molecular mechanism, and interplay with other organelles. Philos. Trans. R. Soc. Lond. B Biol. Sci. 377, 20210325 (2022).
Avellaneda, J. et al. Myofibril and mitochondria morphogenesis are coordinated by a mechanical feedback mechanism in muscle. Nat. Commun. 12, 2091 (2021).
Wickramasinghe, N. M. et al. PPARdelta activation induces metabolic and contractile maturation of human pluripotent stem cell-derived cardiomyocytes. Cell Stem Cell 29, 559–576 (2022).
Tokuyama, T., Ahmed, R. E., Chanthra, N., Anzai, T. & Uosaki, H. Disease modeling of mitochondrial cardiomyopathy using patient-specific induced pluripotent stem cells. Biology (Basel) 10, 981 (2021).
Kageyama, Y. et al. Parkin-independent mitophagy requires Drp1 and maintains the integrity of mammalian heart and brain. EMBO J. 33, 2798–2813 (2014).
Guo, Y. et al. Hierarchical and stage-specific regulation of murine cardiomyocyte maturation by serum response factor. Nat. Commun. 9, 3837 (2018).
Zhang, D. et al. Mitochondrial cardiomyopathy caused by elevated reactive oxygen species and impaired cardiomyocyte proliferation. Circ. Res. 122, 74–87 (2018).
Dupays, L. et al. Sequential binding of MEIS1 and NKX2-5 on the Popdc2 gene: a mechanism for spatiotemporal regulation of enhancers during cardiogenesis. Cell Rep. 13, 183–195 (2015).
Bailey, S. D. et al. Noncoding somatic and inherited single-nucleotide variants converge to promote ESR1 expression in breast cancer. Nat. Genet. 48, 1260–1266 (2016).
Lu, X. et al. Global discovery of lupus genetic risk variant allelic enhancer activity. Nat. Commun. 12, 1611 (2021).
Deplancke, B., Alpern, D. & Gardeux, V. The genetics of transcription factor DNA binding variation. Cell 166, 538–554 (2016).
Reddy, T. E. et al. Effects of sequence variation on differential allelic transcription factor occupancy and gene expression. Genome Res. 22, 860–869 (2012).
Menon, S. et al. Spatial control of the TSC complex integrates insulin and nutrient regulation of mTORC1 at the lysosome. Cell 156, 771–785 (2014).
Demetriades, C., Doumpas, N. & Teleman, A. A. Regulation of TORC1 in response to amino acid starvation via lysosomal recruitment of TSC2. Cell 156, 786–799 (2014).
Manning, B. D., Tee, A. R., Logsdon, M. N., Blenis, J. & Cantley, L. C. Identification of the tuberous sclerosis complex-2 tumor suppressor gene product tuberin as a target of the phosphoinositide 3-kinase/Akt pathway. Mol. Cell 10, 151–162 (2002).
Hudson, C. C. et al. Regulation of hypoxia-inducible factor 1α expression and function by the mammalian target of rapamycin. Mol. Cell. Biol. 22, 7004–7014 (2002).
Duvel, K. et al. Activation of a metabolic gene regulatory network downstream of mTOR complex 1. Mol. Cell 39, 171–183 (2010).
Hu, D. et al. Metabolic maturation of human pluripotent stem cell-derived cardiomyocytes by inhibition of HIF1α and LDHA. Circ. Res. 123, 1066–1079 (2018).
Etard, C., Roostalu, U. & Strahle, U. Shuttling of the chaperones Unc45b and Hsp90a between the A band and the Z line of the myofibril. J. Cell Biol. 180, 1163–1175 (2008).
Martin, T. G. & Kirk, J. A. Under construction: the dynamic assembly, maintenance, and degradation of the cardiac sarcomere. J. Mol. Cell. Cardiol. 148, 89–102 (2020).
Srikakulam, R. & Winkelmann, D. A. Chaperone-mediated folding and assembly of myosin in striated muscle. J. Cell Sci. 117, 641–652 (2004).
Giulino-Roth, L. et al. Inhibition of Hsp90 suppresses PI3K/AKT/mTOR signaling and has antitumor activity in Burkitt lymphoma. Mol. Cancer Ther. 16, 1779–1790 (2017).
Ranek, M. J., Stachowski, M. J., Kirk, J. A. & Willis, M. S. The role of heat shock proteins and co-chaperones in heart failure. Philos. Trans. R. Soc. Lond. B Biol. Sci. 373, 20160530 (2018).
Zhao, X. H., Peng, Y. Z., Wang, Y. Y. & Huang, Y. S. [Influence of heat shock protein 90 on protein serine threonine kinases expression in hypoxic cardiomyocytes]. Zhonghua Shao Shang Za Zhi 23, 265–268 (2007).
Bartha, E. et al. Regulation of kinase cascade activation and heat shock protein expression by poly(ADP-ribose) polymerase inhibition in doxorubicin-induced heart failure. J. Cardiovasc. Pharmacol. 58, 380–391 (2011).
Joung, J. et al. Genome-scale CRISPR–Cas9 knockout and transcriptional activation screening. Nat. Protoc. 12, 828–863 (2017).
Tian, R. et al. CRISPR interference-based platform for multimodal genetic screens in human iPSC-derived neurons. Neuron 104, 239–255 (2019).
Tampakakis, E. et al. Heart neurons use clock genes to control myocyte proliferation. Sci. Adv. 7, eabh4181 (2021).
Ackers-Johnson, M. et al. A simplified, Langendorff-free method for concomitant isolation of viable cardiac myocytes and nonmyocytes from the adult mouse heart. Circ. Res. 119, 909–920 (2016).
Parekh, S., Ziegenhain, C., Vieth, B., Enard, W. & Hellmann, I. zUMIs—a fast and flexible pipeline to process RNA sequencing data with UMIs. Gigascience 7, giy059 (2018).
Hofbauer, P. et al. Cardioids reveal self-organizing principles of human cardiogenesis. Cell 184, 3299–3317 (2021).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Kim, D., Langmead, B. & Salzberg, S. L. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360 (2015).
Liao, Y., Smyth, G. K. & Shi, W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014).
Anders, S., Pyl, P. T. & Huber, W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015).
Koleini, N. et al. Elimination or neutralization of endogenous high-molecular-weight FGF2 mitigates doxorubicin-induced cardiotoxicity. Am. J. Physiol. Heart Circ. Physiol. 316, H279–H288 (2019).
Sanson, K. R. et al. Optimized libraries for CRISPR–Cas9 genetic screens with multiple modalities. Nat. Commun. 9, 5416 (2018).
Acknowledgements
The authors would like to thank all members of the Tampakakis, Kwon and Kass laboratories and M. Ranek at Johns Hopkins University for insightful comments and recommendations. We would also like to thank A. Cammarato for kindly providing Drosophila melanogaster flies. E.T. is supported by grants from the National Heart, Lung, and Blood Institute (NHLBI) (HL-145135), the American Heart Association (AHA) (CDA34660077), the W. W. Smith Charitable Trust, the Magic That Matters Fund, the Johns Hopkins University Catalyst Award and the Maryland Stem Cell Research Fund (MSCRF) (2023-MSCRFL-5984). S.M., E.C. and C.K. were supported by grants from the National Institutes of Health (NIH)/NHLBI (R01HL156947, T32HL007227), the AHA (TPA1058685) and the MSCRF (MSCRFD-6139). M.A. was supported by the NIH/NHLBI (K08 HL166690). B.L.L was supported by the NIH/NHLBI (K99 HL15584).
Author information
Authors and Affiliations
Contributions
M.H. designed, carried out and supervised this work and wrote the manuscript. H.G., S.L., S.B. and A.Z. assisted with experimental work. M.A. performed bioinformatics analyses. B.L.L. assisted with cell isolation and functional analyses. S.M. performed bioinformatics analyses. E.C. assisted with in vitro free fatty acid treatments and bioinformatics. N.K. assisted with mouse cardiomyocyte isolation, Seahorse analyses and co-immunoprecipitation. C.K. assisted with experimental design. E.T. designed and supervised this work, assisted with experimental work and wrote the manuscript. All authors read and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Cardiovascular Research thanks Kailong Li, Sean Wu and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
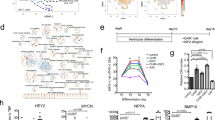
Extended Data Fig. 2 ACTN2 enhancer deletion does not disrupt the differentiation of hPSC-CMs or gene splicing.
a. Gating strategy used to detect differentiated hPSC-CMs. b. Flow cytometry analysis of ACTN2 enh del hPSC-CMs vs isogenic controls after immunostaining for cardiac Troponin T. The gating strategy used to detect hPSC-CMs for flow cytometry analysis is represented. The differentiation efficiency of both hPSC lines is similar. c. Map illustrating the alternatively spliced ACTN2 variants, and the RT-PCR products based on the different primer sets. d. RT-PCR using cDNA generated from ACTN2 enh del hPSC-CMs vs isogenic controls showing no difference in the size of alternatively spliced ACTN2 transcripts. Similar results were obtained after repeating the experiment 4 independent times.
Extended Data Fig. 3 ACTN2 enhancer deletion increases the transcriptomic entropy of hPSC-CMs.
Entropy score calculated from hPSC-CM our single cell RNA-Seq data (one differentiation batch) shows increased entropy in ACTN2 enh del cardiomyocytes suggestive of disrupted maturation. The center line of the box corresponds to the 50th percentile. The lower and upper bounds of the boxes correspond to the 25th and the 75th percentiles. The upper whisker marks the largest value within 1.5 times interquartile range above the 75th percentile. The lower whisker marks the smallest value within 1.5 times interquartile range below the 25th percentile. The interquartile range is defined as the distance between the first and the third quartiles.
Extended Data Fig. 4 Luciferase assay to validate the ACTN2 enhancer conserved region.
a. Illustration of the different luciferase vectors tested. b. Luciferase analysis of hPSC-CMs transfected with minimal promoter vs the conserved ACTN2 enhancer sequence with and without the rs535411 variant, showing increased luciferase activity in the presence of ACTN2 enhancer. This effect was not observed in cells transfected with the mutant variant. c-d. Overexpression of MEF2A and MEF2C transcription factors through modified mRNA transfection in hPSC-CMs resulted in markedly increased luciferase activity in both enhancer vectors. However, this induction was significantly lower in cells with ACTN2 rs535411 enhancer variant. All graphs report Luciferase compared to minimal promoter. (4 independent experiments were analyzed). Data are presented as mean values +/− SEM. All replicates are biological. The Shapiro-Wilk test was performed to assess normal distribution, and one-way ANOVA with Bonferroni’s multiple comparisons test or Kruskal-Wallis with Dunn’s multiple comparisons test were used as appropriate. Only P values < 0.1 are reported.
Extended Data Fig. 5 Temporal ACTN2 enhancer activation in mice.
a. Analysis of ENCODE ChIP-Seq datasets for H3K27Ac and H3K4me1 from mouse hearts at different developmental stages showed increased ACTN2 enhancer activation in older mice supporting a role in the temporal regulation of ACTN2 gene expression. b. Analysis of P300 occupancy data from mouse hearts at different developmental stages, similarly, showed an increased in P300 peaks in postnatal hearts. c. Mef2a ChIP-Seq data showing binding of Mef2a in the ACTN2 enhancer region in adult mouse hearts.
Extended Data Fig. 6 Temporal activation of Actn enhancer regions in Drosophila melanogaster.
a. H3K27Ac ChIP-Sequencing analysis in Drosophila melanogaster reveals an extra active enhancer region upstream of the actinin (Actn) gene promoter that is only present in adult flies. b. Alternatively spliced Actn gene variants (Actn-RB, RC, RD) are expressed at different developmental stages (larva vs fly), likely temporarily regulated by different transcriptional enhancers such as the one revealed by H3K27Ac ChIP-Seq. (5 independent experiments were analyzed). Data are presented as mean values +/− SEM. All replicates are biological.
Extended Data Fig. 7 Actn2 expression is reduced in Actn2 enh del mouse hearts.
Actn2 protein is decreased in both heterozygous and homozygous Actn2 enh del hearts. Data are presented as mean values +/− SEM. The Shapiro-Wilk test was performed to assess normal distribution, and one-way ANOVA with Bonferroni’s multiple comparisons test was used to compare all groups with control. (4 samples per group were analyzed). All replicates are biological. Only P values < 0.1 are reported.
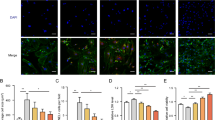
Extended Data Fig. 8 Suppression of mTORC1 improves the maturation of ACTN2 enh del hPSC-CMs.
a. Gene expression of RAPTOR and 3-phosphoinositidine-dependent protein kinase-1 (PDK1) as part of mTORC1 pathway were upregulated, while RICTOR levels which is part of the mTORC2 complex remained unchanged. (3 independent samples per group). b. Both ACTN2 enh del hPSC-CMs and isogenic controls showed very similar cell size after treatment with everolimus (total ~147 cells/group, 4 independent treatments). c. The morphology of ACTN2 enh del hPSC-CMs did not show any difference as compared to controls after treatment with everolimus. (4 independent samples per group) (bar graph: 25μm) d. ACTN2 enh del hPSC-CMs treated with everolimus (25 μM, mTORC1 inhibitor) normalized their calcium handling properties as compared to treated control hPSC-CMs. Time to peak, peak height and time to 50% to baseline are not different between control and ACTN2 enh del hPSC-CMs. (n = 8–10 batches per group). e. Seahorse analysis of cultured hPSC-CMs show reduced maximum oxygen consumption rate (OCR) and ECAR to OCR ratio (consistent with increased glycolysis and reduced oxidative phosphorylation) for ACTN2 enh del hPSC-CMs compared to isogenic controls. Treatment with everolimus, improved oxygen consumption for both for ACTN2 enh del hPSC-CMs and isogenic controls and normalized glycolysis for ACTN2 enh del hPSC-CMs. (4 independent samples per group). f. Both ACTN2 enh del hPSC-CMs and isogenic controls developed very similar gene expression of maturation related genes after everolimus treatment (3 independent samples per group). Data are presented as mean values +/− SEM. All replicates are biological. The Shapiro-Wilk test was performed to assess normal distribution, and the parametric student t-test or ANOVA with Bonferroni’s multiple comparisons test or the non-parametric Mann-Whitney (two-tailed) or Kruskal Willis with Dunn’s multiple comparisons tests were used as appropriate. Only P values < 0.1 are reported.
Extended Data Fig. 9 ACTN2 directly interacts with HSP90A.
a. Co-immunoprecipitation of ACTN2 with HSPA90A in both control and ACTN2 enh del hPSC-CMs, supporting the direct interaction between the two proteins. Despite the reduced ACTN2 levels in ACTN2 enh del hPSC-CMs, the ACTN2 immunoprecipitate appears more enriched with the inducible chaperone HSPA90A. b. Transfection of hPSC-CMs with HSP90AA1 siRNAs suppressed HSP90A protein levels in control hPSC-CMs. (3 independent experiments were analyzed) c. siRNA downregulation of HSP90A in ACTN2 enh del hPSC-CMs. (3 independent experiments were analyzed). Data are presented as mean values +/− SEM. All replicates are biological. The Shapiro-Wilk test was performed to assess normal distribution, and paired student parametric t-test (two-tailed) was used for all comparisons. Only P values < 0.1 are reported.
Extended Data Fig. 10 Leveraging enCRISPRa to upregulate ACTN2.
Transfection of hPSC-CMs with sgRNAs targeting the 5’ border of the conserved ACTN2 enhancer region, upregulated ACTN2 protein. Relative protein expression compared to sgRNA1 is presented. (3 independent experiments were analyzed). Data are presented as mean values +/− SEM. All replicates are biological. The Shapiro-Wilk test was performed to assess normal distribution, and paired student parametric t-test (two-tailed) was used for all comparisons.
Supplementary information
Supplementary Table 1
List of all gRNAs used in this study
Supplementary Table 2
List of all primers used for hPSC-CM qPCR
Supplementary Table 3
List of all primers used for mouse and Drosophila qPCR
Supplementary Table 4
List of primers used for ChIP qPCR
Supplementary Table 5
List of all antibodies
Source data
Source Data Fig.1
Unprocessed western blots for Fig. 1
Source Data
Statistical Source Data
Source Data
Source data for extended data figures
Source Data Fig. 7
Unprocessed western blots for Fig. 7
Source Data Extended Data Fig./Table 7
Source data for Extended Data Fig. 7
Source Data Extended Data Fig./Table 9
Source data for Extended Data Fig. 9
Source Data Extended Data Fig.10
Source data for Extended Data Fig. 10
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Htet, M., Lei, S., Bajpayi, S. et al. A transcriptional enhancer regulates cardiac maturation. Nat Cardiovasc Res 3, 666–684 (2024). https://doi.org/10.1038/s44161-024-00484-2
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s44161-024-00484-2
This article is cited by
-
Molecular gatekeepers of endogenous adult mammalian cardiomyocyte proliferation
Nature Reviews Cardiology (2025)
-
Cardiac ACTN2 enhancer regulates cardiometabolism and maturation
Nature Cardiovascular Research (2024)