Abstract
Understanding the timely regulation of plant growth and phenology is crucial for assessing a terrestrial ecosystem’s productivity and carbon budget. The circadian clock, a system of genetic oscillators, acts as ‘Master of Ceremony’ during plant physiological processes. The mechanism is particularly elusive in trees despite its relevance. The primary and secondary tree growth, leaf senescence, bud set, and bud burst timing were investigated in 68 constructs transformed into Populus hybrids and compared with untransformed or transformed controls grown in natural or controlled conditions. The results were analyzed using generalized additive models with ordered-factor-smooth interaction smoothers. This meta-analysis shows that several genetic components are associated with the clock. Especially core clock-regulated genes affected tree growth and phenology in both controlled and field conditions. Our results highlight the importance of field trials and the potential of using the clock to generate trees with improved characteristics for sustainable silviculture (e.g., reprogrammed to new photoperiodic regimes and increased growth).
Similar content being viewed by others
Introduction
Even under stable conditions, net ecosystem exchange of contrasting biomes displays diurnal oscillations due to endogenous processes1,2. Wood formation is the primary responsible for carbon (C) allocation in woody plants, an important ecosystem C sink3. Trees have a seasonal time window for radial growth with unclear drivers, hampering forest productivity and C budget assesments4,5,6,7,8. Lockhart9 theorized cell expansion and division, allowing radial growth, only occur after exceeding a meristem turgor pressure threshold. Indeed, tree growth is limited by species-specific vapor pressure deficit (VPD) and soil water potential (SWP) ranges explaining most diurnal growth variation10. Since VPD and SWP affect transpiration, and transpiration affects water potential and cell turgor, it is implied tree growth is controlled by the C source and tree water relations (i.e., source-limitation hypothesis)10,11,12,13. Since VPD increases rapidly with daylength, wood growth is inhibited during sunny days6,10. Nevertheless, CO2 assimilation can still occur, even when cambial activity is restrained by unfavorable conditions6,14. Consequently, a temporal decoupling occurs between wood growth (C sink) and C assimilation (C source) which dominate during rainy cloudy days and night-time, and sunny days and daytime, respectively10. The number of growth days might more strongly determine annual growth than the growth period (i.e., growing season)6,15. Trees’ time window for radial growth is thus determined by the C source (i.e., photosynthesis) and limited by tree water relations (i.e., meristem turgor pressure). In addition, it is suggested growth has greater environmental sensitivities than photosynthesis, and these sensitivities are temporally separated on a species-specific basis explained by species’ capacity to timely regulate physiology6,14,16.
The circadian clock (hereinafter “clock”) allows plants to track time and influences plant physiology and phenology (i.e., the timing, duration, and magnitude of life history phases)6,10,17,18,19,20,21,22,23,24,25,26. Not only does the clock explain species-specific growth patterns (i.e., diurnal growth rate and number of growth days) but also phenological responses (i.e., growth period)6,23,24. The clock likely correlates the number of growth days and growth period, determining annual tree growth and C uptake6,15. The extent to which the clock affects trees’ life history phases, related ecosystem processes, a species’ environmental sensitivity (i.e., through resonance, matching internal and external rhythms), and diurnal and annual growth pattern remains unclear27,28.
The clock is a system of genetic oscillators composed of interconnected transcription-translation negative feedback loops (TTNFLs) reset by signals known as zeitgebers. The mechanism generates cyclic endogenous rhythms adapted to changing environments29,30,31,32,33,34,35,36. The clock shows consistent rhythmicity over a broad temperature range (i.e., temperature compensation), is involved in winter hardiness and freezing tolerance, and sensitizes plants to temperature changes32,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55. Many clock-regulated genes co-regulate plant physiology32,56. For example, the circadian period shortens with leaf age suggesting involvement of TIMING OF CAB EXPRESSION 1 (TOC1) during leaf senescence57. TOC1 is associated with expression of FLOWERING LOCUS T (FT) genes, and involved in xylogenesis (i.e., xylem cell formation)58,59,60. Edwards et al.61 showed that expression of LATE ELONGATED HYPOCOTYL 1 and 2 (LHY1 & 2; LHYs, morning-expressed, light-responsive and repressing TOC1) and TOC1 (evening-expressed and repressing LHYs) results in increased biosynthesis of growth regulators cytokinins and auxins during day and night, respectively62,63. Through fluctuating Cyclin D3 (CycD3) concentrations, cell division, and proliferation tend to be initiated during the evening, while cell elongation and xylem differentiation tend to occur at night. The clock thus gates (i.e., temporally restricts) DNA replication and mitosis to the night when favorable conditions prevent DNA damage and VPD is below radial growth thresholds9,19,61,64.
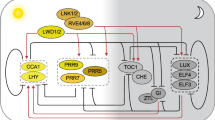
In general, the plant clock is thought to have a conserved architecture65,66. Various components make up the Populus clock (Fig. 1). Photoreceptors are the most prominent input pathway feature and allow daylight perception. Phytochromes A, B1, and B2 (phyA, phyB1 and phyB2) are dimers allowing daylength tracking67,68,69,70,71,72,73,74. Other photoreceptors, like cryptochromes, become phosphorylated upon interaction with blue light75. The ZEITLUPE protein family, containing F-box domains and LOV\PAS-domains with KELCH repeats (ZTL and LKP2), their homologs FLAVIN-BINDING KELCH REPEAT, F-BOX1 (FKF1), and phototropins likewise perceive blue light and play an active role in light-dependent protein degradation.
Relevant proteins comprising or pertaining to the Populus clock are schematically grouped and colored-coded based on their major physiological function. Proteins related to the photoreception of far-red, red, or blue light are highlighted in pink, red, or light blue, respectively. Key components of the central clock oscillator are encircled or highlighted in white, while other clock-associated proteins are highlighted in light green. Termination arrows indicate negative effects. Proteins known to regulate growth or phenology are highlighted in dark green, and those linked to stress or growth hormones are represented in gray or orange, respectively. The shaded blue outer perimeter denotes key traits influenced by the clock.
The core clock pathway consists of the TTNFL between LHY1 and LHY2 and TOC1, with LHYs encoding for v-MyB myeloblastosis viral oncogene homolog (MYB) transcription factors important in secondary plant metabolite biosynthesis76,77. TOC1 (aka PRR1) belongs to the PSEUDO-RESPONSE REGULATOR (PRR) gene family78,79,80.
GIGANTEA (GI) and ZTL are output pathway components and regulate physiological events81. In Arabidopsis thaliana, ZTL affects gene expression, helps degrades TOC1 and PRR5 proteins, and resetting the clock81,82,83,84. GI, GIGANTEA-LIKE (GIL), and FKF1 proteins interact in a complex mediating degradation of CYCLING DOF FACTOR (CDF) proteins85,86. These proteins repress CONSTANS (CO) and FT2 gene transcription by binding to CO and FT promotors87,88. GI and GIL genes regulate photoperiodic response, short-day (SD) induced growth cessation, and bud set85,88,89,90,91,92. Two Populus CO orthologues exist: CO1 and CO259,93. When CO is expressed in A. thaliana, it induces FT expression under long-day (LD) conditions94. FT then travels from phloem to shoot apex and initiates flowering93,95. TEMPRANILLO1 and 2 (TEM1 & 2) prevent flowering, and the CO and TEM balance determines FT levels94,96. A similar regulatory module may control growth cessation and bud set under SD conditions in Populus59. Populus FT paralogs are expressed at different times in different tissues97. For example, FT1 is involved in release of winter dormancy bud flush, while FT2 is involved in vegetative growth and bud set59,85,97,98. Likewise, FT1 and FT2 are mainly expressed in stems and buds, and leaves, respectively93,99.
EARLY BIRD 1 (EBI1) and its Populus homolog EBI2 encode, like their A. thaliana homologs NFXL1 and NFXL2, for zinc finger proteins with putative transcription factor activity involved in abiotic stress responses100,101,102. EBI may bind to ZTL affecting transcriptional activation of LHY and TOC1103. EBI/NFXL2 thus affects A. thaliana’s clock free-running period and speed32. Genes like LEAFY (LFY), SHORT VEGETATIVE PHASE (SVP), MULTICOPY SUPRESSOR OF IRA 1 (MSI1), REGULATORY COMPONENT OF ABSCISIC ACID RECEPTOR 1 (RCAR1), 12-OXOPHYTODIENOATE REDUCTASE 3 (OPR3) and ALLENE OXIDE SYNTHASE (AOS) also relate to the clock, growth and phenology104,105,106,107,108,109. The floral meristem identity gene LFY controls the inflorescence-to-floral meristem transition110,111,112. Likewise, SVP regulates floral meristem specifications and transitions113. SVP affects temperature-responsive regulation of bud break after vernalization and represses expression of FT1 and GA20-OXIDASE1 (GA20ox1)106,114,115,116,117. GA20ox1 is mainly expressed in photosynthesizing tissue and encodes for an enzyme catalyzing the stepwise conversion of C20 gibberellin growth and developmental hormones (GAs)116,117,118,119,120. MSI1 encodes for a WD-40 repeat protein and is associated with flowering, gametophyte, and seed development121,122,123,124,125,126,127. Like SVP, RCAR1 represses bud break while affecting GA20ox1 expression. In addition, RCAR1 regulates abscisic acid (ABA), which inhibits growth and regulates stress104,105. As elucidated in A. thaliana, the OPR3 and AOS genes encode enzymes regulating the biosynthesis of the growth and stress resistance hormone jasmonic acid (JA)107,108,109,128,129,130,131,132,133.
The effects of gene modification regarding plant yield performance are seldom assessed thoroughly in a real-world environment134. This meta-study examines the growth and phenology of transgenic trees exposed to both natural and controlled conditions in Sweden.
The growth and phenology of hybrid WT Populus T89 (WTT89) trees were compared with independently transformed transgenic and WT Elite864012 (WTElite) trees (SI). Their silviculture potential was screened in two large field trials (FEs), with further characterization in five growth chamber experiments (GCEs) and a phenotyping platform experiment (PPE; Fig. 2). To assess statistical significance of growth or phenological changes, we applied generalized additive models (GAMs) allowing determination of when growth or phenological differences become significant and estimation of thresholds at which bud phenological stages occur.
Results
Primary and secondary growth effects
The significance of growth changes in transgenotypes (i.e., independent antibiotic-resistant transgenic lines grouped by the same gene construct) is always reported in contrast to WTT89 unless indicated (Table 1, SI). The deviation of difference smooth’s Bayesian Wabha/Silverman credible interval above or below the horizontal zero-line in a term plot (i.e., representing WTT89 growth) visually indicates significant (p < 0.05) differences in transgenotypes’ growth compared to WTT89 (Fig. 3)135,136,137.
Example of the primary growth of P. tremula L. × P. tremuloides trees during growth chamber experiment 1(A). The black solid line represents the primary growth of the reference WTT89 while the colored solid lines represent the primary growth of two transgenotypes ztl-5 (A; light-green) and ztl-7 (B; yellow). Dots and error bars represent mean and standard error values, respectively. Term plots of a generalized additive (mixed) model with ordered-factor-smooth interaction smoothers modeling the mean primary growth of the respective trees during the experiment (B, C). The horizontal red dotted line represents the mean primary growth of the reference WTT89. In contrast, the mean primary growth of ztl-5 and ztl-7 are represented by colored solid lines. The difference smooth’s upper Bayesian Wabha/Silverman credible interval (shaded band) is below the horizontal red zero-line in the term plot (i.e., representing the growth of WTT89) indicating that the growth of ztl-5 is significantly (p < 0.05) different from the growth of WTT89, whereas the growth of ztl-7 does not differ significantly135,136,137.
The growth of photoreceptor-related transgenotypes generally differed significantly. The primary and secondary growth of pAtCCR2::LUC PHYA1-1 (aPttPHYA-1; knockdown construct targeting endogenous PHYA) and pAtCCR2::LUC PHYA22-2 (oatPHYAox-22; i.e., overexpressor construct of oatPHYA) carrying transgene A. thaliana promoter COLDCIRCADIANRHYTHMN RNA BINDING2 fused to firefly LUCIFERASE (AtCCR2pro:LUC) was usually significantly slower (Figs. S9–S12)138. The primary growth of PHYB-12 and PHYB2KO-6 with downregulated or knocked out the expression of PHYTOCHROME B1 and 2 (PHYB1 and PHYB2), respectively, was usually significantly faster (Fig. S5, S6, S51 and S52). The primary growth of ztl-5 and 7 was significantly slower and faster, respectively (Fig. S1, S2, S9 and S10). The primary and secondary growth of fkf1-11 was significantly slower (Figs. S9–S12).
The growth patterns of core clock-related transgenotypes were condition dependent as lhy-10 displayed both significantly slower (in GCEs) and faster primary and secondary growth (in FEs; Figs. S1–S6, S9–S12, S51 and S52). The growth of toc1-5 and TOC1_OX-7 was usually significantly faster (Figs. S9–S12).
The primary growth of gi-6, gi-13, GIOX-12, and GILOX12 was, except gi-13, always significantly slower (Figs. S5–S8, S51 and S52). In contrast, primary and secondary growth of AtGA20ox1 genotypes under the promoters AINTEGUMENTALIKE1 (MEE1) or RIBULOSE-1,5-BIPHOSPHATE CARBOXYLASE/OXYGENASE SMALL SUBUNIT (RBCS) cloned from Eucalyptus grandis genomic DNA (MEE2) was occasionally significantly faster (Figs. S5–S8, S31, S32, S34, S35, S51 and S52)139,140. When compared against the growth of WTElite in GCE4, the primary and secondary growth of AtGA20ox1 transgenotypes did not consistently grow significantly slower or faster. For example, primary growth of pMEE1::AtGA20ox1-4Elite and pMEE2::AtGA20ox1-10Elite, and pMEE2::AtGA20ox1-5Elite and pMEE2::AtGA20ox1-6Elite was significantly slower and faster, respectively than WTElite (Figs. S33, S36, S53 and S56).
We observed contrasting primary growth patterns between CO1OX-13 and CO2OX-11 in FE2. The former and latter transgenotypes had primary growth that was significantly faster and slower, respectively (Figs. S51 and S52). Primary growth of ft-7 and ft-16 in FE1 and FE2 was usually significantly slower (Figs. S9, S10, S18–S21, S51 and S52). Secondary growth of ft-7 and ft-16 in FE2 was significantly faster, but not for ft-16 in FE1 (Figs. S11, S12, S53 and S54). All EBI1-related transgenotypes (ebi1) and ebi2-6 displayed faster primary and secondary growth, except ebi1-14 in FE2 (Figs. S1, S12, and S51–S64). In contrast, primary and secondary growth of ebi2-7 and ebi2-8 was slower (Figs. S5–S12).
Almost all bud development- or flowering-related transgenotypes had a faster primary growth. From the 12 OPR3-related transgenotypes, including opr3-11 and opr3-7, only opr3-15 had significantly faster primary growth. With an FT RNAi background, opr3_ft-3 and opr3_ft-12, but not opr3_ft-7, explore OPR3 dependence on FT (both FT1 and FT2) and had significantly slower primary growth. Opr3_ft-3 had significantly faster secondary growth in FE2 (Figs. S5, S6, S18 and S19). A similar pattern was observed in the primary growth of AOS-related transgenotypes. aos-10 and aos-13, but not aos-1, had significantly faster primary or secondary growth. aos_ft-10, aos_ft-1, and aos_ft-13 had both significantly slower as faster primary and secondary growth (Fig. S20, S21, and S51–S54). Primary and secondary growth of WTElite was usually significantly faster (Figs. S5, S6, S31 and S32).
Phenological alterations
Differences in chlorophyll content index (CCI), bud set, and bud burst between transgenotypes and WTT89 were analyzed using an ordered-factor-smooth interaction approach and cumulative threshold models (Fig. 4)141,142. The phenological behavior of transgenotypes is always reported in contrast to WTT89 (Tables 1 and 2; Fig. 5; SI).
Example of the lateral bud burst scoring of P. tremula L. × P. tremuloides trees during growth chamber experiment 3. The black solid line represents the bud burst of the reference WTT89, while the colored solid lines represent the bud burst of two transgenotypes aos_ft-10 (A; blue) and ft-16 (B; yellow; A). Dots and error bars represent mean and standard error values, respectively. The lateral bud burst was scored following UPOV193 using the following scoring values: dormant buds enveloped by scales (0), swelling buds with diverging scales (1), sprouting buds (2), opened buds with leaves clustered (3), diverging leaves with rolled op blades (4), and completely unfolded leaves (5). Term plots of a generalized additive (mixed) model with ordered-factor-smooth interaction smoothers modeling the bud burst of the respective trees during the experiment (B, C, D). The colored solid lines, given for the reference WTT89, aos_ft-10, and ft-16, represent the predicted probability of having a bud in a specific lateral bud burst stage at a particular moment. The shaded bands around the colored solid lines represent 95% pointwise confidence intervals. The bud burst categories follow UPOV193 using the following scoring values: dormant buds enveloped by scales (1; red), swelling buds with diverging scales (2; orange), sprouting buds (3; light-green), opened buds with leaves clustered (4; petroleum-green), diverging leaves with rolled op blades (5; blue), and completely unfolded leaves (6; purple). Let Tref. represent a rapid increase in the predicted probability of a bud to have completely unfolded leaves. It can be observed that this rapid increase (Tnew) is delayed substantially in transgenotype A, as opposed to transgenotype B.
Examples of lateral bud burst development in P. tremula L. × P. tremuloides trees during growth chamber experiment 3. The black solid lines represent the development of the lateral bud burst in two transgenotypes aos_ft-10 (A; subplot B) and ft-16 (B, subplot A), and the WTT89 (subplot C), through the linear predictor (i.e., the expected value of a latent variable following a logistic distribution). The black shaded bands around these black solid lines represent 95% pointwise confidence intervals. More specifically, the black solid lines show the factor-smooth interaction effect between Day of the Year and Transgenotype on the linear predictor. Each bud burst category, scored following UPOV193, is shown here as colored vertical bars in each subplot: dormant buds enveloped by scales (0; gray), swelling buds with diverging scales (1; red), sprouting buds (2; orange), opened buds with leaves clustered (3; light-green), diverging leaves with rolled op blades (4; blue), and completely unfolded leaves (5; beige). The cumulative threshold model estimates cut points on the latent variable which allows to predict when buds, given a specific transgenotype, are likely to transition from one bud burst category to the next. The first threshold (i.e., the boundary between the first and second bud burst phases; dormant buds enveloped by scales and swelling buds with diverging scales) is by definition always set to −1. In this example, it can be observed that transitions in the lateral bud burst development of transgenotype A occur substantially slower than similar transitions in transgenotype B, or the reference WTT89. The lateral bud burst development of transgenotype B, on the other hand, is remarkably similar to the reference WTT89.
pAtCCR2::LUC PHYA22-2 had significantly delayed bud set and significantly higher CCI values (Figs. S13–S16). The delay in bud set and leaf senescence suggests pAtCCR2::LUC PHYA22-2 has an extended growth period without faster primary growth. During GCE1, ztl-5 and ztl-7 had significantly lower and higher CCI values, respectively (Figs. S15 and S16). fkf1-11 was observed to have significantly advanced bud set and lower CCI suggesting faster leaf senescence (Figs. S13–S16). Only lhy-10 had a significantly later bud set from core clock-related transgenotypes (Figs. S13 and S14).
Clock-output-related transgenotypes showed significant phenological differences. gi-13 had significantly delayed bud set and higher CCI, suggesting a prolonged growth period (Figs. S13–S16). Differences were observed in bud set between lines with ectopic expression of AtGA20ox1, and WTT89 or WTElite in the background. The former and latter transgenotypes had significantly delayed or advanced bud set, and advanced or delayed bud burst, respectively (Figs. S37, S38, S41, 42, S45, and S46). Compared to the bud phenology of WTElite, only pMEE1::AtGA20ox1-4Elite and pMEE2::AtGA20ox1-10Elite differed significantly (Figs. S39, S40, S43, S44, S47, and S48). CO\FT-regulatory-module-related transgenotypes did not differ significantly in their phenology. Only ft-16 had significantly advanced bud set, delayed apical bud burst, and lower CCI, suggesting a shortened growth period (Figs. S13–S16, S25, and S26).
The phenological differences observed in transgenotypes targeting EBI in FE1 are the significantly higher CCI of ebi1-3, suggesting slower leaf senescence, and advanced bud set of ebi2-7 and ebi2-8 (Figs. S13–S16). The bud set in aos-1, aos-10, and aos-13, and aos_ft-1, aos_ft-10 and aos_ft-13 were significantly delayed and advanced, respectively. The apical bud burst in aos-13, and lateral bud burst in aos_ft-10 and aos_ft-13 was also significantly delayed, suggesting aos_ft transgenotype trees have a shortened growth period dependent on FT. We observed a significantly delayed apical and lateral bud burst in WTElite (Figs. S22, S23, S25, S26, S28 and S29).
Discussion
Clock-associated genes significantly affected tree growth and phenology across various experimental levels. Our findings highlight the potential of modifying clock-associated genes for silviculture. For example, lhy-10 and toc1-5 grew faster in the field (likely due to their sped-up core-clock TTNFL) indicating trees can be reprogrammed to new photoperiodic regimes by altering their critical daylength or growth period. By safeguarding DNA replication licensing timing through gating and optimizing resonance between cell cycle phases and external environmental rhythms, LHY and TOC1 regulate growth. Downregulation of these genes may lead to a respective decrease and increase in cold hardiness and tolerance, and critical day length143,144,145,146.
Increasing the growth of trees cultivated for biofuel may increase bioenergetic yields147,148,149. EBI1 appears key to tree growth. ebi1 transgenotypes generally had faster primary and secondary growth. EBI1, associated with stress and photosynthesis protection in A. thaliana, affects the clock period and expression of LHY and TOC1101,103,150. EBI downregulation may even increase primary growth and early flowering103. The only phenological difference observed in the ebi1 transgenotype is that ebi1-3 had a higher CCI, suggesting slower leaf senescence. The growth functions of EBI2 appear similar to EBI1 in Populus albeit with expression at different times and tissues101,103,150.
Transgenic trees with increased secondary growth have an adaptive advantage under drought conditions and enhanced post-drought recovery8. In this light, primary and secondary growth of transgenotypes with AtGA20ox1 expression were generally faster regardless of background line or reporter-promoter construct. The genetic background of AtGA20ox1 transgenotype trees affected tree phenology showing the background can be used as an alternative to promotors for altering growth. MEE1 and MEE2 did not strongly affect growth. AtGA20ox1 overexpression increases growth elongation and period, biomass, and fiber length115,117. Tree growth even occasionally increased regardless of growth period shortening117. The growth period thus appears less relevant than diurnal growth. The background line strongly affected the bud set irrespective of the promoter.
Trees’ photoperiodic growth constraints can be amended. For example, slower primary growth, delayed bud set, and higher CCI values found in pAtCCR2::LUC PHYA22-2 suggest overexpression of oatPHYA results in a prolonged growth period but hampered primary growth73,151. Given the lack of differences in pCCR2::LUCT89 growth, we conclude the pCCR2::LUC construct did not affect growth. The upregulation of oatPHYA, unlike PHYA1, not only positively affected leaf and bud phenology but also secondary growth. Downregulation or knocking out PHYB in PHYB-12 and PHYB2KO-6 generally resulted in trees with faster primary and secondary growth. PHYB is involved in day length, temperature perception, and growth cessation, which might explain why the primary growth of PHYB-12 was slower in FE2 than the PPE151. Primary growth of ztl-5 and ztl-7 was, respectively, slower and faster in GCE1 and FE1. Primary growth of fkf1-11 was slower and leaf senescence in fkf1-11 progressed faster. No changes were observed in PRR728.
Modification of clock output-associated genes affected tree growth and phenology. All GI-overexpressing transgenotypes had slower primary growth. gi-13 had delayed bud set and significantly higher CCI, suggesting slower leaf senescence. These results contrast the literature suggesting moderate GI downregulation leads to pleiotropic effects in the field85,152,153,154. Trees with GI overexpression also displayed delayed growth cessation and bud set under SD conditions in GCEs. CO1OX-13 and CO2OX-11 in FE2 had faster and slower primary growth, respectively. Likewise, FT downregulation resulted in contrasting growth patterns but generally slower growth. These results suggest CO1, CO2, and FT expression impact Populus growth in natural conditions59,85,155. In line with Böhlenius et al.59, ft-16 had significantly advanced bud set, delayed apical bud burst, and lower CCI. Except for SVPOX-7, all lfy, svp, SVPOX, rcar, or RCAROX transgenotypes had faster primary growth. Our results, suggesting LFY downregulation leads to faster primary growth but no effect on bud phenology, contrast the literature stating upregulation of LFY generally leads to faster primary growth and advanced flowering110,111,156. Due to its role in temperature regulation of flowering and bud burst, SVP downregulation was expected to delay bud set and advance bud burst106. RCAR downregulation likewise advanced bud burst104,106. As expected, msi transgenotype trees lacked a clear phenotype and WTElite transgenotype trees generally had a faster primary growth and advanced bud set125,126,127.
We observed positive growth in trees with downregulation of JA biosynthesis-related genes108,109,128. OPR3 downregulation positively affected primary growth but FT’s reduced expression overruled this response and generally led to slower primary growth59. Primary growth of aos and aos_ft transgenotypes was generally faster and unclear, respectively. Bud set in aos_ft transgenotype was advanced, while apical and lateral bud burst was typically delayed. The implied shortened growth period in aos_ft transgenotype trees is expected due to reduction of FTs in the background59.
Phenological differences were sometimes unexpected or lacking. For example, bud burst in lhy-10 was later but no phenological differences were observed in toc1-5. It remains unclear why TOC1 overexpression, nor ZTL downregulation always altered bud phenology59,84,143,157,158.
Tissue-specific clocks exist that differ in their sensitivity to light or temperature compensation mechanism28,37,159,160,161,162,163,164. Roots, for example, have clocks assumed to be slave versions of shoot clocks159,161,165,166,167. Tissue-specific clocks’ sensitivity to environmental changes and importance in tree growth and phenology therefore affects the source-sink limitation paradigm28,37,159,160,161,162,163,164,165,166,167,168,169,170. Dynamic global vegetation models, implicitly assuming plant productivity is C source-limited and underestimating the sensitivity of growth processes to environmental conditions other than photosynthesis, thus need accounting for the clock16,171,172,173,174,175.
New Populus transgenotypes with promising characteristics were introduced here. ebi1-2, ebi1-3, or ebi1-14, for example, generally had faster primary and secondary growth compared to WTT89 suggesting EBI1 is key to tree growth. Likewise, the effect of different promotors and genetic backgrounds on AtGA20ox1 genotype trees was tested. Indeed, the latter affected phenology. Transgenotypes, such as pMEE1::AtGA20ox1-5Elite or pMEE1::AtGA20ox1-8Elite, may therefore both grow fast and survive cold climates, confirming the importance of exposing transgenic trees to real-world conditions to strengthen laboratory research115,134. It is common forestry practice to transfer southern natural accessions up North. It is now shown core-clock genes’ expression (i.e., LHY1, LHY2, or TOC1) can be used to reprogram trees biotechnologically to a new photoperiodic regime (i.e., latitudinal adaptation) allowing the growth period of targeted accessions to increase. Statistical learning approaches, such as cumulative threshold models, enrich current tree breeding, cultivation, or forestry practices with substantial benefits for chronosilviculture.
The essential takeaway is that transgenic perturbations of genes comprising or pertaining to the Populus clock substantially affected tree physiology across various experimental levels. The clock thus orchestrates tree growth and phenology as if it were Nature’s ‘Master of Ceremony’. Combined with the observation that angiosperm deciduous tree species do not necessarily mirror, or halt, fine- or coarse roots growth in temperature tree species, this confirms the essentiality of Körner’s16 proposed paradigm shift in plant growth control175,176.
Methods
Plant material, constructs, and transformations
Transformable wild type P. tremula L. × P. tremuloides Michx. CV. T89 plants were used to perform Agrobacterium tumefaciens-mediated transformations at the Umeå Plant Science Centre (UPSC)177,178. The transformed plants resulted in circa 68 RNAi transgenic lines targeting 26 genes associated with the clock mechanism (Table S2)179,180,181. The trees were propagated using in vitro culture and grown in greenhouses until the experiments started. Independent antibiotic-resistant transgenic lines grouped by the same gene construct are hereafter called a transgenotype. The transformation procedures of 31 transgenotypes previously described in the relevant literature are briefly summarized in Table S3 (SI). The transformation procedures for 39 remaining transgenotypes are discussed in detail below. Table S2 provides information on the design, base pairs, gene models, and primer sequences. Following Ibanez et al.143 and Jurca et al.158, plasmids were first constructed by obtaining gene-specific fragments from Populus cDNA. These fragments were amplified with DNA polymerase and primers with specific primer sequences. The gene fragments were then cloned into an entry vector and, using an enzyme mix, recombined into a plant destination vector182. The resulting plasmids were transformed into A. tumefaciens C58 strain GV3101 and used to transform WTT89 trees115,180,181. Independent and stable transgenotypes were selected after exposing the trees to an antibiotic selection marker. Gene expression analysis of c. ten transgenotypes of antibiotic-resistant plants was used to select transgenotypes for further analysis.
Transgenotypes targeting photoreceptor and morning-loop genes
Twelve transgenotypes targeted photoreceptor-related genes. Following Kozarewa et al.138, PHYA1 was antisense inhibited in anti-sense transgenotype pAtCCR2::LUC PHYA1-1. pAtCCR2::LUC PHYA22-2* included an over-expression cassette of oatPHYA (*construct number). The antisense transgenotype (pAtCCR2::LUC PHYA1-1) expresses a full-length PHYA cDNA fragment flipped 3′-5′ under the control of the cauliflower mosaic virus (CaMV) 35 s promoter while the oatPHYA (i.e., pAtCCR2::LUC PHYA22-2) was constitutively expressed by 35 s CaMV73. Another promoter-reporter construct of COLD CIRCADIAN RHYTHMN RNA BINDING 2 (pAtCCR2) fused to firefly LUCIFERASE (LUC) was also introduced to these transgenotypes, as well as in a control transgenotype with only the transgene pAtCCR2::LUC T89-562. The transgenotype PHYB-12 and PHYB2KO-6 downregulate and knock out the PHYB gene, respectively, responsible for a plant’s red-light sensitivity. PHYB-12, however, is the result of RNAi, with the gene fragment introduced as two inverted copies with an intron, while PHYB2KO-6 is generated using CRISPR/Cas9-mediated gene editing (i.e., CRISPR, Clustered Regularly Interspaced Short Palindromic Repeats; Cas9, CRISPR-associated protein 9) following Ding et al.151,183,184. ztl-3, ztl- 5 and ztl-7 are transgenic RNAi transgenotypes constructed following Jurca et al.158 which repress ZTL expression and hence affect blue-light sensitivity, clock control and the impulse to reset the clock. The transgenotypes targeting the ZTL homolog FKF1, fkf1-10 and fkf1-11, likewise repress FKF1 affecting blue-light reception and flowering. prr7-5, on the other hand, downregulates PRR7, which directly affects the core-loop of the clock. The gene fragments in fkf1-10, fkf1-11, and prr7-5 are designed as two inverted copies of the respective amplified gene-specific fragment with an intron in between.
Transgenotypes targeting core clock-regulating genes
Four transgenotypes included in this study have been described in Ibanez et al.143 and Edwards et al.61. lhy-10 is a transgenotype targeting LHY1 and LHY2. This transgenotype represses the expression of LHY. Likewise, the expression of LHY’s antipodal core clock-regulated gene TOC1 is downregulated in toc1-5. In transgenotypes TOC1_OX-4 and TOC1_OX-7, however, TOC1 is upregulated. The entire TOC1 cDNA in these transgenotypes is constitutively upregulated with varying degrees. In all the above transgenotypes, the core loop of the clock is disrupted with severe alterations in the rhythmicity of the clock as a consequence. Both LHY and TOC1 are key to the clock mechanism.
Transgenotypes targeting genes controlling growth, bud development, and flowering
GI is a prominent gene in the output loop of the clock. Similar to Ding et al.85, four transgenotypes were constructed targeting GI and GIL. gi-6 and gi-13 were constructed so that GI is repressed185. The transgenotypes GIOX-12 and GILOX-12 upregulate GI and GIL, respectively. All four transgenotypes, through the effect of GI on CO and FT, are supposed to have altered timings in their growth cessation, bud set, and flowering89. Twelve transgenotypes had ectopic AtGA20ox1 expression driven by two specific promoters. This should lead to increased levels of GAs, as the gene coding for the multifunctional enzyme is relevant in synthesizing GAs120. These transgenotypes have WTT89 or WTElite in the background. In addition, the transgenotypes pMEE1::AtGA20ox1-3T89, pMEE1::AtGA20ox1-4Elite, pMEE1::AtGA20ox1-5Elite, pMEE1::AtGA20ox1-8T89, pMEE1::AtGA20ox1-8Elite, pMEE1::AtGA20ox1-10T89 and pMEE1::AtGA20ox1-10Elite have AINTEGUMENTALIKE1 (AIL1; MEE1 or pAIL1) as promotor (i.e., mainly expressed in meristem and young leaves)139,186. In contrast, pMEE2::AtGA20ox1-2T89, pMEE2::AtGA20ox1-5T89, pMEE2::AtGA20ox1-5Elite, pMEE2::AtGA20ox1-6Elite, pMEE2::AtGA20ox1-10Elite, and pMEE2::AtGA20ox1-11T89 have RIBULOSE-1,5-BIPHOSPHATE CARBOXYLASE/OXYGENASE SMALL SUBUNIT (RBCS; MEE2 or pEL1.2) cloned from Eucalyptus grandis genomic DNA as promoter (i.e., mainly expressed in photosynthesizing tissue)139.
Like FKF1, GI and GIL in Populus may regulate the transcription of CO85,88,90,91. Two transgenotypes, CO1OX-13 and CO2OX-11, were constructed using RNAi following Hsu et al.155 and upregulate either CO1 or CO2, respectively. On the other hand, the transgenotype tem1B-5 constructed following Castillejo and Pelaz94 upregulates TEM, which repressed the expression of FT. Since the balance between CO and TEM regulates the expression of FT, these upregulating transgenotypes will display different bud burst and flowering times compared to the WTT89 93. ft-16 and ft-7 are two transgenotypes targeting the FT gene59,97,98. The FT and FT1 gene fragments were introduced in these transgenotypes as two inverted copies with an intron. As a result, the FT gene is downregulated, severely affecting the regulation of bud set and flowering by the CO\FT regulatory module59.
Following Eriksson et al.187, six transgenotypes were constructed with either downregulation of EBI1 or EBI2188. ebi1-2, ebi1-3 and ebi1-14 repress the former gene, while ebi2-6, ebi2-7 and ebi2-8 repress the latter gene. In A. thaliana, EBI (PttEBI1 orthologue) is associated with ZTL and hence regulates the transcriptional activity of LHY and TOC1. It can be expected that these transgenotypes differ significantly in their growth and phenology from the WTT89101,102.
More transgenotypes target genes involved in bud development and flowering (e.g., LFY, SVP, MSI1, or RCAR1). lfy-4, for example, a transgenotype with a fragment of the LFY gene, which is involved in the flower-meristem identity, is introduced as two inverted copies with an intron. The LFY gene is downregulated in lfy-4, which should lead to changes in the timing of tree vegetative growth and flowering. Using a similar design, SVP is introduced into svp-3. In this transgenotype, SVP is also downregulated, supposedly affecting the timing of bud development and the temperature-responsive regulation of flowering. SVP is, on the other hand, upregulated in SVPOX-6 and SVPOX-7. In these transgenotypes, the construct consists of a CaMV 35S promoter that drives the expression of the SVP cDNA as a translational fusion to an N-terminal human influenza hemagglutinin (HA) sequence. The upregulation of SVP in these transgenotypes should likewise affect temperature-responsive regulation of bud break after vernalization. The transgenotype msi-1A is a construct for CaMV 35 s promoter-driven upregulation of RNAi to downregulate MSI1 constructed following Englund127. Because of MSI1 involvement in WD-40 repeat proteins, the trees likely display altered regulation of flowering, and gametophyte and seed development121,122,123,124,125,126,127. Following Singh et al.106, RCAR1 is downregulated and upregulated in transgenotypes rcar-10 and RCAROX-18, respectively. Although RCAR1 represses bud break, it also regulates ABA104,105. Like the transgenotypes targeting RCAR1, twelve other transgenotypes target genes related to JA levels in the trees. Six of these transgenotypes have a fragment introduced of the OPR3 gene encoding for the OPR3 enzyme regulating the biosynthesis of JAs87. opr3-7, opr3-11, and opr3-15 simply have the OPR3 fragment introduced as two inverted copies with an intron leading to downregulation of OPR3. Opr3_ft-3, opr3_ft-7 and opr3_ft-12 have the same design but also have FTRNAi in the background, which normally affects the regulation of flowering. Six other transgenotypes target AOS, encoding for a cytochrome P450 protein regulating JA biosynthesis, and are also split into three transgenotypes that downregulate AOS (aos-1, aos-10, and aos-13) or downregulate AOS and have FTRNAi in the background (aos_ft-1, aos_ft-10 & aos_ft-13). Two other wild-type transgenotypes were investigated as alternatives to the reference WTT89. Both WTElite and WTElite-884056, described in Stener and Westin189, are of particular interest to clonal forestry practices in Scandinavia.
Quantitative reverse transcription polymerase chain reactions
This study characterizes transgenotypes related to the aos, aos_ft, fkf1, opr3, opr3_ft, AtGA20ox, prr7, gi and TOC1_OX transgenotypes found in Table S2 (Figs. S57–S67). Gene expression data for most transgenotypes (SI) was determined using quantitative reverse transcription polymerase chain reactions (RT-qPCR) following Jurca et al.158. Eight hours after dawn (ZT8), we sampled leaves from trees grown in a greenhouse under constant light conditions (i.e., 18:6 h light/dark cycles with a light intensity of 250 μmol m−2 s−1) and shock froze them in liquid nitrogen. RNA was subsequently extracted from each leaf following the classical cetyltrimethylammonium method of le Provost et al.190. After treatment with DNase (TURBO DNA-free kit; Ambion, Austin, US), cDNA was synthesized from 1 μg RNA using an iScript cDNA Synthesis Kit (Bio-Rad Laboratories, California, US). RT-qPCR was performed using a CFX96 Real-Time detection system (Bio-Rad Laboratories), gene-specific primers (SI), three to four biological replicates, and two technical replicates. The relative expression of the genes of interest was normalized against the expression of reference housekeeping genes ELONGATION FACTOR 1 ALPHA (EF1a) or 18S rRNA. Further calculations were done relative to WTT89 or the lowest expressing transgenotype using the 2−ΔΔCT method of Livak and Schmittgen191 and Pfaffl192. qPCR results were inconclusive for aos and aos_ft transgenotype trees, as well as opr3_ft-12.
Growth chamber experiments
We conducted five growth chamber experiments (GCE1 - GCE5), measuring the growth and phenology of transgenotypes in growth chambers at UPSC according to standard experimental setups outlined in Ibanez et al.143, Edwards et al.61, and Jurca et al.158 (Fig. 2; Table 2). Following Nilsson et al. 177, cuttings of P. tremula L. × P. tremuloides were first grown in vitro for 4 weeks. The rooted and in vitro-cultivated transgenic trees were then potted in a 3:1 mix of fertilized peat and perlite and grown for another four weeks under constant LD light, temperature, and relative humidity conditions (i.e., 18:6 h light/dark cycles; 18 °C; and 80% relative humidity, respectively) in the greenhouses at UPSC. The light intensity during this period was 200 μmol m−2 s−1 (Osram Powerstar HQI-T 400W/D lamps; Osram, Munich, Germany). After the first month, each tree received weekly nutrients (SuperbaS, Supra Hydro AB, Landskrona, Sweden) and water (1.5 l). After a period (Table 2), the trees in GCE2, GCE3, GCE4, and GCE5 were exposed to the same temperature, relative humidity, and irradiance conditions. Still, the light conditions were changed to SD conditions (i.e., 15:9 h light/dark cycles), and dusk time remained unchanged. During each GCE, the primary and secondary growth of the trees was measured weekly. Phenological observations were made on the bud set, and the apical and lateral bud burst in GCE3 and GCE4. The development of the bud set was scored weekly following UPOV193 and Ibanez et al.143 using the following (opposite) scoring values: a still actively growing shoot and uninitiated bud set (3), initiation of the bud set and cessation of growth (2), formation of the buds (1), and completed bud set (0)98,143,194. The development of the apical and lateral bud burst was scored following UPOV193 and Ibanez et al.143 using the following scoring values: dormant buds enveloped by scales (0), swelling buds with diverging scales (1), sprouting buds (2), opened buds with leaves clustered (3), diverging leaves with rolled up blades (4), and completely unfolded leaves (5).
Phenotyping platform experiment
The growth of the transgenotypes was also investigated in a trial at the UPSC tree phenotyping platform (WIWAM Conveyor, SMO, Eeklo, Belgium; Fig. 2; Table 2 in SI). In this trial, hereafter called the phenotyping platform experiment (PPE), the trees automatically move around on a conveyor belt, allowing automatic daily watering, fertilization, and monitoring, and recording of growth parameters. The trees in the PPE were first in vitro-cultivated. The trees were then potted in a commercial mix of fertilized peat and soil (Yrkes Plantjord, Weibulls Horto, Hammenhög, Sweden) and grown for another four weeks under constant LD light, temperature, and relative humidity conditions (i.e., 18:6 h light/dark cycles; 18 °C; and 80% relative humidity, respectively). Afterward, the trees were brought into the phenotyping platform to expose them to similar light and temperature conditions (i.e., 18:6 h light/dark cycles and 20:18 °C warm/cold cycles, respectively) but altered relative humidity conditions (i.e., a relative air humidity of 60%). The red-to-far-red light ratio of the artificial LED lights on the phenotyping platform had an approximate value of 0.9 and an irradiance of 150 to 200 μmol m-2s−1. The soil in the pots was kept automatically at a target humidity of 1.9 (i.e., a value close to the experimentally determined water capacity), meaning that 1.9 l of water is added for 1 kg of dry soil. All trees were watered and fertilized based on the procedure in Wang et al.195. After three weeks in the greenhouse, the trees received a weekly dose of 200 ml of 1% Rika-S fertilization (7:1:5 N/P/K; Weibulls Horto, Hammenhög, Sweden). Whilst in the phenotyping platform experiment, the trees were watered twice daily (i.e., according to the target humidity) and fertilized every alternate day with 50 ml of 0.6% Rika-S. In the 7th week, the fertilization dose was increased to 75 ml of 0.6% Rika-S. Additionally, in the 4th and 7th weeks, the trees were treated with Nemasys C insecticide (BASF, Ludwigshafen, Germany). Primary growth of the transgenic trees was measured automatically every alternate day using a light curtain. Secondary growth was also measured every alternate day. However, this was done by automatic photometry of the trees’ sides, top and bottom using three Imperx B4820 RGB cameras. The data were subsequently recorded on a WIWAM computer and made available using the PIPPA web interface (https://pippa.psb.ugent.be/, UGent, Belgium).
Field experiments
Populus transgenotypes were amplified every fourth week until an adequate number of trees was reached. Subsequently, apical shoots of circa five centimeters were cut under sterile conditions and transferred into sterile plastic jars of one liter containing 130 mL MS growth medium (Duchefa Biochemie, Haarlem, Netherlands), adjusted to pH 5.6. Two shoots were transferred to each jar and exposed to cyclic LD light and temperature conditions (i.e., 18:6 h light/dark and 22:18 °C warm/cold cycles, respectively). After roots developed, each tree of c. 15 cm was replanted in a 1 l plastic pot containing a pre-fertilized 3:1 mix of fertilized peat and perlite (Yrkes Plantjord, Weibulls Horto, Hammenhög, Sweden). The potted trees were transferred to a greenhouse with similar environmental settings and treated as described in Johansson et al.196. After a month, the trees were put outside during the daytime to promote acclimatization to field conditions. Subsequently, they were transported and potted at the field site in Våxtorp.
The growth and phenology of the transgenotypes were tested in two transgenic field trials in Våxtorp (56°25′N, 13°47′E; 39 m.a.s.l.; Laholm municipality, Halland county, Sweden; permits 22-2655|12 and 18-3494|16), which are hereafter called field experiment 1 and 2 (FE1 and 2; Fig. 2; Wang et al.98 and Table 2). Before the start of the measurements in 2014 (FE1) and 2018 (FE2), the similarly sized and aged trees were planted in a randomized block design of 18 blocks with 3 × 2.3 m of spacing. In June 2014 (FE1), trees were planted in blocks containing 42 trees, including six WTT89 individuals and 18 transgenic individuals each representing an unique transgenotype. In October 2016 (FE2), trees were planted in blocks containing 40 trees, including four WTT89 individuals and 34 transgenic individuals each representing an unique transgenotype. Each time, a row of WTT89 individuals was planted around the randomized field design. Full-grown larch hedges flanked the long sides of the plots. Stem primary and secondary growth was measured in FE1 and FE2 every alternate month and half a year, respectively. The primary growth of each tree was measured from the base to the top axillar bud using a millimeter-scaled measuring pole. Only in FE1, were weekly phenological observations made on the development of the bud set and leaf senescence. As in the GCEs, the development of the bud set was scored using the scoring scheme of UPOV193 and Ibanez et al.143. The development of leaf senescence was assessed by measuring the chlorophyll content index (CCI) of five randomly chosen leaves per tree with a chlorophyll content meter (CCM 200 plus, Opti-Sciences). Chlorophyll detoxification is the most prominent feature of leaf senescence and allows trees to remobilize nutrients from their leaves towards more vital plant organs197,198. Since the CCI is a proxy for chlorophyll concentrations that have, within limits, a close to linear relationship with chlorophyll concentrations, the decline in the CCI can be used as a proxy for the development of the leaf senescence process199,200. CCI measurements were made at approximately the same day period and using the same leaf side199,201,202. Further details of the number of transgenotypes per experiment, the (approximate) number of tree individuals per transgenotype, and the duration and frequency of the measurement sampling in the GCEs, PPE, and FEs can be found in Table 2.
Statistical analyses
All growth and phenology data of the trees were analyzed using generalized additive (mixed) models141,203. These interpretable models assume that the relationship between the response and explanatory variables can be modeled using (non-)linear smooth functions203,204,205,206. The use of smoothers, as well as integrated smoothness selection methods, allows GAMs to have an a-priori-unknown but flexible predictor function of which the complexity is completely determined by the data. As a result, GAMs can unravel hidden relationships in the data whilst accounting for numerous statistical obstacles (e.g., overfitting, non-linear relationships, bias/variance tradeoffs)204,207,208,209. Because it is possible to implement random effects or specified correlation structures within the GAM framework, GAMs can also be used to model spatiotemporal data gathered from repeated observations in multiple individuals and locations over time210,211. One of the limitations of GAMs, unlike even more flexible statistical models (e.g., GAMLSS; generalized additive models for location, scale, and shape), is that it only focuses on the exponential distribution family and its location parameter μ (i.e., alternatively interpretable as the mean). However, few GAM distributions also allow us to model the distribution parameters σ (i.e., the shape of the response variable’s distribution; alternatively interpretable as the variance).
Generalized additive models with ordered-factor-smooth interactions
To test for significant differences between the growth and phenology of each transgenotype and the growth of WTT89 in the GCEs, PPE, and FEs, we adopted the GAM with ordered-factor-smooth interaction approach outlined in Wood140 and Rose et al. 212. In this pair-wise comparison of smoothers approach, separate difference smoothers are generated by the GAM for each factor level minus one reference level. In our study, we therefore used GAMs where the growth or phenology of each transgenotype was modeled as difference smoothers, which compare against a reference smoother (i.e., the growth of WTT89; additional models were made with the growth of WTElite as reference smoother solely for the AtGA20ox1 transgenotypes with WTElite in the background). Given that all the assumptions of the GAM are met, the deviation of a difference smooth’s upper or lower Bayesian Wabha/Silverman credible interval above or below the horizontal zero-line in a term plot would then visually indicate significant differences in the growth of the transgenotype compared to the growth of WTT89 (Fig. 3)135,136,137. R/mgcv’s summary function also provides a quantitative indication of the significance of the (ordered) difference smoothers.
To model the primary and secondary growth of the transgenic trees as a function of their covariates, we used the gam function in R/mgcv209,211,213. Several GAMs with similar construction but using different distributions were tested. The GAMs with Gaussian location-scale distribution (i.e., gaulss) consistently had the lowest Akaike Information Criterion (AIC), and the Gaussian location-scale distribution was therefore selected to be used in the GAMs modeling tree growth142. The most suited monotonic link function (i.e., the function linking the distribution parameter to the predictor; “identity”, “log” or “logb” for the mean; “logb” for the variance) varied case by case. For each experiment, and both the mean and variance, primary or secondary growth was modeled with the primary or secondary growth of the individual tree as the response variable. The fixed covariates of the primary and secondary growth were the time (continuous), transgenotype (categorical with up to 34 levels), and tree individual (categorical with up to 20 levels). The interaction term was modeled as an (ordered) random factor-smooth interaction between the covariates time and transgenotype. Random factor-smooth interaction smoothers were chosen because a (difference) smoother was required for each of a large number of transgenotypes and because these smoothers required the same smoothing parameter field141,214,215. The dependency among observations of the same tree individual was finally incorporated by using the individual as a random intercept (i.e., thus introducing a coefficient for each tree). To reduce overfitting to a minimum, we chose the restricted maximum likelihood (REML) argument as the smoothness selection method213,216. To test for significant differences between the development of leaf senescence in each transgenotype and the development of leaf senescence in WTT89 during FE1, a Gaussian GAM was made like those used to test tree growth (Eq. 1). The response variable was, in this case, the chlorophyll content index.
The GAMs modeling primary and secondary growth and leaf senescence are formulated in Eq. 1, where both location and scale parameters are considered. Yi represents the independent response variable observations on each individual indexed by i, with i = 1,…, n. D represents the distribution of the response variable, and g is the monotonic link function relating the predictor η to the distribution parameters µi (location) and σi (scale)217,218,219,220. Let t and x denote the covariates time and transgenotype, while \({\boldsymbol{f}}\left({{\boldsymbol{t}}}_{{\boldsymbol{i}}}\right)\), \({{\boldsymbol{f}}}_{{\boldsymbol{1}}}^{{\boldsymbol{o}}}\left({{\boldsymbol{t}}}_{{\boldsymbol{i}}},\,{{\boldsymbol{x}}}_{{\boldsymbol{i}}}\right),\) and \({\boldsymbol{\zeta }}\) represent the smooth function of the covariate ti, the ordered factor-smooth interaction function of the covariates ti and xi, and the random effect (i.e., the random effect in the mean for each individual i), respectively.
Cumulative threshold models
Cumulative threshold models, or more specifically ordered categorical family GAMs, were made to test whether the bud phenology of the transgenic trees differed significantly from the bud phenology of WTT89. We tested specifically for differences in the timing of the bud set between the transgenic and WTT89 trees grown during GC3, GC4, and FE1. We also tested for differences in the timing of the apical and lateral bud burst between the transgenic and WTT89 trees grown during GC3 and GC4. To test for true significance in the differences, we again used the GAM with an ordered-factor-smooth interaction approach. This time, we modeled ordered categorical data. In an additional step, we computed the predicted probabilities of a bud to be in a specific bud set or bud burst stage at each moment in time (Fig. 4)141,142. Wood141; p. 176, in fact, notes that “the linear predictor [in this kind of ordered categorical family GAMs] provides the expected value of a latent variable according to a logistic distribution. The probability of this latent variable to be in between certain cut-points (i.e., the categories or stages) then provides the probability of the ordered categorical variable to be of the corresponding stage”.
To model the bud set and bud burst of the transgenic trees as a function of their covariates, we again used the gam function in R/mgcv209,211,213. Due to the ordered and categorized data, GAMs with ordered categorical distribution (i.e., ocat) and “identity” monotonic link function were subsequently made to model the bud set and bud burst. The bud phenology was modeled with the respective bud set or bud burst for each tree as the response variable. The fixed covariates of the bud set and bud burst were the time (continuous), transgenotype (categorical with up to 19 levels), and tree individual (categorical with up to 12 levels). As for the GAMs modeling growth, the interaction term was modeled as an (ordered) random factor-smooth interaction smoother between the covariates time and transgenotype. A reference smoother was introduced in the ordered categorical GAMs. Random factor-smooth interaction smoothers were also chosen because a (difference) smooth was required for each of the large number of transgenotypes and required the same smoothing parameter141,214,215. The dependency among observations of the same tree individual was again incorporated using the individual as a random intercept. Term plots, generated with the help of the data_slice and fitted_values functions from R/gratia, provide a visual indication of substantial differences between the predicted probabilities of a bud to be in a specific bud set or bud burst stage at a particular moment221. Equations (2) and (3) provide the cumulative threshold models modeling the bud set and bud burst.
Model assumptions
The interpretation of the GAMs, their output, and the extent of the potentially significant differences between their smoothers depends on the degree to which their underlying parametric assumptions are met and reported222,223,224,225. Following Zuur et al.226 these are, in order of importance, the homogeneity, normality, concurvity, nonlinear dependency, and temporal dependency of the model residuals. The assumption of independent and identically distributed residuals (IID) combines aspects of heterogeneity and dependency and requires assessment. The zero adjusting assumption is not applicable to this study. Failure to meet, or deviations from, the model assumptions might increase type I or II errors or affect the effect size estimation or its significance227,228. However, a quantitative analysis of the potential error is not straightforward.
All model residuals were extracted using the residuals functions from R/mgcv140. Several other functions from the same package provided a first indication of the degree to which many of the parametric assumptions are met. The summary function provides approximate p-values on a significant trend in the smoothers and an idea of the model deviance. The model deviance generally proved to be high, adding consideration to the notion that although the hypothesis complexity may increase (i.e., the number of parameters), one can still have low bias and variance (i.e., the interpolation threshold) and that infinite overparameterization can be preferable to any finite number of parameters229,230,231. Deviations from homogeneity or normality of the residuals could be assessed visually using four diagnostic plots of the normalized quantile residuals provided by the gam.check function232. The homoscedasticity in the residuals was also further investigated using the check1D, l_densCheck and l_gridCheck1D functions in R/mgcViz233. In light of the central limit theory and the rather small sample sizes typical for biological experiments, we used the provided histograms and quantile-quantile plots to assess whether the residuals were normally distributed. The gam.check function, which provides the k-indices and p-values, was used to check the required basis dimensions for the smoothers234. Low p-values in combination with k-indices lower than one would suggest that the basis dimension of the smoothers was too low. The shapiro.test (Shapiro-Wilk test), ad.test (Anderson-Darling test) and cvm.test (Cramer-Von-Mises test) functions in the R/base, R/nortest, and R/goftest packages were also used to test whether the model residuals followed a normal distribution235,236,237. Non-linearity in the model residuals was assessed with the bdsTest function (Brock–Dechert–Scheinkmand and LeBaron statistic test) in R/fNonlinear238. The IID characteristics of the model residuals were investigated by testing for white noise in the error vector using the whitenoise.test function in R/normwhn.test and the standard Box.test function (Ljung-Box test)239,240,241. Residual temporal autocorrelation was visually assessed using the acf and pacf functions in R/mgcv. Only the raw residual ACF and pACF plots could be provided. Indications of temporal autocorrelation were further investigated by looking for trends or difference stationarity in the residuals. Four tests were used from the R/urca package242. The KPSS (Kwiatkowski-Phillips-Schmidt-Shin), PP (Phillips-Perron), ADF (Augmented-Dickey-Fuller), ERS (Elliot-Rothenberg and Stock Point Optimal), and ADF-GLS tests for the presence of a unit root (i.e., a stochastic trend) in a time series test, were performed using the ur.kpss, ur.pp, ur.df and ur.ers functions, respectively243,244,245. The additional NP (Ng and Perron) unit root test, giving valid results even when an unknown ARMA process is present, was implemented using the CADFtest function in R/CADFtest246. The potential need to add autoregressive or moving average orders to the GAMs was tested by running the auto.arima function in R/forecast247. Tests were also done to characterize the data further, given that the parametric model assumptions were, according to a strict interpretation, often not met. The Hurst coefficient for long-range dependence and randomness in a system was performed using the WhittleEst function from the R/longmemo package248,249,250,251,252. The skewness (i.e., the asymmetry around the mean of the probability distribution) and kurtosis (i.e., the magnitude in which the tails of a distribution differ from the tails of a normal distribution) were calculated using the skewness, kurtosis and describe functions in the packages R/e1071, R/sur and R/pscyh253,254,255. Indicative for a “good” model fit is a mean, variance, skewness, kurtosis, and Filliben correlation coefficient of 0,1,0,3 and 1, respectively256. The modality of the data distributions was tested using the dip.test function (i.e., unimodal test) in R/diptest, bimodality_amplitude function (i.e., bimodal test) in R/modes and modetest function (i.e., multimodal test) in R/multimode257,258,259. Graphical output was made mainly using R/ggplot2 and R/dplyr260,261,262,263,264,265.
Data availability
Extra information is provided in the supplementary information files. These include also data and code.
References
de Dios, V. R. et al. Endogenous circadian regulation of carbon dioxide exchange in terrestrial ecosystems. Glob. Change Biol. 18, 1956–1970, https://doi.org/10.1111/j.1365-2486.2012.02664.x (2012).
Doughty, C. E. et al. Circadian rhythms constrain leaf and canopy gas exchange in an Amazonian forest. Geophysical Research Letters 33. https://doi.org/10.1029/2006GL026750 (2006).
Pugh, T. A. M. et al. Role of forest regrowth in global carbon sink dynamics. Proc. Natl Acad. Sci. 116, 4382–4387, https://doi.org/10.1073/pnas.1810512116 (2019).
Huang, J. G. et al. Photoperiod and temperature as dominant environmental drivers triggering secondary growth resumption in Northern Hemisphere conifers. Proc. Natl Acad. Sci. USA 117, 20645–20652, https://doi.org/10.1073/pnas.2007058117 (2020).
Richardson, A. D. et al. Influence of spring and autumn phenological transitions on forest ecosystem productivity. Philos. Trans. R. Soc. Lond. B Biol. Sci. 365, 3227–3246, https://doi.org/10.1098/rstb.2010.0102 (2010).
Etzold, S. et al. Number of growth days and not length of the growth period determines radial stem growth of temperate trees. Ecol. Lett. 25, 427–439, https://doi.org/10.1111/ele.13933 (2022).
Lehnebach, R. et al. High-resolution X-ray computed tomography: a new workflow for the analysis of xylogenesis and intra-seasonal wood biomass production. Front. Plant Sci. 12. https://doi.org/10.3389/fpls.2021.698640 (2021).
Zlobin, I. E. Tree post-drought recovery: scenarios, regulatory mechanisms and ways to improve. Biol. Rev. Camb. Philos. Soc. 99, 1595–1612, https://doi.org/10.1111/brv.13083 (2024).
Lockhart, J. A. An analysis of irreversible plant cell elongation. J. Theor. Biol. 8, 264–275, https://doi.org/10.1016/0022-5193(65)90077-9 (1965).
Zweifel, R. et al. Why trees grow at night. N. Phytol. 231, 2174–2185, https://doi.org/10.1111/nph.17552 (2021).
Körner, C. Carbon limitation in trees. J. Ecol. 91, 4–17 (2003).
Zweifel, R. et al. TreeNet–the biological drought and growth indicator network. Front. For. Glob. Chang. 4. https://doi.org/10.3389/ffgc.2021.776905 (2021).
Peters, R. L. et al. Turgor–a limiting factor for radial growth in mature conifers along an elevational gradient. N. Phytol. 229, 213–229, https://doi.org/10.1111/nph.16872 (2021).
Fatichi, S. et al. Modelling carbon sources and sinks in terrestrial vegetation. N. Phytol. 221, 652–668, https://doi.org/10.1111/nph.15451 (2019).
Rossi, S. et al. A meta-analysis of cambium phenology and growth: linear and non-linear patterns in conifers of the northern hemisphere. Ann. Bot. 112, 1911–1920, https://doi.org/10.1093/aob/mct243 (2013).
Körner, C. Paradigm shift in plant growth control. Curr. Opin. Plant Biol. 25, 107–114, https://doi.org/10.1016/j.pbi.2015.05.003 (2015).
de Dios, V. R. et al. Processes driving nocturnal transpiration and implications for estimating land evapotranspiration. Sci. Rep. 5, 10975, https://doi.org/10.1038/srep10975 (2015).
Burki, T. Nobel Prize awarded for discoveries in circadian rhythm. Lancet 390, e25, https://doi.org/10.1016/S0140-6736(17)32661-2 (2017).
Wulund, L. et al. A brief history of circadian time: The emergence of redox oscillations as a novel component of biological rhythms. Perspect. Sci. 6, 27–37, https://doi.org/10.1016/j.pisc.2015.08.002 (2015).
Yanovsky, M. J. et al. Molecular basis of seasonal time measurement in Arabidopsis. Nature 419, 308–312, https://doi.org/10.1038/nature00996 (2002).
Kumar, V. Biological Timekeeping: Clocks, Rhythms and Behaviour. (Springer, 2017).
Mancuso, S. et al. Rhythms in Plants: Dynamic Responses in a Dynamic Environment. (Springer, 2015).
Mariën, B. et al. Timing leaf senescence: a generalized additive models for location, scale and shape approach. Agric. For. Meteorol. 315 (2022). https://doi.org/10.1016/j.agrformet.2022.108823
Post, E. Time in Ecology https://doi.org/10.2307/j.ctv3s8sns (Princeton University Press, 2019).
Borchert, R. et al. Photoperiodic induction of synchronous flowering near the Equator. Nature 433, 627–629, https://doi.org/10.1038/nature03259 (2005).
de Dios, V. R. et al. Genetic variation in circadian regulation of nocturnal stomatal conductance enhances carbon assimilation and growth. Plant Cell Environ. 39, 3–11, https://doi.org/10.1111/pce.12598 (2016).
Dodd, A. N. et al. Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science 309, 630–633, https://doi.org/10.1126/science.1115581 (2005).
Johansson, M. The circadian clock in annuals and perennials: coordination of Growth with Environmental Rhythms. Ph.D Thesis, (Umeå University, 2010).
Hubbard, K. et al. Rhythms of life: the plant circadian clock - (By Katherine Hubbard and Antony Dodd). Plant Cell 28, tpc.116.tt0416. https://doi.org/10.1105/tpc.116.tt0416 (2016).
Merrow, M. et al. The circadian cycle: daily rhythms from behaviour to genes. EMBO Rep. 6, 930–935, https://doi.org/10.1038/sj.embor.7400541 (2005).
de Montaigu, A. et al. Plant development goes like clockwork. Trends Genet. 26, 296–306, https://doi.org/10.1016/j.tig.2010.04.003 (2010).
Johansson, M. et al. in Plant Circadian Networks: Methods and Protocols (eds D. Staiger, S. Davis, & A. M. Davis) 227–242 (Springer US, 2022).
Locke, J. C. et al. Extension of a genetic network model by iterative experimentation and mathematical analysis. Mol. Syst. Biol. 1, 0–13, https://doi.org/10.1038/msb4100018 (2005).
Locke, J. C. W. et al. Experimental validation of a predicted feedback loop in the multi-oscillator clock of Arabidopsis thaliana. Mol. Syst. Biol. 2, 59, https://doi.org/10.1038/msb4100102 (2006).
Millar, A. J. The intracellular dynamics of circadian clocks reach for the light of ecology and evolution. Annu. Rev. Plant Biol. 67, 595–618, https://doi.org/10.1146/annurev-arplant-043014-115619 (2016).
Solomon, O. L. et al. Diurnal and circadian patterns of gene expression in the developing xylem of Eucalyptus trees. South Afr. J. Bot. 76, 425–439, https://doi.org/10.1016/j.sajb.2010.02.087 (2010).
Gould, P. D. et al. The molecular basis of temperature compensation in the Arabidopsis circadian clock. Plant Cell 18, 1177–1187, https://doi.org/10.1105/tpc.105.039990 (2006).
Thomashow, M. F. Plant cold acclimation: freezing tolerance genes and regulatory mechanisms. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 571–599, https://doi.org/10.1146/annurev.arplant.50.1.571 (1999).
Jones, M. A. et al. Jumonji domain protein JMJD5 functions in both the plant and human circadian systems. Proc. Natl. Acad. Sci. 107, 21623–21628, https://doi.org/10.1073/pnas.1014204108 (2010).
Helfer, A. et al. LUX ARRHYTHMO encodes a nighttime repressor of circadian gene expression in the Arabidopsis core clock. Curr. Biol. 21, 126–133, https://doi.org/10.1016/j.cub.2010.12.021 (2011).
Ruoff, P. et al. Modeling temperature compensation in chemical and biological oscillators. Chronobiol. Int. 14, 499–510, https://doi.org/10.3109/07420529709001471 (1997).
Weiser, C. J. Cold resistance and injury in woody plants. Science 169, 1269–1278 (1970).
Dong, M. A. et al. Circadian clock-associated 1 and late elongated hypocotyl regulate expression of the C-repeat binding factor (CBF) pathway in Arabidopsis. Proc. Natl. Acad. Sci. USA 108, 7241–7246, https://doi.org/10.1073/pnas.1103741108 (2011).
Benedict, C. et al. The CBF1-dependent low temperature signalling pathway, regulon and increase in freeze tolerance are conserved in Populus spp. plant. Cell Environ. 29, 1259–1272, https://doi.org/10.1111/j.1365-3040.2006.01505.x (2006).
Arora, R. et al. Cold acclimation and alterations in dehydrin-like and bark storage proteins in the leaves of sibling deciduous and evergreen peach. J. Am. Soc. Hortic. Sci. 121. https://doi.org/10.21273/JASHS.121.5.915 (1996).
Stockinger, E. J. et al. Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc. Natl Acad. Sci. USA 94, 1035–1040, https://doi.org/10.1073/pnas.94.3.1035 (1997).
Fowler, S. G. et al. Low-temperature induction of Arabidopsis CBF1, 2, and 3 is gated by the circadian clock. Plant Physiol. 137, 961–968, https://doi.org/10.1104/pp.104.058354 (2005).
Liu, Q. et al. Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 10, 1391–1406, https://doi.org/10.1105/tpc.10.8.1391 (1998).
Harmer, S. L. et al. Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science 290, 2110–2113, https://doi.org/10.1126/science.290.5499.2110 (2000).
Kim, H. J. et al. Light signalling mediated by phytochrome plays an important role in cold-induced gene expression through the C-repeat/dehydration responsive element (C/DRE) in Arabidopsis thaliana. Plant J. 29, 693–704, https://doi.org/10.1046/j.1365-313x.2002.01249.x (2002).
Welling, A. et al. Independent activation of cold acclimation by low temperature and short photoperiod in hybrid aspen. Plant Physiol. 129, 1633–1641, https://doi.org/10.1104/pp.003814 (2002).
Schrader, J. et al. Cambial meristem dormancy in trees involves extensive remodelling of the transcriptome. Plant J. 40, 173–187, https://doi.org/10.1111/j.1365-313X.2004.02199.x (2004).
Puhakainen, T. et al. Short-day potentiation of low temperature-induced gene expression of a C-repeat-binding factor-controlled gene during cold acclimation in silver birch. Plant Physiol. 136, 4299–4307, https://doi.org/10.1104/pp.104.047258 (2005).
Yanovsky, M. J. et al. Signaling networks in the plant circadian system. Curr. Opin. Plant Biol. 4, 429–435, https://doi.org/10.1016/S1369-5266(00)00196-5 (2001).
Ramos, A. et al. Winter disruption of the circadian clock in chestnut. Proc. Natl. Acad. Sci. USA 102, 7037–7042, https://doi.org/10.1073/pnas.0408549102 (2005).
de Dios, V. R. et al. Woody clockworks: circadian regulation of night-time water use in Eucalyptus globulus. N. Phytol. 200, 743–752, https://doi.org/10.1111/nph.12382 (2013).
Kim, H. et al. Age-associated circadian period changes in Arabidopsis leaves. J. Exp. Bot. 67, 2665–2673, https://doi.org/10.1093/jxb/erw097 (2016).
Schultz, T. F. et al. Circadian clocks in daily and seasonal control of development. Science 301, 326–328, https://doi.org/10.1126/science.1085935 (2003).
Böhlenius, H. et al. CO/FT regulatory module controls timing of flowering and seasonal growth cessation in trees. Science 312, 1040–1043, https://doi.org/10.1126/science.1126038 (2006).
Ramos-Sánchez, J. M. et al. LHY2 integrates night-length information to determine timing of poplar photoperiodic growth. Curr. Biol. 29, 2402–2406.e2404, https://doi.org/10.1016/j.cub.2019.06.003 (2019).
Edwards, K. D. et al. Circadian clock components control daily growth activities by modulating cytokinin levels and cell division-associated gene expression in Populus trees. Plant Cell Environ. 41, 1468–1482, https://doi.org/10.1111/pce.13185 (2018).
Strayer, C. et al. Cloning of the Arabidopsis clock gene TOC1, an autoregulatory response regulator homolog. Science 289, 768–771, https://doi.org/10.1126/science.289.5480.768 (2000).
Schaffer, R. et al. The late elongated hypocotyl mutation of Arabidopsis disrupts circadian rhythms and the photoperiodic control of flowering. Cell 93, 1219–1229, https://doi.org/10.1016/s0092-8674(00)81465-8 (1998).
Takeuchi, T. et al. Light and the circadian clock mediate time-specific changes in sensitivity to UV-B stress under light/dark cycles. J. Exp. Bot. 65, 6003–6012, https://doi.org/10.1093/jxb/eru339 (2014).
McClung, C. R. Sem. Cell Dev. Biol. 430–436 (Elsevier).
Linde, A.-M. et al. Early evolution of the land plant circadian clock. N. Phytol. 216, 576–590, https://doi.org/10.1111/nph.14487 (2017).
Legris, M. et al. Phytochrome B integrates light and temperature signals in Arabidopsis. Science 354, 897–900, https://doi.org/10.1126/science.aaf5656 (2016).
Franklin, K. A. et al. Phytochrome functions in Arabidopsis development. J. Exp. Bot. 61, 11–24, https://doi.org/10.1093/jxb/erp304 (2010).
Mancinelli, A. L. et al. The “High Irradiance Responses” of plant photomorphogenesis. Bot. Rev. 44, 129–180 (1978).
Poorter, H. et al. A meta-analysis of plant responses to light intensity for 70 traits ranging from molecules to whole plant performance. N. Phytol. 223, 1073–1105, https://doi.org/10.1111/nph.15754 (2019).
Neff, M. M. et al. Light: an indicator of time and place. Genes Dev. 14, 257–271, https://doi.org/10.1101/gad.14.3.257 (2000).
Harmer, S. L. The circadian system in higher plants. Annu. Rev. Plant Biol. 60, 357–377, https://doi.org/10.1146/annurev.arplant.043008.092054 (2009).
Olsen, J. E. et al. Ectopic expression of oat phytochrome A in hybrid aspen changes critical daylength for growth and prevents cold acclimatization. Plant J. 12, 1339–1350, https://doi.org/10.1046/j.1365-313x.1997.12061339.x (1997).
Nozue, K. et al. Rhythmic growth explained by coincidence between internal and external cues. Nature 448, 358–361, https://doi.org/10.1038/nature05946 (2007).
Banerjee, R. et al. Plant blue-light receptors. Planta 220, 498–502, https://doi.org/10.1007/s00425-004-1418-z (2005).
Kim, J.-Y. et al. Light-regulated translation mediates gated induction of the Arabidopsis clock protein LHY. EMBO J. 22, 935–944, https://doi.org/10.1093/emboj/cdg075 (2003).
Takata, N. et al. Phylogenetic footprint of the plant clock system in angiosperms: evolutionary processes of Pseudo-Response Regulators. BMC Evolut. Biol. 10, 126, https://doi.org/10.1186/1471-2148-10-126 (2010).
Murakami, M. et al. Circadian-associated rice pseudo response regulators (OsPRRs): insight into the control of flowering time. Biosci. Biotechnol. Biochem. 69, 410–414, https://doi.org/10.1271/bbb.69.410 (2005).
Eriksson, M. E. et al. Response regulator homologues have complementary, light-dependent functions in the Arabidopsis circadian clock. Planta 218, 159–162, https://doi.org/10.1007/s00425-003-1106-4 (2003).
Alabadi, D. et al. Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science 293, 880–883, https://doi.org/10.1126/science.1061320 (2001).
Kiba, T. et al. Targeted degradation of pseudo-response regulator5 by an SCFZTL complex regulates clock function and photomorphogenesis in Arabidopsis thaliana. Plant Cell 19, 2516–2530, https://doi.org/10.1105/tpc.107.053033 (2007).
Fujiwara, S. et al. Post-translational regulation of the Arabidopsis circadian clock through selective proteolysis and phosphorylation of pseudo-response regulator proteins. J. Biol. Chem. 283, 23073–23083, https://doi.org/10.1074/jbc.M803471200 (2008).
Harmon, F. et al. CUL1 regulates TOC1 protein stability in the Arabidopsis circadian clock. Plant J. 55, 568–579, https://doi.org/10.1111/j.1365-313X.2008.03527.x (2008).
Kim, W. Y. et al. ZEITLUPE is a circadian photoreceptor stabilized by GIGANTEA in blue light. Nature 449, 356–360, https://doi.org/10.1038/nature06132 (2007).
Ding, J. et al. GIGANTEA-like genes control seasonal growth cessation in Populus. N. Phytol. 218, 1491–1503, https://doi.org/10.1111/nph.15087 (2018).
Song, Y. H. et al. Distinct roles of FKF1, Gigantea, and Zeitlupe proteins in the regulation of Constans stability in Arabidopsis photoperiodic flowering. Proc. Natl. Acad. Sci. USA 111, 17672–17677, https://doi.org/10.1073/pnas.1415375111 (2014).
Imaizumi, T. et al. FKF1 F-box protein mediates cyclic degradation of a repressor of CONSTANS in Arabidopsis. Science 309, 293–297 (2005).
Song, Y. H. et al. FKF1 conveys timing information for CONSTANS stabilization in photoperiodic flowering. Science 336, 1045–1049, https://doi.org/10.1126/science.1219644 (2012).
Park, D. H. et al. Control of circadian rhythms and photoperiodic flowering by the Arabidopsis GIGANTEA gene. Science 285, 1579–1582, https://doi.org/10.1126/science.285.5433.1579 (1999).
Jung, J. H. et al. The GIGANTEA-regulated microRNA172 mediates photoperiodic flowering independent of CONSTANS in Arabidopsis. Plant Cell 19, 2736–2748, https://doi.org/10.1105/tpc.107.054528 (2007).
Sawa, M. et al. GIGANTEA directly activates flowering locus T in Arabidopsis thaliana. Proc. Natl. Acad. Sci. 108, 11698–11703, https://doi.org/10.1073/pnas.1106771108 (2011).
Baudry, A. et al. F-box proteins FKF1 and LKP2 act in concert with ZEITLUPE to control Arabidopsis clock progression. Plant Cell 22, 606–622, https://doi.org/10.1105/tpc.109.072843 (2010).
Triozzi, P. M. et al. Photoperiodic regulation of shoot apical growth in poplar. Front. Plant Sci. 9. https://doi.org/10.3389/fpls.2018.01030 (2018).
Castillejo, C. et al. The balance between CONSTANS and TEMPRANILLO activities determines FT expression to trigger flowering. Curr. Biol. 18, 1338–1343, https://doi.org/10.1016/j.cub.2008.07.075 (2008).
Corbesier, L. et al. FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316, 1030–1033, https://doi.org/10.1126/science.1141752 (2007).
Suárez-López, P. et al. CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature 410, 1116–1120, https://doi.org/10.1038/35074138 (2001).
Andre, D. et al. FLOWERING LOCUS T paralogs control the annual growth cycle in Populus trees. Curr. Biol. 32, 2988–2996 e2984, https://doi.org/10.1016/j.cub.2022.05.023 (2022).
Wang, J. et al. A major locus controls local adaptation and adaptive life history variation in a perennial plant. Genome Biol. 19, 72, https://doi.org/10.1186/s13059-018-1444-y (2018).
Pin, P. A. et al. The multifaceted roles of FLOWERING LOCUS T in plant development. Plant Cell Environ. 35, 1742–1755, https://doi.org/10.1111/j.1365-3040.2012.02558.x (2012).
Bernhardsson, C. et al. Molecular population genetics of elicitor-induced resistance genes in European Aspen (Populus tremula L., Salicaceae). PloS One 6, e24867, https://doi.org/10.1371/journal.pone.0024867 (2011).
Lisso, J. et al. The AtNFXL1 gene encodes a NF-X1 type zinc finger protein required for growth under salt stress. FEBS Lett. 580, 4851–4856, https://doi.org/10.1016/j.febslet.2006.07.079 (2006).
Ashelford, K. et al. Full genome re-sequencing reveals a novel circadian clock mutation in Arabidopsis. Genome Biol. 12, R28, https://doi.org/10.1186/gb-2011-12-3-r28 (2011).
Johansson, M. et al. Partners in time: EARLY BIRD associates with ZEITLUPE and regulates the speed of the Arabidopsis clock. Plant Physiol. 155, 2108–2122, https://doi.org/10.1104/pp.110.167155 (2011).
Ma, Y. et al. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 324, 1064–1068, https://doi.org/10.1126/science.1172408 (2009).
Park, S.-Y. et al. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324, 1068–1071, https://doi.org/10.1126/science.1173041 (2009).
Singh, R. K. et al. A genetic network mediating the control of bud break in hybrid aspen. Nat. Commun. 9, 4173, https://doi.org/10.1038/s41467-018-06696-y (2018).
Liang, H. et al. Poplar allene oxide synthase 1 gene promoter drives rapid and localized expression by wounding. Biotechnol. J. Int. 16–28. https://doi.org/10.9734/bji/2021/v25i530151 (2021).
Sasaki-Sekimoto, Y. et al. Coordinated activation of metabolic pathways for antioxidants and defence compounds by jasmonates and their roles in stress tolerance in Arabidopsis. Plant J. 44, 653–668, https://doi.org/10.1111/j.1365-313X.2005.02560.x (2005).
Song, Y. et al. Comparison of the physiological effects and transcriptome responses of Populus simonii under different abiotic stresses. Plant Mol. Biol. 86, 139–156, https://doi.org/10.1007/s11103-014-0218-5 (2014).
Weigel, D. et al. A developmental switch sufficient for flower initiation in diverse plants. Nature 377, 495–500, https://doi.org/10.1038/377495a0 (1995).
Rottmann, W. H. et al. Diverse effects of overexpression of LEAFY and PTLF, a poplar (Populus) homolog of LEAFY/FLORICAULA, in transgenic poplar and Arabidopsis. Plant J. 22, 235–245, https://doi.org/10.1046/j.1365-313x.2000.00734.x (2000).
Weigel, D. et al. LEAFY controls floral meristem identity in Arabidopsis. Cell 69, 843–859, https://doi.org/10.1016/0092-8674(92)90295-n (1992).
Gregis, V. et al. Identification of pathways directly regulated by SHORT VEGETATIVE PHASE during vegetative and reproductive development in Arabidopsis. Genome Biol. 14, R56, https://doi.org/10.1186/gb-2013-14-6-r56 (2013).
Eriksson, M. The Role of Phytochrome A and Gibberellins in Growth under Long and Short Day Conditions Studies in Hybrid Aspen. Ph.D Thesis, (Swedish University of Agricultural Sciences, 2000).
Eriksson, M. E. et al. Increased gibberellin biosynthesis in transgenic trees promotes growth, biomass production and xylem fiber length. Nat. Biotechnol. 18, 784–788, https://doi.org/10.1038/77355 (2000).
Eriksson, M. E. et al. Daylength and spatial expression of a gibberellin 20-oxidase isolated from hybrid aspen (L. x Michx). Planta 214, 920–930, https://doi.org/10.1007/s00425-001-0703-3 (2002).
Eriksson, M. E. et al. Transgenic hybrid aspen trees with increased gibberellin (GA) concentrations suggest that GA acts in parallel with FLOWERING LOCUS T2 to control shoot elongation. N. Phytol. 205, 1288–1295, https://doi.org/10.1111/nph.13144 (2015).
Zawaski, C. et al. Roles of gibberellin catabolism and signaling in growth and physiological response to drought and short-day photoperiods in Populus trees. PloS One 9, e86217, https://doi.org/10.1371/journal.pone.0086217 (2014).
Hedden, P. et al. Manipulation of hormone biosynthetic genes in transgenic plants. Curr. Opin. Biotechnol. 11, 130–137, https://doi.org/10.1016/s0958-1669(00)00071-9 (2000).
Kende, H. et al. The five “classical” plant hormones. Plant Cell 9, 1197–1210, https://doi.org/10.1105/tpc.9.7.1197 (1997).
Hennig, L. et al. Arabidopsis MSI1 is required for epigenetic maintenance of reproductive development. Development 130, 2555–2565, https://doi.org/10.1242/dev.00470 (2003).
Ausín, I. et al. Regulation of flowering time by FVE, a retinoblastoma-associated protein. Nat. Genet. 36, 162–166, https://doi.org/10.1038/ng1295 (2004).
Guitton, A.-E. et al. Loss of function of MULTICOPY SUPPRESSOR OF IRA 1 produces nonviable parthenogenetic embryos in Arabidopsis. Curr. Biol. 15, 750–754, https://doi.org/10.1016/j.cub.2005.02.066 (2005).
Guitton, A.-E. et al. Identification of new members of Fertilisation Independent Seed Polycomb Group pathway involved in the control of seed development in Arabidopsis thaliana. Development 131, 2971–2981, https://doi.org/10.1242/dev.01168 (2004).
Kohler, C. et al. Arabidopsis MSI1 is a component of the MEA/FIE Polycomb group complex and required for seed development. EMBO J. 22, 4804–4814, https://doi.org/10.1093/emboj/cdg444 (2003).
Steinbach, Y. et al. Arabidopsis MSI1 functions in photoperiodic flowering time control. Front. Plant Sci. 5. https://doi.org/10.3389/fpls.2014.00077 (2014).
Englund, M. Molecular Control of Activity–Dormancy Transitions in Populus. Ph.D. Thesis, (Swedish University of Agricultural Sciences, 2010).
Sasaki, Y. et al. Monitoring of methyl jasmonate-responsive genes in Arabidopsis by cDNA macroarray: self-activation of jasmonic acid biosynthesis and crosstalk with other phytohormone signaling pathways. DNA Res. 8, 153–161, https://doi.org/10.1093/dnares/8.4.153 (2001).
Mueller, M. J. Enzymes involved in jasmonic acid biosynthesis. Physiol. Plant. 100, 653–663, https://doi.org/10.1111/j.1399-3054.1997.tb03072.x (1997).
Sivasankar, S. et al. Expression of allene oxide synthase determines defense gene activation in tomato. Plant Physiol. 122, 1335–1342, https://doi.org/10.1104/pp.122.4.1335 (2000).
Tijet, N. et al. Allene oxide synthases and allene oxides. Prostaglandins Other Lipid Mediat. 68-69, 423–431, https://doi.org/10.1016/S0090-6980(02)00046-1 (2002).
Placido, D. F. et al. Downregulation of an allene oxide synthase gene improves photosynthetic rate and alters phytohormone homeostasis in field-grown guayule. Ind. Crops Prod. 153, 112341 (2020).
Siqueira-Junior, C. L. et al. Wound response in passion fruit (Passiflora f. edulis flavicarpa) plants: gene characterization of a novel chloroplast-targeted allene oxide synthase up-regulated by mechanical injury and methyl jasmonate. Plant Cell Rep. 27, 387–397, https://doi.org/10.1007/s00299-007-0451-3 (2008).
Khaipho-Burch, M. et al. Genetic modification can improve crop yields - but stop overselling it. Nature 621, 470–473, https://doi.org/10.1038/d41586-023-02895-w (2023).
Marra, G. et al. Coverage properties of confidence intervals for generalized additive model components. Scand. J. Stat. 39, 53–74, https://doi.org/10.1111/j.1467-9469.2011.00760.x (2012).
Wahba, G. Bayesian confidence-intervals for the cross-validated smoothing spline. J. R. Stat. Soc. Ser. B-Methodol. 45, 133–150, https://doi.org/10.1111/j.2517-6161.1983.tb01239.x (1983).
Nychka, D. Bayesian confidence-intervals for smoothing splines. J. Am. Stat. Assoc. 83, 1134–1143, https://doi.org/10.2307/2290146 (1988).
Kozarewa, I. et al. Alteration of PHYA expression change circadian rhythms and timing of bud set in Populus. Plant Mol. Biol. 73, 143–156, https://doi.org/10.1007/s11103-010-9619-2 (2010).
Jonsén, D. et al. Plants with improved growth. 2018/5/3, Patent application WO2018080389A1 (2018).
Sjölander, J. Timing is Everything: The role of the circadian clock in plant growth and adaptation. Ph.D. Thesis, (Umeå University, 2024).
Wood, S. N. Generalized Additive Models (Chapman and Hall/CRC, 2017) https://doi.org/10.1201/9781315370279.
Wood, S. N. et al. Smoothing parameter and model selection for general smooth models. J. Am. Stat. Assoc. 111, 1548–1563, https://doi.org/10.1080/01621459.2016.1180986 (2016).
Ibáñez, C. et al. Circadian clock components regulate entry and affect exit of seasonal dormancy as well as winter hardiness in Populus trees. Plant Physiol. 153, 1823–1833, https://doi.org/10.1104/pp.110.158220 (2010).
Gutierrez, C. The Arabidopsis cell division cycle. Arabidopsis Book 7, e0120, https://doi.org/10.1199/tab.0120 (2009).
Castellano, M. M. et al. Expression and stability of Arabidopsis CDC6 are associated with endoreplication. Plant Cell 13, 2671–2686, https://doi.org/10.1105/tpc.010329 (2001).
Fung-Uceda, J. et al. The circadian clock sets the time of DNA replication licensing to regulate growth in Arabidopsis. Dev. Cell 45, 101–113.e104, https://doi.org/10.1016/j.devcel.2018.02.022 (2018).
Nordborg, M. et al. Energy analysis of poplar production for bioenergy in Sweden. Biomass. Bioenergy 112, 110–120, https://doi.org/10.1016/j.biombioe.2018.01.021 (2018).
Djomo, S. N. et al. Energy and greenhouse gas balance of bioenergy production from poplar and willow: a review. Glob. Change Biol. Bioenergy 3, 181–197, https://doi.org/10.1111/j.1757-1707.2010.01073.x (2011).
Böhlenius, H. et al. Biomass production and fuel characteristics from long rotation poplar plantations. Biomass. Bioenergy 178, 106940, https://doi.org/10.1016/j.biombioe.2023.106940 (2023).
He, F. et al. Genome-wide investigation of the NF-X1 gene family in Populus trichocarpa expression profiles during development and stress. Int. J. Mol. Sci. 22. https://doi.org/10.3390/ijms22094664 (2021).
Ding, J. et al. Phytochrome B and PHYTOCHROME INTERACTING FACTOR8 modulate seasonal growth in trees. N. Phytol. 232, 2339–2352, https://doi.org/10.1111/nph.17350 (2021).
Fowler, S. et al. GIGANTEA: a circadian clock-controlled gene that regulates photoperiodic flowering in Arabidopsis and encodes a protein with several possible membrane-spanning domains. EMBO J. 18, 4679–4688, https://doi.org/10.1093/emboj/18.17.4679 (1999).
Webb, A. A. R. et al. Understanding circadian regulation of carbohydrate metabolism in arabidopsis using mathematical models. Plant Cell Physiol. 56, 586–593, https://doi.org/10.1093/pcp/pcv033 (2015).
Flis, A. et al. Multiple circadian clock outputs regulate diel turnover of carbon and nitrogen reserves. Plant Cell Environ. 42, 549–573, https://doi.org/10.1111/pce.13440 (2019).
Hsu, C. Y. et al. Overexpression of CONSTANS homologs CO1 and CO2 fails to alter normal reproductive onset and fall bud set in woody perennial poplar. PloS One 7, e45448, https://doi.org/10.1371/journal.pone.0045448 (2012).
Klocko, A. L. et al. RNAi suppression of LEAFY gives stable floral sterility, and reduced growth rate and leaf size, in field-grown poplars. Plants 10. https://doi.org/10.3390/plants10081594 (2021).
Más, P. et al. Dual role of TOC1 in the control of circadian and photomorphogenic responses in Arabidopsis. Plant Cell 15, 223–236 (2003).
Jurca, M. et al. ZEITLUPE Promotes ABA-induced stomatal closure in Arabidopsis and Populus. Front. Plant Sci. 13. https://doi.org/10.3389/fpls.2022.829121 (2022).
Bordage, S. et al. Organ specificity in the plant circadian system is explained by different light inputs to the shoot and root clocks. N. Phytol. 212, 136–149, https://doi.org/10.1111/nph.14024 (2016).
Uemoto, K. et al. Root PRR7 improves the accuracy of the shoot circadian clock through nutrient transport. Plant Cell Physiol. 64, 352–362, https://doi.org/10.1093/pcp/pcad003 (2023).
James, A. B. et al. The circadian clock in Arabidopsis roots is a simplified slave version of the clock in shoots. Science 322, 1832–1835, https://doi.org/10.1126/science.1161403 (2008).
Greenwood, M. et al. The circadian clock coordinates plant development through specificity at the tissue and cellular level. Curr. Opin. Plant Biol. 53, 65–72, https://doi.org/10.1016/j.pbi.2019.09.004 (2020).
Hall, A. et al. Distinct regulation of CAB and PHYB gene expression by similar circadian clocks. Plant J. 32, 529–537, https://doi.org/10.1046/j.1365-313x.2002.01441.x (2002).
Kusakina, J. et al. A fast circadian clock at high temperatures is a conserved feature across Arabidopsis accessions and likely to be important for vegetative yield. Plant Cell Environ. 37, 327–340, https://doi.org/10.1111/pce.12152 (2014).
Gould, P. D. et al. Coordination of robust single-cell rhythms in the Arabidopsis circadian clock via spatial waves of gene expression. Elife 7. https://doi.org/10.7554/eLife.31700 (2018).
Chen, W. W. et al. A mobile ELF4 delivers circadian temperature information from shoots to roots. Nat. Plants 6, 416–426, https://doi.org/10.1038/s41477-020-0634-2 (2020).
Perez-Garcia, P. et al. The nature of the root clock at single cell resolution: principles of communication and similarities with plant and animal pulsatile and circadian mechanisms. Curr. Opin. Cell Biol. 77, 102102, https://doi.org/10.1016/j.ceb.2022.102102 (2022).
Holmes, R. et al. in Hubbard Brook: The Story of a Forest Ecosystem (ed T. Fahey) 9, 288 (Yale University Press, 2016).
Huang, Y. et al. A global map of root biomass across the world’s forests. Earth System Science Data 13 https://doi.org/10.5194/essd-13-4263-2021 (2021).
Díaz, S. Encyclopedia of Biodiversity (ed S. A. Levin) 72–89 (Academic Press, 2013).
Friend, A. D. et al. On the need to consider wood formation processes in global vegetation models and a suggested approach. Ann. For. Sci. 76, 49, https://doi.org/10.1007/s13595-019-0819-x (2019).
Lloyd, J. et al. The CO2 dependence of photosynthesis, plant growth responses to elevated atmospheric CO2 concentrations and their interaction with soil nutrient status. I. General principles and forest ecosystems. Functional Ecology, 4–32. https://doi.org/10.1046/j.1365-2435.1999.00350.x (1996).
Fatichi, S. et al. Moving beyond photosynthesis: from carbon source to sink-driven vegetation modeling. N. Phytol. 201, 1086–1095, https://doi.org/10.1111/nph.12614 (2014).
Rademacher, T. et al. Insights into source/sink controls on wood formation and photosynthesis from a stem chilling experiment in mature red maple. N. Phytol. 236, 1296–1309, https://doi.org/10.1111/nph.18421 (2022).
Marchand, L. J. et al. No winter halt in below-ground wood growth of four angiosperm deciduous tree species. Nat. Ecol. Evol. https://doi.org/10.1038/s41559-024-02602-6 (2025).
Mariën, B. et al. On the below- and aboveground phenology in deciduous trees: observing the fine-root lifespan, turnover rate, and phenology of Fagus sylvatica L., Quercus robur L., and Betula pendula roth for two growing seasons. Forests 12, 1680, https://doi.org/10.3390/f12121680 (2021).
Nilsson, O. et al. Spatial pattern of cauliflower mosaic virus 35S promoter-luciferase expression in transgenic hybrid aspen trees monitored by enzymatic assay and non-destructive imaging. Transgenic Res. 1, 209–220, https://doi.org/10.1007/BF02524751 (1992).
Takata, N. et al. A simple and efficient transient transformation for hybrid aspen (Populus tremula × P. tremuloides). Plant Methods 8, 30, https://doi.org/10.1186/1746-4811-8-30 (2012).
Fire, A. et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391, 806–811, https://doi.org/10.1038/35888 (1998).
Fladung, M. et al. RNAi for Plant Improvement and Protection 54–71 (CABI, 2021).
Liu, S. et al. RNAi technology for plant protection and its application in wheat. aBIOTECH 2, 365–374, https://doi.org/10.1007/s42994-021-00036-3 (2021).
Helliwell, C. A. et al. Isolation and functional characterization of cytochrome P450s in gibberellin biosynthesis pathway. Methods Enzymol. 357, 381–388, https://doi.org/10.1016/S0076-6879(02)57696-5 (2002).
Karimi, M. et al. GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7, 193–195, https://doi.org/10.1016/s1360-1385(02)02251-3 (2002).
Lampropoulos, A. et al. GreenGate—a novel, versatile, and efficient cloning system for plant transgenesis. PloS One 8, e83043, https://doi.org/10.1371/journal.pone.0083043 (2013).
Böhlenius, H. Control of flowering time and growth cessation in Arabidopsis and Populus trees. PhD Thesis. (Swedish University of Agricultural Science, 2007).
Rigal, A. et al. The AINTEGUMENTA LIKE1 homeotic transcription factor PtAIL1 controls the formation of adventitious root primordia in poplar. Plant Physiol. 160, 1996–2006, https://doi.org/10.1104/pp.112.204453 (2012).
Eriksson, M. E. et al. Plants having improved growth properties. 2015/01/14, patent application EP2823046A1 (2015).
Miki, D. et al. Simple RNAi vectors for stable and transient suppression of gene function in rice. Plant Cell Physiol. 45, 490–495, https://doi.org/10.1093/pcp/pch048 (2004).
Stener, L.-G. et al. Resultat från klontester av hybridasp och poppel efter två års tillväxt. 30 (Skogforsk, 2018).
le Provost, G. et al. A micromethod for high throughput RNA extraction in forest trees. Biol. Res. 40, 291–297, https://doi.org/10.4067/S0716-97602007000400003 (2007).
Livak, K. J. et al. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25, 402–408, https://doi.org/10.1006/meth.2001.1262 (2001).
Pfaffl, M. W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29, e45, https://doi.org/10.1093/nar/29.9.e45 (2001).
UPOV. Guidelines for the Conduct of Test for Distinctiveness, Uniformity and Stability Populus L. 9 (International Union for the Protection of New Varieties of Plants, 1981).
Robinson, K. M. et al. Variation in non-target traits in genetically modified hybrid aspens does not exceed natural variation. N. Biotechnol. 64, 27–36, https://doi.org/10.1016/j.nbt.2021.05.005 (2021).
Wang, W. et al. Aspen growth is not limited by starch reserves. Curr. Biol. 32, 3619–3627.e3614, https://doi.org/10.1016/j.cub.2022.06.056 (2022).
Johansson, M. et al. Monitoring seasonal bud set, bud burst, and cold hardiness in Populus. Methods Mol. Biol. 2398, 215–226, https://doi.org/10.1007/978-1-0716-1912-4_17 (2022).
Matile, P. Biochemistry of Indian summer: physiology of autumnal leaf coloration. Exp. Gerontol. 35, 145–158, https://doi.org/10.1016/s0531-5565(00)00081-4 (2000).
Matile, P. et al. Chlorophyll degradation. Annu Rev. Plant Physiol. Plant Mol. Biol. 50, 67–95, https://doi.org/10.1146/annurev.arplant.50.1.67 (1999).
Bresson, J. et al. A guideline for leaf senescence analyses: from quantification to physiological and molecular investigations. J. Exp. Bot. 69. https://doi.org/10.1093/jxb/erx246 (2017).
Michelson, I. H. et al. Autumn senescence in aspen is not triggered by day length. Physiol. Plant. 162, 123–134, https://doi.org/10.1111/ppl.12593 (2018).
Parry, C. et al. In situ measurement of leaf chlorophyll concentration: analysis of the optical/absolute relationship. Plant Cell Environ. 37. https://doi.org/10.1111/pce.12324 (2014).
Mariën, B. et al. Detecting the onset of autumn leaf senescence in deciduous forest trees of the temperate zone. N. Phytol. 224, 166–176, https://doi.org/10.1111/nph.15991 (2019).
Hastie, T. et al. Generalized additive models. Stat. Sci. 1, 297–310, https://doi.org/10.1214/ss/1177013604 (1986).
Pedersen, E. J. et al. Hierarchical generalized additive models in ecology: an introduction with MGCV. PeerJ 7, e6876, https://doi.org/10.7717/peerj.6876 (2019).
Ruppert, D. et al. Semiparametric Regression https://doi.org/10.1017/CBO9780511755453 (Cambridge University Press, 2003).
Yee, T. W. et al. Generalized additive models in plant ecology. J. Veg. Sci. 2, 587–602, https://doi.org/10.2307/3236170 (1991).
Simpson, G. L. Modelling palaeoecological time series using generalised additive models. Front. Ecol. Evol. 6. https://doi.org/10.3389/fevo.2018.00149 (2018).
Larsen, K. GAM: The Predictive Modeling Silver Bullet (2015).
Zuur, A. et al. Analysing Ecological Data. 75. https://doi.org/10.1017/CBO978051175545310.1007/978-0-387-45972-1(Springer, 2007).
Linear and Nonlinear Mixed Effects Models v. 3.1-131 (2017).
Wood, S. N. Stable and efficient multiple smoothing parameter estimation for generalized additive models. J. Am. Stat. Assoc. 99, 673–686, https://doi.org/10.1198/016214504000000980 (2004).
Rose, N. L. et al. An assessment of the mechanisms for the transfer of lead and mercury from atmospherically contaminated organic soils to lake sediments with particular reference to Scotland, UK. Geochim. Cosmochim. Acta 82, 113–135, https://doi.org/10.1016/j.gca.2010.12.026 (2012).
Wood, S. N. Fast stable restricted maximum likelihood and marginal likelihood estimation of semiparametric generalized linear models.J. R. Stat. Soc.: Ser. B (Stat. Methodol.) 73, 3–36, https://doi.org/10.1111/j.1467-9868.2010.00749.x (2011).
Eilers, P. H. C. et al. Twenty years of P-splines. Sort-Stat. Oper. Res. Trans. 39, 149–186 (2015).
Eilers, P. H. C. et al. Flexible smoothing with B-splines and penalties. Stat. Sci. 11, 89–121, https://doi.org/10.1214/ss/1038425655 (1996).
Reiss, P. et al. Smoothing parameter selection for a class of semiparametric linear models. J. R. Stat. Soc. Ser. B 71, 505–523, https://doi.org/10.1111/j.1467-9868.2008.00695.x (2009).
Stasinopoulos, D. et al. Generalized additive models for Location Scale and Shape (GAMLSS) in R. J. Stat. Softw. 23. https://doi.org/10.18637/jss.v023.i07 (2007).
Rigby, R. et al. Distributions for Modeling Location, Scale, and Shape: Using GAMLSS in R. https://doi.org/10.1201/9780429298547 (2019).
Stasinopoulos, M. D. et al. GAMLSS: a distributional regression approach. Stat. Model. 18, 248–273, https://doi.org/10.1177/1471082X18759144 (2018).
Stasinopoulos, D. et al. Flexible regression and smoothing: Using GAMLSS in R. https://doi.org/10.1201/b21973 (2017).
gratia: Graceful {ggplot}-Based Graphics and Other Functions for {GAM}s Fitted using {mgcv} v. 0.8.2 (development version) (2024).
Burnham, K. et al. Model Selection and Multimodel Inference. A Practical Information-theoretic Approach. https://doi.org/10.1007/978-0-387-22456-5_5 (2004).
Barker, L. E. et al. Best (but oft-forgotten) practices: checking assumptions concerning regression residuals. Am. J. Clin. Nutr. 102, 533–539, https://doi.org/10.3945/ajcn.115.113498 (2015).
Schielzeth, H. et al. Robustness of linear mixed-effects models to violations of distributional assumptions. Methods Ecol. Evol. 11, 1141–1152, https://doi.org/10.1111/2041-210x.13434 (2020).
Harrison, X. et al. A brief introduction to mixed effects modelling and multi-model inference in ecology. PeerJ 6, e4794, https://doi.org/10.7717/peerj.4794 (2018).
Zuur, A. F. et al. A protocol for data exploration to avoid common statistical problems. Methods Ecol. Evol. 1, 3–14, https://doi.org/10.1111/j.2041-210X.2009.00001.x (2010).
Zuur, A. F. et al. A protocol for conducting and presenting results of regression-type analyses. Methods Ecol. Evol. 7, 636–645, https://doi.org/10.1111/2041-210x.12577 (2016).
Osbourne, J. W. et al. Four assumptions of multiple regression that researchers should always test. Pract. Assess., Res. Eval. 8, 2, https://doi.org/10.7275/r222-hv23 (2002).
Belkin, M. Fit without fear: remarkable mathematical phenomena of deep learning through the prism of interpolation. Acta Num. 30, 203–248, https://doi.org/10.1017/S0962492921000039 (2021).
Belkin, M. et al. Reconciling modern machine-learning practice and the classical bias–variance trade-off. Proc. Natl. Acad. Sci. 116, 15849–15854, https://doi.org/10.1073/pnas.1903070116 (2019).
Belkin, M. et al. Reply to Loog et al.: looking beyond the peaking phenomenon. Proc. Natl. Acad. Sci. 117, 10627–10627, https://doi.org/10.1073/pnas.2003206117 (2020).
Dunn, P. K. et al. Randomized quantile residuals. J. Comput. Graph. Stat. 5, 236–244, https://doi.org/10.2307/1390802 (1996).
Fasiolo, M. et al. Scalable visualization methods for modern generalized additive models. J. Comput. Graph. Stat. 29, 78–86, https://doi.org/10.1080/10618600.2019.1629942 (2019).
Pya, N. et al. A note on basis dimension selection in generalized additive modelling. https://doi.org/10.48550/arXiv.1602.06696 (2016).
Shapiro, S. S. et al. An analysis of variance test for normality (Complete Samples). Biometrika 52, 591, https://doi.org/10.2307/2333709 (1965).
nortest: Tests for Normality v. R package version 1.0-4 (2015).
goftest: Classical Goodness-of-Fit Tests for Univariate Distributions v. R package version 1.2-2 (2019).
fNonlinear: Rmetrics - Nonlinear and Chaotic Time Series Modelling v. R package version 3042.79 (2017).
normwhn.test: Normality and White Noise Testing v. 1.0 (2012).
Box, G. E. P. et al. Distribution of residual autocorrelations in autoregressive-integrated moving average time series models. J. Am. Stat. Assoc. 65, 1509–1526, https://doi.org/10.1080/01621459.1970.10481180 (1970).
Ljung, G. M. et al. On a measure of lack of fit in time series models. Biometrika 65, 297–303, https://doi.org/10.1093/biomet/65.2.297 (1978).
Pfaff, B. Analysis of Integrated and Cointegrated Time Series with R. Second edn. (Springer, 2008).
Phillips, P. C. B. et al. Testing for a unit-root in time-series regression. Biometrika 75, 335–346, https://doi.org/10.1093/biomet/75.2.335 (1988).
Bhargava, A. On the theory of testing for unit roots in observed time series. Rev. Econ. Stud. 53, 369–384, https://doi.org/10.2307/2297634 (1986).
Elliott, G. et al. Efficient tests for an autoregressive unit root. Econometrica 64, 813–836, https://doi.org/10.2307/2171846 (1996).
Ng, S. et al. LAG length selection and the construction of unit root tests with good size and power. Econometrica 69, 1519–1554, https://doi.org/10.1111/1468-0262.00256 (2001).
Hyndman, R. J. et al. Automatic time series forecasting: the forecast package for R. J. Stat. Softw. 27, 1–22, https://doi.org/10.18637/jss.v027.i03 (2008).
Beran, J. Statistics for Long-Memory Processes. 315 (Chapmann & Hall/CRC, 1994).
longmemo: Statistics for Long-Memory Processes (Book Jan Beran), and Related Functionality v. R package version 1.1-2 (2020).
Hurst, H. E. The long-term storage capacity of reservoir. Trans. Am. Soc. Civ. Eng. 16, 770–799, https://doi.org/10.1061/TACEAT.0006518 (1951).
Kleinow, T. Testing Continuous Time Models in Financial Markets (Humboldt-Universität zu Berlin, Wirtschaftswissenschaftliche Fakultät, 2002).
Mandelbrot, B. B. et al. The (Mis)Behavior of Markets: a Fractal View of Risk, Ruin, and Reward (Basic Books, 2004)
e1071: Misc Functions of the Department of Statistics, Probability Theory Group (Formerly: E1071), TU Wien v. version 1.7.3. (2019).
psych: Procedures for Personality and Psychological Research v. Version = 1.9.12. (Northwestern University, 2019).
sur: Companion to “Statistics Using R: An Integrative Approach” v. version 1.0.2. (2019).
Hohberg, M. et al. Treatment effects beyond the mean using distributional regression: methods and guidance. PloS One 15, e0226514, https://doi.org/10.1371/journal.pone.0226514 (2020).
diptest: Hartigan’s Dip Test Statistic for Unimodality - Corrected v. version 0.75-77 (2016).
modes: Find the Modes and Assess the Modality of Complex and Mixture Distributions, Especially with Big Datasets v. version 0.7.0. (2016).
Ameijeiras-Alonso, J. et al. Multimode: An R Package for Mode Assessment (2018). https://doi.org/10.18637/jss.v097.i09
R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, 2023).
Wickham, H. ggplot2: Elegant Graphics for Data Analysis (Springer-Verlag, 2009)
cowplot: Streamlined Plot Theme and Plot Annotations for ‘ggplot2’ v. 1.0.0 (2019).
viridis: Default Color Maps from ‘matplotlib’ v. 0.5.1 (2018).
ggpubr: ‘ggplot2’ Based Publication Ready Plots v. 0.2.4 (2019).
dplyr: A Grammar of Data Manipulation v. 0.7.4 (2018).
Bendix, C. et al. Circadian clock genes universally control key agricultural traits. Mol. Plant 8, 1135–1152, https://doi.org/10.1016/j.molp.2015.03.003 (2015).
Fogelmark, K. et al. Rethinking transcriptional activation in the Arabidopsis circadian clock. PLoS Comput. Biol. 10, e1003705 (2014).
Singh, R. K. et al. Growing in time: exploring the molecular mechanisms of tree growth. Tree Physiol. 41, 657–678, https://doi.org/10.1093/treephys/tpaa065 (2020).
Acknowledgements
We express our gratitude to Assoc. Prof. Dr. Natalya Pya Arnqvist, Prof. Dr. Gavin Simpson and Prof. Dr. Rishikesh P. Bhalerao for their helpful advice and support. B.M. received funding from the Kempe Stiftelserna. Other activities were supported by the VINNOVA VR Center, Knut and Alice Wallenberg Foundation, and Trees and Crops for the Future provided funds for the field trials and construction of transgenic trees. We additionally thank the staff of UPSC’s greenhouse and transgenic facility, the field trial team headed by Ulf Johansson, and Dr. Ioana Gaboreanu.
Author information
Authors and Affiliations
Contributions
B.M.: Conceptualization, Methodology, Formal analysis, Writing-Original Draft, Writing-Review and Editing; K.M.R.: Investigation, Writing-Review and Editing; M.J.: Investigation and Methodology; I.H.M.: Investigation, Writing-Review and Editing; N.T.: Investigation, Methodology, Writing-Review and Editing; I.K.: Investigation and Methodology; P.A.P.: Investigation, Methodology, Writing-Review and Editing; P.K.I.: Investigation; T.M.: Methodology, Writing-Review and Editing; C.I.: Methodology, Writing-Review and Editing, O.N.: Methodology, Writing-Review and Editing; S.J.: Conceptualization, Methodology, Writing-Review and Editing, and Project administration; S.P.: Methodology, Writing-Review and Editing; J.Y.: Conceptualization, Methodology, Formal analysis, Writing-Review and Editing, Supervision, Project administration and Funding acquisition; M.E.E.: Conceptualization, Investigation, Methodology, Writing-Review and Editing, Supervision, Project administration and Funding acquisition.
Corresponding authors
Ethics declarations
Competing interests
Conflicting Statement: O.N., S.J., T.M. and M.E.E. are members of the holding company Woodheads AB, a part-owner of SweTree Technologies AB (STT). M.E.E. is the non-paid CEO of Woodheads AB. O.N. and M.E.E. are also board members of STT.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Mariën, B., Robinson, K.M., Jurca, M. et al. Nature’s Master of Ceremony: The Populus Circadian Clock as Orchestrator of Tree Growth and Phenology. npj Biol Timing Sleep 2, 16 (2025). https://doi.org/10.1038/s44323-025-00034-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s44323-025-00034-4
This article is cited by
-
The circadian clock of Populus affects physiological, transcriptional and metabolomic responses to osmotic and ionic components of salt stress
npj Biological Timing and Sleep (2025)