Abstract
In response to stress, the body attempts to maintain protein homeostasis through intron retention (IR). By identifying the genes that undergo IR in response to stress, possibly we can identify what type of stress the body is experiencing. Using this principle, we reanalyzed RNA sequencing data of patients with treatment-resistant depression and examined IR genes to investigate their characteristics to ketamine. The ketamine nonresponders were characterized by a state of elevated viral infection. Several IR genes related to viral infection were restored to a healthy state by ketamine, regardless of whether the individual was a nonresponder or a responder. Accordingly, it was more reasonable to describe nonresponders not as individuals for whom ketamine is ineffective but rather as individuals for whom ketamine is insufficient to overcome the extremely elevated inflammatory state of these individuals. This study is pioneering in its exploration of the molecular underpinnings that differentiate between nonresponders and responders.
Similar content being viewed by others
Introduction
Major depressive disorder (MDD) is a debilitating disorder that causes high individual morbidity and a significant socioeconomic burden, and it has a profound impact on both the mental and physical health of affected individuals1,2. Research into new and more effective treatments for depression has been limited by an incomplete understanding of its underlying pathophysiology. Despite the availability of many psychopharmacologic and psychotherapeutic treatment options, approximately one-third of patients do not achieve remission after multiple treatment attempts. The majority of patients with MDD who do not adequately respond to these medications are commonly referred to as having treatment-resistant depression (TRD). TRD can impose economic burdens and cause human suffering; for instance, TRD can result in high medical costs, increased use of medical resources, reduced quality of life, increased risk of relapse, and increased mortality rates3,4,5,6,7. Unsurprisingly, one of the most serious complications associated with TRD is an increased risk of suicide8. Although TRD patients account for the majority of MDD patients, the mechanisms underlying treatment resistance are poorly understood; thus, understanding the pathophysiology of TRD and identifying alternative treatment strategies are important research goals. Therefore, increasing the understanding of the biology of TRD and identifying new treatment targets and blood biomarkers that can be used to monitor baseline treatment responses are necessary9.
Accumulating evidence suggests that bidirectional interactions between the peripheral immune system and the brain contribute to the etiology of neuropsychiatric disorders10,11,12,13,14. Indeed, some patients with MDD are known to have chronic inflammation, as indicated by elevated levels of CRP or inflammatory cytokines in the blood. In his book “The Inflamed Mind”, Bullmore suggested that stress causes inflammation and that inflammation causes depression15. In addition, research has shown that changes in peripheral cytokine levels are associated with the response of MDD patients to antidepressant treatment and that inflammatory states are associated with treatment resistance. Cathomas et al.16 made a breakthrough in understanding the molecular mechanisms underlying TRD by showing that TRD patients have a blood transcript signature that indicates higher immune pathway activation at baseline than healthy controls. These authors hypothesized that this signature could be reversed by ketamine treatment, and they compared the baseline transcriptional signatures of these groups before treatment to investigate differences between ketamine responders and nonresponders; however, no differences were found in their analysis16.
Intron retention (IR), which is a type of alternative splicing, was previously thought to simply reflect errors in pre-mRNA splicing; however, it has recently been suggested that IR is a biologically meaningful phenomenon because an increase or decrease of introns in a given transcript is associated with specific biological phenomena, such as cell differentiation17,18,19,20, aging21, and carcinogenesis22. Using mouse models, we previously showed that the degree of IR increases in response to stress in aging mice, such as Klotho mice23 and SAMP8 mice24, and that the degree of IR can be restored to a healthy state by the administration of Japanese herbal medicine23,24. On the basis of these data, we hypothesize that IR is a regulatory mechanism that maintains protein expression homeostasis via alterations that occur during RNA processing25. Notably, gene expression is regulated mainly at the transcription level via the functions of transcription factors; however, stress conditions require the regulation of gene expression via IR even before any obvious changes in transcription occur. Acceleration is expressed as a decrease in introns (called DecIR), and deceleration is expressed as an increase in introns (called IncIR). The genes that require fine-tuning during stress are determined by the particular type of stress. Not all genes are regulated by IR, but of the 20,000 to 30,000 genes in the entire genome, 10 to 20% require the most stringent expression regulation during stress, and therefore, these genes are likely to be selected for IR. As our previous studies and those of others21,23 have shown, the loci of genes that undergo IR are relatively GC-rich and have short intron lengths. In other words, the set of genes that undergo IR during stress is genetically predetermined and evolutionarily conserved26,27, and the total set of genes that undergo IR (IRGs) can be considered to define the nature of the stress that causes IR.
Based on the context provided above, it is possible that genes that are affected by IR (hereafter referred to as IRGs) may function as sensors that detect disruptions in cell homeostasis25. Therefore, by analyzing IRGs in patients with depression, it would be possible to identify the stress factors that are experienced by these patients, and it has been suggested that such regulatory genes may underlie depression. Given this background, we explored the possibility of using the incidence of IR to investigate the etiology of depression, and indeed, we discovered that IRGs are excellent markers of depression in our recent study9.
In the present study, we sought to clarify what TRD is as an extension of that study9. Previous studies from several laboratories have reported that inflammation is significantly increased in TRD patients (see a review by Johnston et al.)28. Consistent with such previous studies, we found that inflammation is likely exacerbated by viral infection in nonresponders of TRD patients by analyzing IR genes in TRD patients using RNA-seq data on TRD reported by Cathomas et al.16. Furthermore, since ketamine is effective not only in responders but also in nonresponders of TRD, it was suggested that nonresponders may remain so because inflammation is exacerbated to a degree that ketamine’s effects cannot keep pace. This interpretation requires further confirmation using various scientific tools. However, given that Cathomas’ DEG analysis did not fully demonstrate the efficacy of ketamine, the IR used in this study is considered a valid method to distinguish responders from nonresponders at the molecular level.
Methods
Downloading and preprocessing of RNA sequencing (RNA-seq) data
The paired-end FASTQ files of whole-blood RNA-seq data from patients with TRD and healthy controls were downloaded from the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) database under accession number GSE18585516. The subjects included 21 healthy controls (CON) and 26 patients with TRD. The TRD patients were divided according to their sensitivity to ketamine, and their sequencing data were recorded as 18 responders (Res-PRE) and eight nonresponders (nonRes-PRE) prior to ketamine administration. Regarding the data after ketamine administration, there were 13 responders (Res-POST) and eight nonresponders (nonRes-POST). However, the data from five responders were not included in GSE185855. The sequencing data were processed by removing Illumina adapter sequences using Cutadapt v1.1629 (options: -j 8 -m 30 -b GATCGGAAGAGCACACGTCTGAACTCCAGTCAC -b AGATCGGAAGAGCGTCGTGTAGGGAAAGAGTGT). Poly(A) tails were trimmed using the FASTX-Toolkit v0.0.14 (http://hannonlab.cshl.edu/fastx_toolkit/). Low-quality bases and sequences were further filtered using fastq_quality_trimmer (options: -t 20 -l 30 -Q 33) and fastq_quality_filter (options: -q 20 -p 80 -Q 33). During this process, reads containing any of the above adapter or poly(A) sequences were additionally filtered using Trimmomatic v0.3830. Reads aligning to human rRNA, tRNA, globin-related gene sequences (HBA1, HBA2, HBB, HBD, HBM, HBG1, HBG2, HBE1, HBQ1, and HBZ), or phiX control sequences were removed using Bowtie2 v2.3.4.131. Subsequently, the reads mapped to the above genes were excluded using bam2fastq (option: --no-aligned --force --strict). The quality-filtered reads (ranging from 13.3 M to 70.7 M) were aligned to the human reference genome (NCBI GRCh38) using HISAT2 v2.1.0 (options: --dta -q -1)32. The resulting BAM files were processed using SAMtools (options: view -q 4 and sort) to remove duplicate-mapped reads and sort the data. Gene-level read counts were obtained using FeatureCounts v2.0.033 (options: -p -t exon -g gene_id) based on the NCBI Homo sapiens Annotation Release 109. Read counts were normalized using the trimmed mean of M-values (TMM) method implemented in the edgeR package (R v4.2.0) and subsequently used for gene expression analysis.
Differentially expressed gene (DEG) analysis
DEG analysis was performed using the edgeR package in R. Significantly DEGs between different groups (CON vs. TRD, CON vs. non-Res-PRE, CON vs. Res-PRE, etc.) were identified using likelihood ratio tests (p < 0.05, fold change >1.2). Gene Ontology (GO) (biological process) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses were performed on the DEGs using the Database for Annotation, Visualization, and Integrated Discovery (DAVID) website (https://doi.org/10.1093/nar/gkac194).
Detection of IR
The rMATS v.4.3.0 program was used to evaluate the differential IR context embedded in the RNA-seq data34,35. To perform our analysis, we used the following parameters for the rMATS program: [-t paired -readLength 150 -variable-read-length]. Statistical significance was evaluated on the basis of the skipping junction counts (SJCs) and inclusion junction counts (IJCs), which were calculated by rMATS, at the respective loci derived from the gene annotation file. The intron ratios at each locus were approximately calculated with the following formula: IJC / (IJC + SJC * 2). A cutoff of p < 0.05 in the likelihood ratio test and an absolute difference in the IR ratio >0.05 (both used to determine statistical significance) were used to identify differential IR events. As with the DEGs, GO and KEGG enrichment analyses were performed on IRGs with significantly different loci.
Protein‒protein interaction network analysis
A protein‒protein interaction network was generated using Cytoscape version 3.9.136. The analysis was performed using StringApp version 1.7.1. The “full STRING network” type was selected for plotting, and a confidence score cutoff of 0.4 was used (default values were used for all the other parameters). The data that are shown in Fig. 5D were generated using proteins that were encoded by IRGs and DEGs.
Results
Innate immunity, including innate immunity to viral infection, is increased in nonresponders, according to DEG analysis
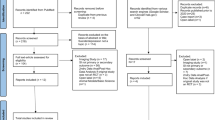
Cathomas et al.16 administered ketamine to patients with TRD and designated those whose depression was reversed as responders (18 patients) and those whose depression was not reversed as nonresponders (8 patients). Additionally, the authors collected blood samples from these patients and 21 healthy controls (HCs), and they performed RNA-seq (Fig. 1A). These RNA-seq data were the first to reveal human responsiveness to ketamine at the RNA level, and thus, these data are extremely valuable. During the process of writing a previous paper9, we noted that these authors did not perform quality filtering on the RNA-seq data (Fig. 1B(ii)). Thus, we performed DEG analyses according to our procedure (Fig. 1B(i)).
A Table showing the data for each group. The numbers of reads that were used for mapping are shown as the mean, minimum, and maximum in each group. B (i) Scheme showing the pipeline for analyzing downloaded raw FASTQ files of the RNA-seq data used in this study. (ii) The pipeline described in the article by Cathomas et al.16.
Figure 2A, B shows the results of GO analysis of the DEGs that were upregulated (546 genes upregulated in nonresponder, 1720 genes upregulated in responders) or downregulated (208 genes downregulated in nonresponders, 342 genes downregulated in responders) in nonresponders or responders compared with the Cons (Supplementary Data 1–6). The results were clear; since nonresponders were enriched for innate immunity and defense responses to viruses, it is suggested that nonresponders were in a state of heightened viral infection (Supplementary Data 2). Similar results were obtained when we made different comparisons, such as a comparison of nonresponders to responders (Fig. 2C(ii)) and a comparison of TRD patients to controls (Supplementary Fig. 1A). Interestingly, few genes with coordinated expression (118 upregulated genes, 37 downregulated genes) were observed between the nonresponders and responders (Supplementary Fig. 1B), suggesting that each group has a unique transcriptome.
A DEG analysis between nonresponders and CONs (baseline). (i) Volcano plots of RNA expression comparisons are shown. The vertical axis shows −log10 p values, and the horizontal axis shows log2 fold-change. Genes with significant changes in expression are shown in red (upregulated) and blue (downregulated) (threshold: p < 0.05 and fold change >1.2). Symbols indicate the top 10 genes with the lowest p values among the up- and downregulated genes. (ii, iii) GO enrichment analysis of upregulated (ii) and downregulated (iii) genes in nonresponders. The horizontal axis indicates −log10 p values. The vertical axis indicates enriched GO terms, and the number of genes corresponding to each term is indicated in parentheses. GO terms related to innate immunity are indicated in bold. B (i, ii, iii) DEG analysis between responders and CONs (baseline). C (i, ii, iii) DEG analysis between nonresponders and responders (baseline).
IR analysis revealed the activation of innate immunity, including innate immunity to viral infection, in nonresponders
Recently, we showed that IR is useful for diagnosing the depressive state of individuals with depression9. On the basis of these findings, we believe that IR might also be useful for describing the differences between nonresponders and responders to ketamine. Figure 3A, B shows the results of the GO analysis of IRGs in nonresponders and responders. As already known37,38,39,40,41, innate immunity is increased in patients with depression, and this can be manifested as changes in the IR (IncIR or DecIR) of innate immunity-related genes9. As expected, changes in the IR of innate immunity-related genes were observed in both nonresponders and responders (Fig. 3A, B). Interestingly, however, far more genes in nonresponders (Supplementary Data 7) than in responders (Supplementary Data 9 and 10) exhibited changes in IR, suggesting greater activation of innate immunity in nonresponders. These data are consistent with the DEG data (Fig. 2). Additionally, changes in IR were observed in many genes related to innate immunity in nonresponders (Fig. 3C, D), including activation of the interferon pathway (Fig. 3A and Supplementary Data 8) and suggesting that nonresponders may be characterized by elevated innate immune responses due to viral infection. Indeed, innate immunity-related genes account for more IRGs in nonresponders than in responders (compare Fig. 3C(ii) with Fig. 3D(ii)).
A Intron retention analysis between nonresponders and CONs (baseline). (i) Scatter plots of IR ratio comparisons are shown. The vertical axis shows the −log10 p values, and the horizontal axis shows the difference in the IR ratio (∆IR). Genes with significant changes are shown in red (IncIR genes) and blue (DecIR genes) (threshold: p < 0.05 and absolute ∆IR > 0.05). Symbols indicate the top 10 genes with the lowest p values among the respective IncIR and DecIR genes. (ii) GO and KEGG enrichment analyses were performed by merging the IncIR and DecIR genes. The horizontal axis indicates −log10 p values. The deep blue bars indicate GO terms, and the orange bars indicate KEGG pathway names. The vertical axis indicates enriched GO/KEGG terms, and the number of genes corresponding to each term is indicated in parentheses. Bold indicates GO/KEGG terms related to innate immunity. B (i, ii) Intron retention analysis between responders and CONs (baseline). C Venn diagram showing inflammation- or immunity-related genes among IncIR/DecIR genes in the nonresponder group. (i) Comparison with gene sets containing the term “GO:0006954 inflammatory response” (508 genes). (ii) Comparison with gene sets containing the term “GO:0045087 innate immune response” (698 genes). (iii) Comparison with gene sets containing the term “GO:0002250 adaptive immune response” (764 genes). D (i, ii, iii) Venn diagram showing inflammation- or immunity-related genes among the IncIR/DecIR genes in the responder group.
Recovery of DEGs or IRGs by ketamine
We first examined the recovery of DEGs after ketamine treatment in nonresponders and responders (Fig. 4A, B). In general, there are two types of recovery: reverse V-shaped recovery (Fig. 4A) and V-shaped recovery (Fig. 4B). Interestingly, the expression of several DEGs related to inflammation or innate immunity was restored by ketamine treatment in both nonresponders and responders (genes shown in bold in Fig. 4A, B). In nonresponders, genes such as CXCL1042,43 and MCAM44 were restored by ketamine, and in responders, genes such as DEFA1B45, IL4I146, LRP147, MUC148, MUC5B49, MUC650, and C1QTNF351 were restored by ketamine. Many of these genes participate in modulating cytokine expression by protecting against viral infection. Notably, indoleamine 2,3-dioxygenase 1 (IDO1) was restored in nonresponders (ref. 52; Fig. 4A); this finding is interesting because IDO1 participates in the first rate-limiting step of the degradation of the essential amino acid tryptophan via the kynurenine (KYN) pathway, and it is currently considered a true immune regulator in autoimmune diseases and chronic inflammation53.
A Venn diagram comparing genes whose RNA expression was upregulated in TRD patients and downregulated by ketamine treatment (reverse V shape). The left panel shows nonresponders, the right panel shows responders, and the gene symbols indicate overlapping (recovered) genes. Bold indicates genes related to innate immunity. B Venn diagram comparing genes whose RNA expression was downregulated in TRD patients and upregulated by ketamine treatment (V shape). C (i) Box and whisker plots showing the relative IR ratios of the 65 IncIR loci (pretreatment) in nonresponders and these loci after ketamine treatment. (ii) Box and whisker plot showing the relative IR ratios of the 145 DecIR loci (pretreatment) in nonresponders and those loci after ketamine treatment. D (i) The 45 IncIR loci in responders. (ii) The 98 DecIR loci in responders. E Venn diagram comparing genes whose IR ratio was increased (IncIR) in TRD patients and decreased (DecIR) by ketamine treatment (reverse V shape). The left panel shows nonresponders, the right panel shows responders, and the gene symbols indicate overlapping (recovered) genes. Bold indicates genes related to innate immunity. F Venn diagram comparing genes whose IR ratio is decreased (DecIR) in TRD patients and increased (IncIR) by ketamine treatment (V shape). The left panel shows the nonresponder case, the right panel shows the responder case, and the gene symbols indicate overlapping (recovered) genes. Bold indicates genes related to innate immunity. G Heatmap showing sites with recovered intron ratios. Relative intron retention ratios are indicated by z scores. (i) Recovery of IR loci (60 loci) in the nonresponder group. (ii) Recovery of IR loci (26 loci) in the responder group. H GO enrichment analysis of the recovered IR genes. The horizontal axis indicates −log10 p values. The dark blue bars are GO terms with p values < 0.05, and the light blue bars are those with no significant difference. Gene symbols indicate genes related to innate immunity. (i) Fifty-eight genes, including the 60 recovered IR loci in nonresponders, were used. (ii) Twenty-six genes, including the 26 recovered IR loci in responders, were used.
Ketamine tends to restore altered degrees of IR to healthy levels in both nonresponders (Fig. 4C) and responders (Fig. 4D). As shown in Fig. 4E, F, some IRGs were indeed statistically significantly restored (p < 0.05), including 60 IRGs in nonresponders (10 by reverse V-shaped recovery and 50 by V-shaped recovery) and 26 IRGs in responders (10 by reverse V-shaped recovery and 16 by V-shaped recovery). A heatmap showing each IRG is shown in Fig. 4G. Innate immunity-related GO terms that were enriched in nonresponders (Fig. 4H(i)) and responders (Fig. 4H(ii)) are shown in bold, and these terms include BCL354, SLC11A155, SLC22A556, CARD957, DDX1758 and PLSCR159, which are involved in the modulation of inflammation.
Differential protein‒protein interactions between nonresponders and responders
On the basis of principles that have been previously described9, we investigated the frequency of protein‒protein interactions between IRGs that were isolated from nonresponders or responders and genes related to innate immunity (Fig. 5A). We hypothesized that the level of interaction would reflect the importance of a particular IRG in innate immunity. Figure 5B shows the ranking of the connections of IRGs that were associated with innate immunity-related genes in nonresponders and responders. The average number of innate immunity-related genes that interacted with each group of IRGs was 12.03 and 14 in nonresponders and responders, respectively, and the difference between the groups was not significant (Fig. 5C). However, the total number of links in nonresponders was 2124, while it was 1882 in responders; thus, it is possible that this difference reflects the fact that innate immunity is more strongly affected in nonresponders than in responders.
A Interaction of protein-coding IR genes against the gene set of the term “GO:0006954 inflammatory response” (505 genes). Networks were generated for interactions with a confidence score of 0.4 or greater in the full STRING network using the STRING database. (Upper) Network of 43 IncIR and 93 DecIR protein-coding and innate immunity-related genes in the nonresponder group. (Lower) Network of 28 IncIR genes, 67 DecIR genes and 1 mixed protein-coding and innate immunity-related gene in the responder group. B Table showing the genes ranked by the number of interactions (links). The left panel shows nonresponders, and the right panel shows responders. C (i) Bar graph showing the mean number of interactions in each group. Student’s t test was used to analyze significant differences. (ii) Bar chart showing the total number of interactions for each group. D Interaction networks focused on innate immunity-related genes. Red filling indicates IncIR genes, and blue filling indicates DecIR genes. Red borders indicate upregulated genes, and blue borders indicate downregulated genes. Squares indicate genes whose IR or expression was recovered by ketamine treatment, and circles indicate genes whose IR or expression were not recovered. (i) Nonresponder group. (ii) Responder group. E Bar graph showing the recovery of the IR ratio or expression of the genes of interest. The error bars indicate the standard deviation. Asterisks indicate p values according to the likelihood test (*p < 0.05, **p < 0.01, ***p < 0.001). (i) Nonresponder group. (ii) Responder group.
Formation of networks involving DEGs and IRGs characteristic of nonresponders or responders
Following our previously published method for constructing networks9, we first constructed protein‒protein interaction networks using all the DEGs and IRGs in both nonresponders and responders (data not shown). From each of these networks, we extracted compact networks containing as many DEGs and IRGs that were restored by ketamine treatment as possible; those DEGs and IRGs in nonresponders and responders are shown in (i) and (ii) of Fig. 5D, respectively. Each of these networks contains inflammation-related genes, including IFN genes, and notably, each network is unique. Figure 5E shows bar graphs of the expression levels of some of the DEGs and IRGs that were restored by ketamine treatment in the network. It appears that ketamine-induced restoration of DEGs and IRFs may be synchronized (see “Discussion”).
After one outlier in the nonresponder group was removed, the DEG data fluctuated greatly, but the IR data did not change
When we performed PCA on the RNA-seq data of all the individuals who were included in the nonresponder group in this study, we found that individual PB100 was an outlier (Supplementary Fig. 2A). We then created a heatmap of the gene expression patterns of all the individuals; this individual, along with an individual who was treated with ketamine, was located at the edge of the dendrogram, and a group of highly expressed genes is indicated in red (Fig. 6A). Then, DAVID was used to perform GO enrichment analysis on the 47 genes that were identified in this study, and the results are shown in Fig. 6B; surprisingly, most of the genes listed here were associated with viral infection.
A Heatmap of genes with high expression variability. Genes with high expression variability were defined as genes whose coefficient of variation (CV) values were greater than 1; 352 such genes were identified. CV values were calculated by dividing the standard deviation by the mean expression. On the left side, the 47 gene symbols that had a z score >3 for nonresponder_PB100 both before and after treatment are listed. B GO enrichment analysis of 47 genes that were highly expressed in the outlier (non-Responder_PB100). The horizontal axis indicates the –log10 p values. The vertical axis indicates the enriched GO term, with the number of genes corresponding to that term indicated in parentheses. Bold indicates GO terms related to innate immunity. C Venn diagram comparing DEGs or IRGs before and after excluding the outlier in the nonresponder group. (i) Comparison of DEGs that were both upregulated on the right and downregulated on the left and the proportions of genes that remained stable after outlier exclusion are shown in the lower panel. (ii) Comparison of IRGs that were both IncIR genes on the right and both DecIR genes on the left, and the proportions of genes that remained stable after outlier exclusion are shown on in the lower panel. D GO enrichment analysis of upregulated (i) and downregulated (ii) genes in nonresponders after excluding the outlier. The horizontal axis indicates the –log10 p values. The vertical axis indicates the enriched GO term, and the number of genes corresponding to that term is indicated in parentheses. E GO and KEGG enrichment analyses were performed by merging the IncIR and DecIR genes in nonresponders after the outlier was excluded. The horizontal axis indicates the –log10 p values. The deep blue bars indicate the GO terms, and the orange bars indicate the KEGG pathway names. The vertical axis indicates the enriched GO/KEGG term, and the number of genes corresponding to that term is indicated in parentheses. Bold indicates GO/KEGG terms related to innate immunity. F Venn diagram showing inflammation- or immunity-related genes among the IncIR/DecIR genes in the nonresponder group after the outlier was excluded. (i) Comparison with gene sets containing the term “GO:0006954 inflammatory response” (508 genes). (ii) Comparison with gene sets containing the term “GO:0045087 innate immune response” (698 genes). (iii) Comparison with gene sets containing the term “GO:0002250 adaptive immune response” (764 genes).
To exclude the possibility that the conclusion that nonresponders are in a state of accelerated viral infection was drawn because of the effect of individual PB100, we performed DEG and IR analyses on the seven remaining individuals after this individual was excluded from the nonresponder group (Fig. 6C). When the DEGs were analyzed without this outlier, only 45% of the upregulated genes and 35% of the downregulated genes remained compared with the numbers of DEGs that were identified when the outlier was included (Fig. 6C(i)). On the other hand, even when the outlier was excluded, 71.4% of the IRGs remained in the IncIR group, and 79.3% remained in the DecIR group (Fig. 6C(ii)). This finding indicates that DEGs are highly susceptible to the effect of the outlier, whereas IRGs are not. Indeed, none of the GO terms that were enriched with the DEGs that were upregulated in nonresponder group after the outlier was removed were related to viral infection or innate immunity (compare Fig. 2A(ii) and Fig. 6D(i)). In contrast, the GO terms that were enriched with the IRGs included terms related to viral infection (Fig. 6E), and indeed, when we examined the IRGs to determine whether they included inflammation-related genes or innate immunity-related genes, it became clear that there was an increase in one inflammation-related gene and no change in the number of innate immunity-related genes (compare Fig. 3C and Fig. 6F). These data clearly indicated that after removal of the outlier, the DEG data fluctuated greatly, but the IR data did not change significantly.
Discussion
The major innate immune cytokines that respond to viral infection are type I IFNs (IFN-α and IFN-β), which activate genes that inhibit viral replication, activate other immune responses to infection, and suppress cell proliferation60. The fact that this pathway is activated in patients with TRD is also mentioned in the paper by Cathomas16, which formed the basis for this paper, and there are many reports of the activation of this pathway in patients with MDD in general61. Our reanalysis of the DEGs also confirmed this point (Fig. 2A(ii)). It is also noteworthy that ketamine treatment restored the expression of several inflammation-related genes to healthy forms in both nonresponders and responders. These genes include IDO1 and EXCL10 (Fig. 5E(i)) in nonresponders and MUC1 and MUC6 (Fig. 5E(ii)) in responders. In the original paper by Cathomas et al.16, since it was noted that ketamine did not restore the expression of inflammation-related genes, we believe that these data reaffirm the validity of our protocol shown in Fig. 1B. It is important to note this point, however, became even clearer when IR was analyzed. The comparison of IRGs between nonresponders and HCs highlighted not only IFN activation but also viral infection in nonresponders (Fig. 3A(ii)).
PCA of the transcriptome data from all the TRD patients revealed that patient PB100 was an outlier among the nonresponders (Supplementary Fig. 2A) and GO analysis of the genes that were activated in this individual revealed that this patient had significant viral infection (Fig. 6A). Even when IR analysis was performed by excluding this individual from the eight nonresponders, GO analysis of the IRGs whose pattern changed compared with those in HCs still revealed viral infection (Fig. 6E), and the pattern of IRGs that are related to innate immunity did not change (rather, they increased) even when the outlier was excluded (Fig. 6F). On the basis of these data, it can be proposed that nonresponders to ketamine treatment are those for whom the effects of ketamine are insufficient to overcome the excessive viral infection.
In the nonresponders, the protein that is encoded by the IRG IRF5 (an IFN-related gene that is involved in maintaining inflammation62) interacts with that of the DEG gene CXCL10 (a chemokine that is involved in inflammation43). These two proteins interact with that encoded by the IRG FCGR3B (involved in autoimmunity63), which interacts with that encoded by NLRC5 (inflammatory gene involved in the regulation of IFN64). All four genes were restored by ketamine administration. In addition, the three genes FCGR3B, IRF5 and NLRC5 are among the top five IRGs that interact with innate immune response-related genes (Fig. 5B, left_panel); thus, the impact of the restoration of these genes by ketamine on general innate immunity is very strong.
In addition, IDO1 is a DEG whose encoded protein interacts with the protein encoded by the CXCL10 gene and whose expression is restored by ketamine administration. IDO1 is activated under chronic inflammatory conditions. The IDO1 enzyme catabolizes L-tryptophan (L-Trp) to generate KYN, thereby stimulating the KYN pathway. Trp depletion is known to inhibit immune cell proliferation in inflamed tissues and reduce the synthesis of serotonin, which is a factor that predisposes patients to the development of psychiatric disorders; thus, the fact that ketamine administration restores the DEG IDO1 in nonresponders is an important finding52,53.
Notably, the IRG EEF1A1 also participates in neuroinflammation65, possibly through the regulation of IL6 production66. eEF1A proteins interact with several proteins that are involved in protein synthesis, such as ribosomal proteins (RPL27 and RPS17), aminoacyl tRNA synthetases (LARS1 and MARS), and protein quality control of nascent proteins (PFDN567), leading to the global regulation of protein synthesis. Notably, eEF1A proteins interact with several viral proteins, resulting in increased viral replication, decreased apoptosis, and increased cellular transformation68. Most of the above IRGs were restored by ketamine administration, providing indirect evidence that nonresponders have severe viral infection.
In contrast, a completely different network was generated between restored DEGs and IRGs in responders. Low-density lipoprotein receptor-related protein 1 (LRP1), which is a protein that is encoded by a restored DEG, participates in various physiological functions, such as blood‒brain barrier permeability69, and the control of many cellular mechanisms that drive physiological and pathological inflammatory responses70; which LRP1 interacts with the restored IRG PLSCR1, which participates in IFN regulation in viral infection71.
The family of cell surface mucins that are encoded by the restored DEG genes MUC148, MUC5B49, and MUC650 are constitutively expressed on the cell surface of nearly all epithelial cells, and they participate in the barrier to viral infection by regulating inflammation72. The fact that MUC1 interacts with TNFRSF1A also suggests the role of mucins in inflammation.
All of the data described above indicate that ketamine administration restores the cellular inflammatory response to viral infection to some extent, regardless of whether the patient is a nonresponder or a responder. The genes that were restored in nonresponders and included in the top 10 links in Fig. 5B are FCGR3B, IRF5, NLRC5, and VCP, and the corresponding genes in the responder group are CD19, CARD9, DDX5, and PLSCR1, with 354 and 266 links, respectively. Assuming that the total number of links represents an effect that reflects the recovery of innate immunity, it appears that the effect of ketamine on the recovery of inflammation is greater in nonresponders than in responders. This fact does not support the hypothesis that nonresponders have a constitution that is resistant to ketamine; rather, it seems more reasonable to hypothesize that nonresponder have such a high degree of inflammation that it cannot be overcome by ketamine treatment.
In our previous paper9, which is a companion paper to this one, we proposed that IRGs are better than DEG in the diagnosis of depression. Many studies have previously shown that DEGs that are related to depression vary from cohort to cohort and are not good markers73. We argue that IRGs are stress-responsive sensor genes and that variation in the IR of these genes more accurately reflects deviation from intracellular protein homeostasis9,23,24,25. The DEG and IR data we analyzed in this study, excluding the outlier data, adequately illustrate the differences in the natures of the DEGs and IRGs. When the outlier PB100 was excluded from the DEG analysis, the DEGs of the remaining nonresponders fluctuated greatly compared with those that were identified when this outlier was not excluded; however, there was almost no change in the IRGs. This fact supports our claim that IRGs may be better markers than DEGs. Individual PB100 was determined to be an outlier due to significant virus infection; however, it is not clear whether it would be better to exclude this individual if we wanted to elucidate the characteristics of the nonresponders, since the nonresponders were characterized as those who were infected with viruses. Our proposal in this paper is that the concept of an outlier is defined according to DEGs, so we do not need to consider this concept when analyzing the IRGs. Rather, it would be better to analyze IR with the outlier included, as this approach will more accurately reflect the characteristics of the cohort.
In this study, based on the fact that even nonresponders showed that several patterns of intron retention can be restored to a healthy type by ketamine, we propose the hypothesis that TRD nonresponders do not possess a constitutional resistance to ketamine, but rather that the effects of ketamine are overwhelmed by severe viral infection. This hypothesis requires rigorous scientific validation using a cohort with a larger sample size in the future. It should be noted that this study has several limitations. As the authors detail in their article, the cohort is well-characterized, but the sample size is relatively small (n = 27). Due to its limited size, it is difficult to reliably include adjustments for age, sex, and other confounding factors in statistical models. There is a notable imbalance in group sizes within this dataset, particularly for the nonresponder group (n = 8), which raises concerns about statistical power and overfitting. Intron retention (IR) analysis is more robust than gene-level differential expression analysis with respect to sample variability. Thus, IR analysis may detect biologically meaningful signals that DEG analysis could overlook. To confirm these potential limitations, we conducted sensitivity analyses in which we excluded the identified outlier (PB100). Notably, the primary conclusions remained unchanged, demonstrating the robustness of our findings. However, these limitations underscore the necessity of further validation in larger cohorts.
Data availability
The raw RNA-seq dataset used in this study, originally reported by Cathomas et al., can be downloaded from the NCBI Gene Expression Omnibus (GEO) under accession number GSE185855.
References
Global Health Estimates: Depression and Other Common Mental Disorders (World Health Organization, 2017).
Drevets, W. C., Wittenberg, G. M., Bullmore, E. T. & Manji, H. K. Immune targets for therapeutic development in depression: towards precision medicine. Nat. Rev. Drug Discov. 21, 224–244 (2022).
Kiraly, D. D. et al. Altered peripheral immune profiles in treatment-resistant depression: response to ketamine and prediction of treatment outcome. Transl. Psychiatry 7, e1065–e1065 (2017).
Akil, H. et al. Treatment resistant depression: a multi-scale, systems biology approach. Neurosci. Biobehav. Rev. 84, 272–288 (2018).
Chamberlain, S. R. et al. Treatment-resistant depression and peripheral C-reactive protein. Br. J. Psychiatry 214, 11–19 (2019).
Sforzini, L. Lost in translation. The quest for definitions of treatment-resistant depression with a focus on inflammation-related gene expression. Brain Behav. Immun. Health 16, 100331 (2021).
Amasi-Hartoonian, N., Pariante, C. M., Cattaneo, A. & Sforzini, L. Understanding treatment-resistant depression using “omics” techniques: a systematic review. J. Affect Disord. 318, 423–455 (2022).
Bhattacharya, A., Derecki, N. C., Lovenberg, T. W. & Drevets, W. C. Role of neuro-immunological factors in the pathophysiology of mood disorders. Psychopharmacology 233, 1623–1636 (2016).
Okada, N. et al. Intron retention as an excellent marker for diagnosing depression and for discovering new potential pathways for drug intervention. Front. Psychiatry 15, 1450708 (2024).
Dantzer, R., O’Connor, J. C., Freund, G. G., Johnson, R. W. & Kelley, K. W. From inflammation to sickness and depression: when the immune system subjugates the brain. Nat. Rev. Neurosci. 9, 46–56 (2008).
Grosse, L. et al. Clinical characteristics of inflammation-associated depression: monocyte gene expression is age-related in major depressive disorder. Brain Behav. Immun. 44, 48–56 (2015).
Grosse, L. et al. Deficiencies of the T and natural killer cell system in major depressive disorder: T regulatory cell defects are associated with inflammatory monocyte activation. Brain Behav. Immun. 54, 38–44 (2016).
Snijders, G. et al. A dynamic course of T cell defects in individuals at risk for mood disorders. Brain Behav. Immun. 58, 11–17 (2016).
Lynall, M. E. et al. Peripheral Blood Cell-Stratified Subgroups of Inflamed Depression. Biol. Psychiatry 88, 185–196 (2020).
Bullmore E. The Inflamed Mind: A Radical New Approach to Depression (Picador, 2018).
Cathomas, F. et al. Whole blood transcriptional signatures associated with rapid antidepressant response to ketamine in patients with treatment resistant depression. Transl. Psychiatry 12, 12 (2022).
Wong, J. J. et al. Orchestrated intron retention regulates normal granulocyte differentiation. Cell 154, 583–595 (2013).
Pimentel, H. et al. A dynamic intron retention program enriched in RNA processing genes regulates gene expression during terminal erythropoiesis. Nucleic Acids Res. 44, 838–851 (2016).
Naro, C. et al. An orchestrated intron retention program in meiosis controls timely usage of transcripts during germ cell differentiation. Dev. Cell 41, 82–93.e4 (2017).
Ullrich, S. & Guigo, R. Dynamic changes in intron retention are tightly associated with regulation of splicing factors and proliferative activity during B-cell development. Nucleic Acids Res. 48, 1327–1340 (2020).
Adusumalli, S., Ngian, Z. K., Lin, W. Q., Benoukraf, T. & Ong, C. T. Increased intron retention is a post-transcriptional signature associated with progressive aging and Alzheimer’s disease. Aging Cell 18, e12928 (2019).
Dvinge, H. & Bradley, R. K. Widespread intron retention diversifies most cancer transcriptomes. Genome Med. 7, 45 (2015).
Okada, N. et al. Intron retention as a new pre-symptomatic marker of aging and its recovery to the normal state by a traditional Japanese multi-herbal medicine. Gene 794, 145752 (2021).
Vu, T.-D. et al. Intron retention is a stress response in sensor genes and is restored by Japanese herbal medicines: a basis for future clinical applications. Gene 830, 146496 (2022).
Okada, N. et al. A potential pathway that links intron retention with the physiological recovery by a Japanese herbal medicine. bioRxiv https://doi.org/10.1101/2023.12.02.569734 (2023).
Morgan, J. T., Fink, G. R. & Bartel, D. P. Excised linear introns regulate growth in yeast. Nature 565, 606–611 (2019).
Parenteau, J. et al. Introns are mediators of cell response to starvation. Nature 565, 612–617 (2019).
Johnston, J. N. et al. Inflammation, stress and depression: an exploration of ketamine’s therapeutic profile. Drug Discov. Today 28, 103518 (2023).
Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnetjournal 17, 10–12 (2011).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Kim, D., Paggi, J. M., Park, C., Bennett, C. & Salzberg, S. L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 37, 907–915 (2019).
Liao, Y., Smyth, G. K. & Shi, W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014).
Shen, S. et al. rMATS: robust and flexible detection of differential alternative splicing from replicate RNA-Seq data. Proc. Natl. Acad. Sci. USA 111, E5593–E5601 (2014).
Wang, Y. et al. rMATS-turbo: an efficient and flexible computational tool for alternative splicing analysis of large-scale RNA-seq data. Nat. Protoc. 19, 1083–1104 (2024).
Shannon, P. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003).
Leonard, B. E. The concept of depression as a dysfunction of the immune system. Curr. Immunol. Rev. 6, 205–212 (2010).
Leday, G. G. R. et al. Replicable and coupled changes in innate and adaptive immune gene expression in two case-control studies of blood microarrays in major depressive disorder. Biol. Psychiatry 83, 70–80 (2018).
Sullivan, P. F. & Geschwind, D. H. Defining the genetic, genomic, cellular, and diagnostic architectures of psychiatric disorders. Cell 177, 162–183 (2019).
Beurel, E., Toups, M. & Nemeroff, C. B. The bidirectional relationship of depression and inflammation: double trouble. Neuron 107, 234–256 (2020).
Nobis, A., Zalewski, D. & Waszkiewicz, N. Peripheral markers of depression. J. Clin. Med. 9, 1–54 (2020).
Proost, P. et al. Coexpression and interaction of CXCL10 and CD26 in mesenchymal cells by synergising inflammatory cytokines: CXCL8 and CXCL10 are discriminative markers for autoimmune arthropathies. Arthritis Res. Ther. 8, R107 (2006).
Lee, E. Y., Lee, Z. H. & Song, Y. W. The interaction between CXCL10 and cytokines in chronic inflammatory arthritis. Autoimmun. Rev. 12, 554–557 (2013).
Charabati, M. et al. MCAM+ brain endothelial cells contribute to neuroinflammation by recruiting pathogenic CD4+ T lymphocytes. Brain 146, 1483–1495 (2023).
Maneerat, Y., Prasongsukarn, K., Benjathummarak, S. & Dechkhajorn, W. PPBP and DEFA1/DEFA3 genes in hyperlipidaemia as feasible synergistic inflammatory biomarkers for coronary heart disease. Lipids Health Dis. 16, 80 (2017).
Molinier-Frenkel, V., Prévost-Blondel, A. & Castellano, F. The IL4I1 enzyme: a new player in the immunosuppressive tumor microenvironment. Cells 8, 753 (2019).
Gorovoy, M., Gaultier, A., Campana, W. M., Firestein, G. S. & Gonias, S. L. Inflammatory mediators promote production of shed LRP1/CD91, which regulates cell signaling and cytokine expression by macrophages. J. Leukoc. Biol. 88, 769–778 (2010).
Wykes, M. et al. MUC1 epithelial mucin (CD227) is expressed by activated dendritic cells. J. Leukoc. Biol. 72, 692–701 (2002).
Roy, M. G. et al. Muc5b is required for airway defence. Nature 505, 412–416 (2014).
Bartman, A. E. et al. The MUC6 secretory mucin gene is expressed in a wide variety of epithelial tissues. J. Pathol. 186, 398–405 (1998).
Micallef, P. et al. C1QTNF3 is upregulated during subcutaneous adipose tissue remodeling and stimulates macrophage chemotaxis and M1-like polarization. Front. Immunol. 13, 914956 (2022).
Salminen, A. Role of indoleamine 2,3-dioxygenase 1 (IDO1) and kynurenine pathway in the regulation of the aging process. Ageing Res. Rev. 75, 101573 (2022).
Pallotta, M. T. et al. Indoleamine 2,3-dioxygenase 1 (IDO1): an up-to-date overview of an eclectic immunoregulatory enzyme. FEBS J. 289, 6099–6118 (2022).
Kreisel, D. et al. Bcl3 prevents acute inflammatory lung injury in mice by restraining emergency granulopoiesis. J. Clin. Invest. 121, 265–276 (2011).
Correa, M. A. et al. Slc11a1 (Nramp-1) gene modulates immune-inflammation genes in macrophages during pristane-induced arthritis in mice. Inflamm. Res. 66, 969–980 (2017).
Dudicova, S., Kacerovsky, M. & Ceckova, M. Effect of inflammatory environment on the gene expression of OCTN1 (SLC22A4) and OCTN2 (SLC22A5) transporters in human trophoblast cell lines BeWo, HTR-8/SVneo and placental villous explants. Authorea (2023).
Liu, X., Jiang, B., Hao, H. & Liu, Z. CARD9 signaling, inflammation, and diseases. Front. Immunol. 13, 880879 (2022).
Wang, S. B. et al. DDX17 is an essential mediator of sterile NLRC4 inflammasome activation by retrotransposon RNAs. Sci. Immunol. 6, eabi4493 (2021).
Sadanari, H. et al. The interferon-inducible human PLSCR1 protein is a restriction factor of human cytomegalovirus. Microbiol. Spectr. 10, e0134221 (2022).
Sangfelt, O., Erickson, S. & Grander, D. Mechanisms of interferon-induced cell cycle arrest. Front. Biosci. 5, D479–D487 (2000).
Mostafavi, S. et al. Type I interferon signaling genes in recurrent major depression: increased expression detected by whole-blood RNA sequencing. Mol. Psychiatry 19, 1267–1274 (2014).
Weiss, M. et al. IRF5 controls both acute and chronic inflammation. Proc. Natl. Acad. Sci. USA 112, 11001–11006 (2015).
Willcocks, L. C. et al. Copy number of FCGR3B, which is associated with systemic lupus erythematosus, correlates with protein expression and immune complex uptake. J. Exp. Med. 205, 1573–1582 (2008).
Kienes, I., Weidl, T., Mirza, N., Chamaillard, M. & Kufer, T. A. Role of NLRs in the regulation of type I interferon signaling, host defense and tolerance to inflammation. Int. J. Mol. Sci. 22, 1301 (2021).
Aisha, Z., Lei, J., Zhang, Y. & Ma, J. EEF1A1 is involved the regulating neuroinflammatory processes in Parkinson’s disease. J. Integr. Neurosci. 22, 122 (2023).
Xu, S. et al. CagA orchestrates eEF1A1 and PKCδ to induce interleukin-6 expression in Helicobacter pylori-infected gastric epithelial cells. Gut Pathog. 12, 31 (2020).
Lee, Y. et al. Prefoldin 5 is required for normal sensory and neuronal development in a murine model. J. Biol. Chem. 286, 726–736 (2011).
Abbas, W., Kumar, A. & Herbein, G. The eEF1A proteins: at the crossroads of oncogenesis, apoptosis, and viral infections. Front. Oncol. 5, 75 (2015).
Yepes, M. et al. Tissue-type plasminogen activator induces opening of the blood-brain barrier via the LDL receptor-related protein. J. Clin. Invest. 112, 1533–1540 (2003).
Wujak, L., Schnieder, J., Schaefer, L. & Wygrecka, M. LRP1: a chameleon receptor of lung inflammation and repair. Matrix Biol. 68-69, 366–381 (2018).
Yang, A. X. et al. Phospholipid scramblase 1 (PLSCR1) regulates interferon-lambda receptor 1 (IFN-λR1) and IFN-λ signaling in influenza A virus (IAV) infection. bioRxiv (2024), https://doi.org/10.1101/2024.11.20.624469.
Dhar, P. & McAuley, J. The role of the cell surface mucin MUC1 as a barrier to infection and regulator of inflammation. Front. Cell Infect. Microbiol. 9, 117 (2019).
Hori, H. et al. Blood-based gene expression signatures of medication-free outpatients with major depressive disorder: integrative genome-wide and candidate gene analyses. Sci. Rep. 6, 18776 (2016).
Acknowledgements
The authors thank Dr. Akinori Nishi for useful discussions on the regulation of protein homeostasis by IR and for discussions with Drs. Masahiro Yamamoto and Akino Wakasugi on the characteristics of nonresponders and responders. This study was supported by a research grant from Tsumura and Co. to N.O.
Author information
Authors and Affiliations
Contributions
N.O.: conceptualization, funding acquisition, project administration, supervision, validation, visualization, writing—review & editing. K.O.: data curation, formal analysis, methodology, writing of methods. A.M.: formal analysis, methodology, Y.K.: project administration.
Corresponding author
Ethics declarations
Competing interests
N.O., K.O., and A.M. were supported by a research grant from Tsumura & Co. The remaining author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as potential conflicts of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Okada, N., Oshima, K., Maruko, A. et al. Intron retention: a novel method for evaluating the response to ketamine in patients with treatment-resistant depression. npj Mental Health Res 4, 44 (2025). https://doi.org/10.1038/s44184-025-00161-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s44184-025-00161-7