Abstract
Because viruses encode only a small number of proteins, all steps of virus infection rely on specific interactions between viruses and hosts. We previously screened several Nicotiana benthamiana (Nb) proteins that interact with the stem-loop 1 (SL1) RNA structure located at the 5′ end of the potato virus X (PVX) genome. In this study, we characterized two of these proteins (NbCPIP2a and NbCPIP2b), which are homologous and are induced upon PVX infection. Electrophoretic mobility shift assay confirmed that both proteins bind to either SL1(+) or SL1(−) RNAs of PVX. The two proteins also interact with the PVX capsid protein (CP) in planta. Overexpression of NbCPIP2a positively regulated systemic movement of PVX in N. benthamiana, whereas NbCPIP2b overexpression did not affect systemic movement of PVX. Transient overexpression and silencing experiments demonstrated that NbCPIP2a and NbCPIP2b are positive regulators of PVX replication and that the effect on replication was greater for NbCPIP2a than for NbCPIP2b. Although these two host proteins are associated with plasma membranes, PVX infection did not affect their subcellular localization. Taken together, these results indicate that NbCPIP2a and NbCPIP2b specifically bind to PVX SL1 RNAs as well as to CP and enhance PVX replication and movement.
Similar content being viewed by others
Introduction
Potato virus X (PVX), which belongs to the genus Potexvirus, is an important pathogen of potato and other solanaceous plants and is considered one of the 10 most important plant viruses1,2,3. The PVX genome, which is approximately 6.4 kb long and is capped and polyadenylated, is composed of a 84-nucleotide (nt) 5′-nontranslated region (NTR), five viral open reading frames (ORFs), and a 72-nt 3′-NTR4. The five ORFs encode an RNA-dependent RNA polymerase (RdRp), triple gene blocks proteins (TGBs), and the capsid protein (CP). In general, RNA elements at these NTRs of RNA viruses include sequences and structural elements important for viral translation and replication; they also function as binding sites for cellular proteins required for maintaining the viral infection cycle5,6,7. The 5′ NTR of the PVX genome contains five repeated ACCA sequences followed by three stem-loop structures (5′ SL1, 5′ SL2, and 5′ SL3), while the 3′ NTR contains three stem-loop structures (3′ SL1, 3′ SL2, and 3′ SL3) and a poly(A) tail8,9. The 5′ SL1 is particularly important for PVX translation, viral RNA replication, and initiation of PVX virion assembly2,5,10. The CP of PVX also plays an important role during virus encapsidation and movement11,12.
Viruses encode only a few proteins, and virus replication and pathogenicity therefore depend on specific interactions between viral elements and host factors13,14,15. Many host factors that interact with viral RNAs and proteins have been identified by yeast-two hybrid (Y2H) assay, bimolecular fluorescence complementation (BiFC), co-immunoprecipitation (Co-IP), and Northwestern blot analysis followed by matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI TOP MS). Several host proteins have been identified that interact with PVX viral proteins including NbPCIP1, TIP, NbMPB2Cb, NbDnaJ, eEF1A, and eEF1Bβ16,17,18,19,20. Moreover, researchers have used Northwestern blot followed by MALDI TOP MS to identify tobacco proteins that bind to PVX 5′ SL RNAs21,22.
The functions of several host proteins in PVX infection have been determined. For example, TGB12K interacting proteins (TIP1 to TIP3) specifically interact with TGB2 protein and are associated with callose degradation and cell-to-cell movement of PVX18. The interaction between NbPCIP1 and CP affects PVX replication16. NbMPB2Cb interacts with four PVX viral proteins as well as with SL1 RNAs and reduces PVX movement19. The binding of NbDnaJ to minus-strand SL1 RNA reduces PVX replication and movement20. Interaction between the WRKY1 transcription factor with SL1 is important for viral RNA accumulation and disease symptom development22. The interaction between eukaryotic translation elongation factor 1 (eEF1) and TGB1 protein facilitates PVX infection17.
In this study, we characterized two homologous host proteins that were previously determined to interact with SL RNAs of PVX21. Electrophoretic mobility shift assay (EMSA) as well as BiFC were used to confirm the interactions of host proteins and PVX viral components. The effects of these host proteins on PVX replication and movement were determined with transient overexpression and silencing experiments.
Results
Characterization of NbCPIP2a and NbCPIP2b interacting with SL1 RNA structures of PVX
As noted, we previously detected 24 N. benthamiana proteins that interact with SL1 RNAs of PVX21. Of the 24 proteins, we selected two that are homologous to known potyviral CP interacting protein 2a (CPIP2a) from N. tabacum23. A blast search using CPIP2a protein as a query against available draft genomes for N. benthamiana and N. tabacum revealed that each Nicotiana genome contains two homologues of these proteins designated as NbCPIP2a and NbCPIP2b. We found that these two proteins are about 34 kDa in size (305 amino acids) and contain two conserved DnaJ domains. We compared the protein sequences of NbCPIP2a and NbCPIP2b as well as of NtCPIP2a and NtCPIP2b. The amino acid sequences of these four proteins differ in only 11 amino acids (Fig. 1A). NbCPIP2 homologous proteins were searched against the Phytozome database to determine the evolutionary origin and divergence of NbCPIP2 in plants. A phylogenetic tree using NtCPIP2a homologs showed that NbCPIP2a and NbCPIP2b were grouped with members of the Solanaceae such as Solanum lycopersicum and S. tuberosum (Fig. 1B). Interestingly, an NbCPIP2a orthologue was also present in marine algae Coccomyxa subellipsoidea and Micromonas pusilla.
(A) Abbreviations for gene names are as follows: Nb2a (NbCPIP2a), Nb2b (NbCPIP2b), Nt2a (NtCPIP2a), and Nt2b (NtCPIP2b). The red line and blue line indicate the conserved DnaJ domain and the DnaJ C terminal domain, respectively. Aligned sequences were visualized with the Megalign program. (B) Phylogenetic relationship of NbCPIP2a-like proteins identified from plants for which draft genome sequences are available. The phylogenetic tree was constructed by protein sequences of NbCPIP2a-like proteins from diverse plants. NbCPIP2a-like proteins were identified by blast search in the Phytozome database. For simplicity, only plant species names instead of protein names are indicated in the phylogenetic tree. The phylogenetic tree was generated by the neighbor-joining method with 1,000 bootstrap replicates and Kimura 2-parameter distance using the MEGA6 program55.
To confirm the interaction between NbCPIP2a and NbCPIP2b with SL1 RNAs of PVX, we conducted EMSA24. The 5′ SL1(−) RNA transcripts generated shifted complexes when incubated with the NbCPIP2a and NbCPIP2b in EMSA and the abundance of the shifted complexes increased as the abundance of RNA transcripts increased (Supplementary Figure S1). RNA transcript containing 5′ SL1(+) bound only to NbCPIP2b (Supplementary Figure S1). Moreover, both host proteins can bind to SL structures at the 3′ SLs of PVX RNA. This result indicates that the two homologous proteins might differ in their binding to the SL RNA structures of PVX. Overall, RNA transcripts containing PVX 5′ SL1 or 3′ SLs were capable of forming RNA–protein complexes with two homologous tobacco proteins, NbCPIP2a and NbCPIP2b.
NbCPIP2a and NbCPIP2b also interact with PVX CP
To determine whether NbCPIP2a and NbCPIP2b directly interact with PVX proteins including RdRp, TGB1, TGB2, TGB3, and CP, we carried out an in planta BiFC assay using the Agrobacterium-infiltration method4. In the BiFC assay, we used divided yellow fluorescence protein (YFP), referred to as YFP-N (N-terminal) and YFP-C (C-terminal). Because the different combinations of YFP-fused proteins might affect the protein-protein interaction25, we generated two different sets of YFP fusion constructs (Fig. 2). For example, host proteins were fused to YFP-N and viral proteins were fused to YFP-C (Fig. 2A,C), and vice versa (Fig. 2B,D). The combination of negative controls (nEYFP, cEYFP, and nEYFP + cEYFP) did not produce any fluorescence. The BiFC assay demonstrated that NbCPIP2a and NbCPIP2b interact with CP but not with the other four PVX proteins (Fig. 2A–D).
To study protein-protein interactions, we used the BiFC system in planta. Two different sets of vectors containing split YFP were used. (A) Interaction of NbCPIP2a fused to nYFP with each viral protein fused to cYFP and (B) vice versa. (C) Interaction of NbCPIP2b fused to nYFP with each viral protein fused to cYFP, and (D) vice versa. (E) Positive controls, including the interaction of NbCPIP2a or NbCPIP2b fused to nYFP with SMV CP fused to cYFP. Negative controls, including nEYFP, cEYFP, and both nEYFP and cEYFP. The white bar indicates 100 μm.
A previous report indicated that the CP of Potato virus Y (PVY), which is a Potyvirus, interacts with NtCPIP2a and NtCPIP2b23. Therefore, we hypothesized that NbCPIP2 can interact with the CPs of other potyviruses. We tested this hypothesis with CPs of the following potyviruses: soybean mosaic virus (SMV), pepper mottle virus (PepMoV), pepper severe mosaic virus, potato virus A (PVA), PVY, turnip mosaic virus (TuMV), and zucchini yellow mosaic virus. The CP of SMV interacted with both NbCPIP2a and NbCPIP2b (Fig. 2E). Although NbCPIP2b bound to the CP of PepMoV, PVA, and TuMV, NbCPIP2a bound only to the CP of TuMV (Supplementary Figure S2). In addition, we found that the interaction between host protein:YFP-C and viral protein:YFP-N was much stronger than the interaction between host protein:YFP-N and viral protein:YFP-C.
NbCPIP2a and NbCPIP2b enhance PVX RNA replication
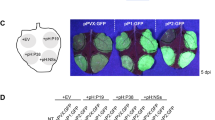
To characterize the functions of the two host proteins in response to PVX infection, we generated vectors for the transient overexpression and silencing of the two genes that encode the proteins. For transient overexpression, we generated vectors containing dual 35S promoters of cauliflower mosaic virus and tagged each host gene with hemagglutinin epitope (HA-tag) (Fig. 3A). At 2 days after infiltration, we extracted total proteins from the N. benthamiana leaves that were infiltrated with the overexpression vector. Western blot analyses using anti-HA confirmed transient overexpression of the two host proteins in N. benthamiana (Fig. 3B). After transient overexpression of each gene, we inoculated the leaves with PVX tagged with green fluorescence protein (GFP). The area with green fluorescence was much wider on the leaves that transiently overexpressed NbCPIP2a or NbCPIP2b than on the leaves that were inoculated only with PVX (the positive control). Movement of GFP-tagged PVX was faster with transient overexpression of NbCPIP2a than with transient overexpression of NbCPIP2b (Fig. 3C).
(A) Schemes of overexpression vectors. (B) Western blot analysis to confirm transient overexpression of the two host proteins in N. benthamiana. (C) Intercellular movement of GFP-tagged PVX in the transiently overexpressed N. benthamiana leaves. (D) PVX viral RNA replication in the extracted protoplasts. (E) Long-distance movement of GFP-tagged PVX in the transiently overexpressed N. benthamiana leaves. NC = negative control (leaves were not inoculated with overexpression vectors or with PVX). PC = positive control (leaves were not inoculated with overexpression vectors but were inoculated with PVX). 1st = the first leaf from the top. 5th = the fifth leaf from the top. NbCPIP2a or NbCPIP2b (leaves were inoculated with overexpression vectors of NbCPIP2a or NbCPIP2b and were also inoculated with PVX).
Next, we examined PVX replication in N. benthamiana protoplasts. We first transiently overexpressed each gene in N. benthamiana leaves. On the next day, we inoculated the leaves with PVX by agro-infiltration. We prepared protoplasts after 12 h, and incubated the protoplasts at 25 °C in a growth chamber for 2 days. We then extracted total RNAs from the protoplasts according to a previous study26. Real-time RT-PCR results showed that PVX replication was about 13-times and 6.5-times higher in the protoplasts overexpressing NbCPIP2a and NbCPIP2b, respectively, than in the positive control (Fig. 3D).
For systemic movement assay, we transiently overexpressed NbCPIP2a and NbCPIP2b, respectively, on N. benthamiana leaf. The fourth leaves from the top at a 5–6 leaves stage N. benthamiana were agro-infiltrated. Two days later, we agro-infiltrated GFP-tagged PVX in the same leaf which overexpressing individual host protein. At 7 days after inoculation of GFP-tagged PVX, we examined the green fluorescence in the upper leaves. In the positive control, only the first leaf from the top showed green fluorescence. Green fluorescence was evident in the first, second, and third leaves from the top in plant which was infiltrated with NbCPIP2a in local leaf. However, we observed green fluorescence in the first and second leaves from the top in the plant which was infiltrated with NbCPIP2b in local leaf (Fig. 3E).
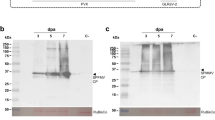
For transient silencing of the two host genes, we generated RNAi silencing vectors (Fig. 4A). To verify the silencing of target gene expression, we carried out real-time RT-PCR using gene-specific primers (Supplementary Table S1). After normalization based on expression of actin and ubiquitin genes, we calculated relative expression for NbCPIP2a and NbCPIP2b in the silenced leaves. Relative to their expression in the negative control leaves (i.e., leaves that were not silenced and were not inoculated with PVX), NbCPIP2a expression was reduced by 32.3% and NbCPIP2b expression was reduced by 28.9% in the silenced leaves (Fig. 4B). After transient silencing of each gene, we performed a challenge inoculation of GFP-tagged PVX on the inoculated leaves using agrobacterium. The area with green fluorescence and the intensity of the fluorescence were much less on the inoculated leaves where NbCPIP2a or NbCPIP2b was transiently silenced than on the non-silenced, positive control leaves, which were only inoculated with GFP-tagged PVX (Fig. 4C). We then examined PVX replication in the protoplasts extracted from silenced leaves for each gene. PVX replication was reduced by 48.4% in the NbCPIP2a-silenced leaves and by 15.1% in the NbCPIP2b-silenced leaves (Fig. 4D). We again examined the long-distance movement of GFP-tagged PVX in the transiently silenced N. benthamiana. Unexpectedly, PVX long-distance movement was similar in the silenced leaves and the positive control leaves (Fig. 4E).
(A) Schemes of silencing vectors. (B) Gene-silencing activities induced by RNAi vectors. (C) Intercellular movement of GFP-tagged PVX in transiently silenced N. benthamiana leaves. (D) PVX viral RNA replication in the extracted protoplasts. (E) Long-distance movement of GFP-tagged PVX in the transiently silenced N. benthamiana leaves. NC = negative control (leaves were not silenced and were not inoculated with PVX). PC = positive control (leaves were not silenced but were inoculated with PVX). 1st = the first leaf of the top. 5th = the fifth leaf from the top. NbCPIP2a-i or NbCPIP2b-i (leaves were infiltrated with the silencing vectors of NbCPIP2a or NbCPIP2b and were inoculated with PVX).
PVX infection does not alter the subcellular localization of NbCPIP2a or NbCPIP2b
To determine the subcellular localization of NbCPIP2a and NbCPIP2b, we infiltrated the GFP-tagged expression vector for each gene; green fluorescence for the two proteins was mostly evident in the plasma membranes (Fig. 5C,G). We then examined the subcellular localization of the two proteins following PVX infection; green fluorescence for the two proteins was once again mostly evident in the plasma membranes (Fig. 5D,H). Free GFP was dispersed in most cellular organelles. As compared to free GFP, we did not observe change of subcellular localization for two host proteins after PVX infection. This result indicates that PVX infection does not affect the subcellular localization of NbCPIP2a or NbCPIP2b.
(A,B,E,F) Leaves were not infiltrated with GFP-tagged expression vectors for NbCPIP2a or NbCPIP2b and were inoculated (B,F) or were not inoculated (A,E) with PVX. (C,D,G,H) Leaves were infiltrated with GFP-tagged expression vectors for NbCPIP2a (C,D) or NbCPIP2b (G,H) and were inoculated (D,H) or were not inoculated (C,G) with PVX.
NbCPIP2 expression is induced by PVX infection but not by CMV or PepMoV infection
To monitor the expression of NbCPIP genes over time in response to infection by PVX, cucumber mosaic virus (CMV), and PepMoV, we conducted real-time RT-PCR analysis using a primer-pair that amplifies both genes (Fig. 6). The expression of NbCPIP2 genes increased steadily for the first 4 days and remained high for the next 3 days in plants inoculated with PVX but not in plants inoculated with CMV or PepMoV (Fig. 6). Expression of NbCPIP2 genes in non-inoculated plants tended to increase for the first 3 days but then did not exhibit a clear pattern. These results suggest that the expression level of NbCPIP2 might be regulated by specific viruses.
N. benthamiana leaves were inoculated with the indicated virus or were not inoculated (mock). Leaf samples were collected 1 to 7 days later, and expression of NbCPIP2a or NbCPIP2b was quantified. For each combination of treatment and time, three independent plants were sampled. Expression of NbCPIP was normalized based on expression of ubiquitin and actin genes. Values are means + SD.
Discussion
Because viruses have small genomes that encode only a few proteins, they rely on host factors for their replication, movement, and pathogenicity15,27. To identify such host factors, researchers have often investigated the interactions between host proteins and virus proteins or between host proteins and viral RNAs. In the current study, we characterized two homologous host proteins that interact with PVX SL1 RNAs and CP.
A previous study characterized the functions of the host protein NtCPIP2a, which interacts with PVY CP in tobacco23. In the current study, we functionally characterized two NtCPIP2a-like proteins, NbCPIP2a and NbCPIP2b, in N. benthamiana in response to PVX infection. A phylogenetic analysis using NbCPIP2a-like proteins from diverse plants showed that NbCPIP2a-like proteins occur in most plant kingdoms, ranging from marine algae to higher plants. NbCPIP2a might have evolved from marine photosynthetic organisms. NbCPIP2a and NbCPIP2b have two conserved domains, i.e., the DnaJ domain (IPR001623) at the N-terminal and the HSP40/DnaJ peptide-binding domain (IPR008971) at the C-terminal regions28,29. Both domains occur in many prokaryotic and eukaryotic genomes. Researchers have often reported that HSP40/DnaJ is required for protein translation, folding/unfolding, trafficking, and secretion as co-chaperone of HSP7028,30. HSP70s and its co-chaperone HSP40/DnaJ are involved in virus replication31,32 and the assembly or disassembly of the virus capsid33. Specific binding between DnaJ/HSP40 proteins and other viral proteins has also been reported23,34.
Interactions between NbCPIP2a and NbCPIP2b with PVX SL1(+) RNAs have previously been shown by Northwestern blot followed by MS/MS analysis21. In contrast to the previous study, the current study demonstrated that NbCPIP2a interacts with SL1(−) but not with SL1(+) RNA. While the previous study identified a partial fragment of NbCPIP2a by MS/MS analysis, the current study used the complete cDNA sequence for cloning NbCPIP2a. Therefore, we believe that the results of the current study are more convincing than those of the previous study21. EMSA confirmed the different binding activities of two homologous proteins to PVX SL1 RNAs. We suspect that the difference in the five amino acids might explain the difference in the binding of the two host proteins to viral RNA. To our knowledge, this is the first report that demonstrates a difference in viral RNA-interaction capacity for two homologous proteins that differ in only five amino acids. The residue(s) within these host proteins that are responsible for controlling the interaction with PVX viral RNA structures should be determined in future research.
CPs of potyviruses are important for virion assembly, cell-to-cell movement, and long-distance movement35. To fulfill multiple functions in the host, CPs must interact with host components36. In this study the homologous host proteins have the same binding affinity for PVX CP; however, they differ in their interaction with SL1 RNAs of PVX. Furthermore, many homologous host proteins have a common conserved domain that is required for binding with CP. For example, a previous study used Y2H and BiFC to document that NtCPIP1 and NtCPIP2a interact with PVY CP23. In our BiFC assay, NbCPIP2a and NbCPIP2b interacted only with PVX CP but not with four other PVX proteins. Moreover, the binding to PVX CP did not differ between the two host proteins. NbCPIP2a and NbCPIP2b also interacted with the CPs of SMV, TuMV, PVA, and PepMoV, which belong to the genus Potyvirus. Although CPs of four potyviruses might interact with two host proteins, interaction ability of two host proteins were different based on BiFC results. For example, CPs of SMV and TuMV strongly interact with two host proteins. However, NbCPIP2a showed very weak interaction with CPs of PepMoV and PVA while NbCPIP2b displayed strong interaction with CPs of PepMoV and PVA. These results indicate binding abilities of two host proteins with potyviral CP might be virus specific. Based on these results, it is likely that CPs of potyviruses might have a conserved domain that is required for interaction with these two host proteins. A previous study using mutational analysis identified the CP core region (including the three highly conserved residues; S125, R157, D201 of PVY CP, which are essential for virion formation and plasmodesmal trafficking) as the interacting domain23,35.
The differences in the binding of the two host proteins to the PVX RNAs and CP suggested that the functions of the two host proteins might differ in response to PVX infection. To investigate that possibility, we performed transient overexpression and silencing experiments using N. benthamiana. Both host proteins were found to enhance PVX replication in experiments using protoplasts; the enhancement of PVX replication, however, was greater for NbCPIP2a than for NbCPIP2b. Long-distance movement PVX was also enhanced by both host proteins, and the effect of NbCPIP2a was slightly greater than that of NbCPIP2b. We also observed a decreased accumulation of PVX RNAs with partial silencing of the NbCPIP2 genes, suggesting that NbCPIP2s are required during the initial stages of PVX RNA replication. Although overexpression of NbCPIP2a and NbCPIP2b increased RNA replication, cell-to-cell movement, and long-distance movement of PVX in N. benthamiana, partial silencing of these two genes reduced RNA replication but not cell-to-cell or long-distance movement of PVX. The intensity of fluorescence, however, was significantly reduced in silenced cells with respect to early cell-to-cell movement. This might result from the significantly reduced level of RNA replication during the early infection of silenced leaves. It is possible that 1) NbCPIP2s-mediated cell-to-cell movement of PVX is complemented by other host factor(s) in N. benthamiana or that 2) the silencing levels (32.3% and 28.9% for NbCPIP2a and NbCPIP2b genes, respectively) were too small to reduce the cell-to-cell movement of PVX. Regarding other host factors that may contribute to cell-to-cell movement, HSP70 proteins may enable viral movement complex to translocate through plasmodesmata (PD) pores27 and could provide the motive force that facilitates intercellular transport of plant virus via PD37. If both NbCPIP2 genes are silenced, it is possible that HSP70 activity related to virus movement might be not affected because of functional complementation by DnaJ-like proteins that function as co-chaperones of HSP70; these include CPIP23, NtDNAJ-M54137, and RME-838. Because HSP70 and its co-chaperone CPIP are required for PVY infection (presumably by interacting with the viral CP23), it is tempting to speculate that NbCPIP2a and NbCPIP2b act as important regulators during PVX infection by interacting with PVX SL1 RNA and CP and possibly by recruiting the HSP70. Therefore, characterization of DnaJ-like proteins involved in HSP70 interaction will increase our understanding of the roles of NbCPIP2a and NbCPIP2b in PVX replication and movement.
Several kinds of evidence indicate that the expression of NbCPIP2s might be specifically regulated by PVX and that NbCPIP2s might be important for PVX infection. First, the NbCPIP2 genes were induced by PVX infection but not by CMV or PepMoV infection (Fig. 6). Second, the NbCPIP2s interacted with SL1 RNA elements of the PVX genomic RNA (gRNA) and CP. Third, over-expression of the NbCPIP2s significantly increased the accumulation of PVX RNAs and PVX movement (Fig. 3). Viral replicases of most plant viruses with plus-strand RNA associate with host cellular membranes by specific interaction with host proteins and form functional complexes for viral replication39,40,41,42. In PVX, RNA elements, including the first of five repeated ACCA sequence elements and the 5′ SL1 of PVX gRNA, are required for viral RNA replication and movement5,43,44,45,46. In addition, Lough et al.47 reported that the SL1(+) RNA or SL1(−) RNA of PVX behaves as a cis-acting element and can affect cell-to-cell movement of PVX. We recently identified host proteins, including NbMPB2Cb and NbDnaJ, that bind to the 5′ SL1 of PVX gRNA and reduce PVX RNA replication19,20. All of the identified host proteins, i.e., NbCPIP2s, NbMPB2Cb, and NbDnaJ, also interact with PVX CP, a protein that is also required for PVX movement. Moreover, subcellular localization of NbCPIP2s in host cellular membranes further indicates that NbCPIP2s may function in the formation of replicase complexes. Together, these results suggest that several host factors increase or decrease PVX RNA replication and movement possibly by the specifically interacting with an SL1 RNA element and by interacting with viral proteins such as CP or MP.
Overall, our study has demonstrated that the homologous host proteins NbCPIP2a and NbCPIP2b differ in their binding to PVX RNA elements but not to PVX CP. In addition, functional studies using transient assay showed that the difference in the binding properties of NbCPIP2a and NbCPIP2b resulted in functional differences, especially in PVX replication. We also found that the impaired functions of a host gene might be complemented by another homologous protein.
Methods
Plant materials and growth condition
N. benthamiana plants used in this study were grown in a growth chamber at 25 °C under a 16 h/8 h (light/dark) photoperiod. We used 4-week-old N. benthamiana plants for BiFC, transient expression, functional study, subcellular localization, and time-course gene expression analysis.
RNA extraction and cloning of the two host genes
We extracted total RNAs from N. benthamiana plants with Isol-RNA Lysis Reagent (5 Prime, Hilden, Germany) according to the manufacturer’s instructions. Complementary DNA (cDNA) was synthesized with the GoScript Reverse Transcription System (Promega, Madison, USA) following the manufacturer’s instructions. Full-length NbCPIP2a and NbCPIP2b were amplified by gene-specific primers (Supplementary Table 1). The amplified PCR products were cloned into the Gateway pENTR vector following the manufacturer’s instructions (Invitrogen, Carlsbad, USA). The generated pENTR containing each gene was further cloned into appropriate vectors.
Recombinant protein expression
Full-length NbCPIP2a and NbCPIP2b were cloned into the pET28 vector by restriction enzyme treatment using EcoRI and BamHI. The cloned vectors were transformed into E. coli BL21(DE3) codon plus RIL strain (Stratagene, La Jolla, USA). Recombinant proteins were expressed with 0.4 mM IPTG at 37 °C and were extracted and then purified based on a previous study47. The purified proteins were subjected to EMSA.
EMSA
Probes were amplified by PCR using probe-specific primers (Supplementary Table S1). After purification, in vitro transcription using T7 RNA polymerase (Takara, Shiga, Japan) and general procedures for EMSA were adapted from a previous study48. In brief, 100 ng of P32-labeled RNA transcripts and recombinant NbCPIP2a protein or NbCPIP2b protein were incubated on ice in 1X EMSA binding buffer (10 μl final volume). The mixtures were supplemented with a 0.2 volume of 5X loading buffer (50% glycerol and 0.05% bromophenol blue) and electrophoresed on a 5% non-denaturing polyacrylamide gel at 100 V (4 °C) for 40 min. The gels were dried with a vacuum dryer and visualized using a Fuji BAS-2500 Phosphor Imager (Fuji Film, Nakanuma, Japan).
BiFC assay
The full-length of each gene in the pENTR vectors was cloned into the modified pPZP-DEST-nEYFP-C1 and pPZP-DEST-cEYFP-C1 vectors for BiFC49,50. The vectors containing NbCPIP2a and NbCPIP2b were transformed into Agrobacterium strain GV2260 by the heat shock method. The transformed bacteria were suspended in MMA buffer containing 10 mM MES salt (pH 5.6), 10 mM MgCl2 and 100 μM acetosyringone. A syringe was used to infiltrate the bacteria into the abaxial sides of N. benthamiana leaves. After 2 days, green fluorescence in the infiltrated leaves was observed with a fluorescence microscope (Carl Zeiss, Oberkochen, Germany).
Transient overexpression and silencing of target host genes
To transiently overexpress the two host genes, we generated two overexpression constructs using pCAMBIA2300 vectors that were modified by tagging with the HA epitope. By StuI enzyme digestion, each full-length cDNA for NbCPIP2a and NbCPIP2b was cloned into the modified pCAMBIA2300 vector. For viral replication and systemic movement experiment influenced by overexpressed NbCPIP2a and NbCPIP2b, we infiltrated vectors expressing NbCPIP2a and NbCPIP2b, respectively. Two days after transient overexpression of each protein, we inoculated the leaves with PVX tagged with green fluorescence protein (GFP) using an agrobacterium mediated infectious clone. As a viral replication observation in infiltrated leaves, we observed the PVX:GFP infiltrated leaves at 2 days after PVX:GFP infiltration. For systemic movement observation for PVX:GFP, we took pictures of PVX:GFP infected plants at 7 days after infiltration under hand UV-lamp. For transient silencing of the two host genes, a partial sequence for an individual host gene was cloned into a hairpin expression vector51, which was constructed by modifying the pPZP vector16. After four days infiltration of silencing agrobacterium clones, we analyzed the silencing level using total RNAs extracted from silenced leaves then inoculated PVX expressing GFP by agro-infiltration. We observed the viral replication in local leaves at 2 days post inoculation and systemic movement of PVX:GFP at 7 days post inoculation of PVX:GFP. All of cloned vectors were transformed into Agrobacterium strain GV3101 by the heat shock method. A syringe was used to infiltrate the transformed bacteria into the abaxial sides of N. benthamiana leaves. Agrobacterium strain GV3101 transformed with an empty vectors expressing only HA-tag (for overexpression experiment) or pdk intron (for silencing experiment) without PVX:GFP were used as negative controls. For positive control, N. benthamiana plants agro-infiltrated with an empty vector followed by infiltration of agrobacteria containing PVX:GFP on the same leaves were used. Experiments for overexpression and silencing were performed with three biological replicates with at least three individual plants for each experiment.
Quantification of PVX replication in protoplasts
To quantify PVX replication, we transiently overexpressed or silenced the target gene. In the case of overexpression, PVX was challenge-inoculated on the infiltrated leaves at 1 day after infiltration. In the case of gene silencing, PVX was challenge-inoculated on the infiltrated leaves at 4 days after infiltration. At 12 h after PVX challenge inoculation, protoplasts were extracted from the inoculated leaves according to previous reports52,53. The extracted protoplasts were cultured in Aoki’s salt medium at 25 °C for 2 days. The total RNA of the cultured protoplasts was extracted as previously described26. PVX replication in protoplasts was quantified by real-time RT-PCR using a CFX384 real-time PCR detection system (Bio-Rad, Hercules, USA). The PCR results were normalized with actin and ubiquitin genes.
Time-course gene expression analysis using real-time RT-PCR
For time-course expression analysis of the target host genes and PVX, we performed real-time RT-PCR using gene-specific primers (Supplementary Table S1). Plant samples were harvested from the leaves that each three viruses inoculated at each time point for 1 week. Three biological replicates were used for this experiment and conducted three times. PVX, CMV, and PepMoV were mechanically inoculated with carborundum.
Subcellular localization of the two host genes
The full-length cDNAs of the two host genes were individually cloned into the pENTR vector. Each pENTR vector was cloned into pSITE-2CA54 containing GFP by LR reaction according to the manufacturer’s instructions (Invitrogen). Cellular markers tagged with RFP were co-infiltrated with vectors expressing GFP-tagged host protein or empty vector expressing free GFP (pSITE-2CA). Two days after agroinfiltration, GFP as well as RFP were observed with a fluorescence microscope (Carl Zeiss, Oberkochen, Germany). We carried out subcellular localization experiments with three independent biological replicates.
Additional Information
How to cite this article: Choi, H. et al. Two homologous host proteins interact with potato virus X RNAs and CPs and affect viral replication and movement. Sci. Rep. 6, 28743; doi: 10.1038/srep28743 (2016).
References
Scholthof, K. B. G. et al. Top 10 plant viruses in molecular plant pathology. Mol. Plant Pathol. 12, 938–954 (2011).
Kim, K.-H. & Hemenway, C. Mutations that alter a conserved element upstream of the potato virus X triple block and coat protein genes affect subgenomic RNA accumulation. Virology 232, 187–197 (1997).
Batten, J. S., Yoshinari, S. & Hemenway, C. Potato virus X: a model system for virus replication, movement and gene expression. Mol. Plant Pathol. 4, 125–131 (2003).
Bercks, R. Potato virus X. CMI/AAB Descriptions of Plant Viruses No. 4. Culross and Son, Ltd. Perthshire, Scotland (1970).
Miller, E. D., Plante, C. A., Kim, K. H., Brown, J. W. & Hemenway, C. Stem-loop structure in the 5’ region of potato virus X genome required for plus-strand RNA accumulation. J. Mol. Biol. 284, 591–608 (1998).
Chiu, W.-W., Hsu, Y.-H. & Tsai, C.-H. Specificity analysis of the conserved hexanucleotides for the replication of bamboo mosaic potexvirus RNA. Virus Res. 83, 159–167 (2002).
Pogue, G. P. & Hall, T. C. The requirement for a 5′stem-loop structure in brome mosaic virus replication supports a new model for viral positive-strand RNA initiation. J. Virol. 66, 674–684 (1992).
Verchot-Lubicz, J., Ye, C.-M. & Bamunusinghe, D. Molecular biology of potexviruses: recent advances. J. Gen. Virol. 88, 1643–1655 (2007).
Park, M.-R., Jeong, R.-D. & Kim, K.-H. Understanding the intracellular trafficking and intercellular transport of potexviruses in their host plants. Front. Plant Sci. 5 (2014).
Kwon, S.-J. et al. cis-Acting sequences required for coat protein binding and in vitro assembly of potato virus X. Virology 334, 83–97 (2005).
Chapman, S., Hills, G., Watts, J. & Baulcombe, D. Mutational analysis of the coat protein gene of potato virus X: effects on virion morphology and viral pathogenicity. Virology 191, 223–230 (1992).
Fedorkin, O. et al. Cell-to-cell movement of potato virus X involves distinct functions of the coat protein. J. Gen. Virol. 82, 449–458 (2001).
Hyodo, K. & Okuno, T. Host factors used by positive-strand RNA plant viruses for genome replication. J. Gen. Plant Pathol. 80, 123–135 (2014).
Ahlquist, P., Noueiry, A. O., Lee, W.-M., Kushner, D. B. & Dye, B. T. Host factors in positive-strand RNA virus genome replication. J. Virol. 77, 8181–8186 (2003).
Whitham, S. A. & Wang, Y. Roles for host factors in plant viral pathogenicity. Curr. Opin. Plant Biol. 7, 365–371 (2004).
Park, M.-R., Park, S.-H., Cho, S.-Y. & Kim, K.-H. Nicotiana benthamiana protein, NbPCIP1, interacting with potato virus X coat protein plays a role as susceptible factor for viral infection. Virology 386, 257–269 (2009).
Hwang, J. et al. Plant translation elongation factor 1Bβ facilitates potato virus X (PVX) infection and interacts with PVX triple gene block protein 1. PloS one 10, e0128014 (2015).
Fridborg, I. et al. TIP, a novel host factor linking callose degradation with the cell-to-cell movement of potato virus X. Mol. Plant Microbe Interact. 16, 132–140 (2003).
Cho, S.-Y., Cho, W. K., Choi, H.-S. & Kim, K.-H. Cis-acting element (SL1) of potato virus X controls viral movement by interacting with the NbMPB2Cb and viral proteins. Virology 427, 166–176 (2012).
Cho, S.-Y., Cho, W. K., Sohn, S.-H. & Kim, K.-H. Interaction of the host protein NbDnaJ with potato virus X minus-strand stem-loop 1 RNA and capsid protein affects viral replication and movement. Biochem. Biophys. Res. Commun. 417, 451–456 (2012).
Cho, S.-Y., Cho, W. K. & Kim, K.-H. Identification of tobacco proteins associated with the stem-loop 1 RNAs of potato virus X. Mol. Cells 33, 379–384 (2012).
Park, S.-H. & Kim, K.-H. Virus-induced silencing of the WRKY1 transcription factor that interacts with the SL1 structure of potato virus X leads to higher viral RNA accumulation and severe necrotic symptoms. Plant Pathol. J. 28, 40–48 (2012).
Hofius, D. et al. Capsid protein-mediated recruitment of host DnaJ-like proteins is required for potato virus Y infection in tobacco plants. J. Virol. 81, 11870–11880 (2007).
Hellman, L. M. & Fried, M. G. Electrophoretic mobility shift assay (EMSA) for detecting protein–nucleic acid interactions. Nat. Protoc. 2, 1849–1861 (2007).
Bracha‐Drori, K. et al. Detection of protein–protein interactions in plants using bimolecular fluorescence complementation. Plant J. 40, 419–427 (2004).
Fabian, M. R. & Andrew White, K. Extracting viral RNAs from plant protoplasts. Curr. Protoc. Microbiol. 16E, 11.11–16E, 11.16 (2007).
Boevink, P. & Oparka, K. J. Virus-host interactions during movement processes. Plant Physiol. 138, 1815–1821 (2005).
Qiu, X.-B., Shao, Y.-M., Miao, S. & Wang, L. The diversity of the DnaJ/Hsp40 family, the crucial partners for Hsp70 chaperones. Cell. Mol. Life Sci. 63, 2560–2570 (2006).
Cheetham, M. E. & Caplan, A. J. Structure, function and evolution of DnaJ: conservation and adaptation of chaperone function. Cell Stress Chaperone. 3, 28 (1998).
Knox, C., Luke, G. A., Blatch, G. L. & Pesce, E.-R. Heat shock protein 40 (Hsp40) plays a key role in the virus life cycle. Virus Res. 160, 15–24 (2011).
Tomita, Y. et al. Mutation of host DnaJ homolog inhibits brome mosaic virus negative-strand RNA synthesis. J. Virol. 77, 2990–2997 (2003).
Serva, S. & Nagy, P. D. Proteomics analysis of the tombusvirus replicase: Hsp70 molecular chaperone is associated with the replicase and enhances viral RNA replication. J. Virol. 80, 2162–2169 (2006).
Ivanovic, T., Agosto, M. A., Chandran, K. & Nibert, M. L. A role for molecular chaperone Hsc70 in reovirus outer capsid disassembly. J. Biol. Chem. 282, 12210–12219 (2007).
Hafrén, A., Hofius, D., Rönnholm, G., Sonnewald, U. & Mäkinen, K. HSP70 and its cochaperone CPIP promote potyvirus infection in Nicotiana benthamiana by regulating viral coat protein functions. Plant Cell 22, 523–535 (2010).
Dolja, V., Haldeman, R., Robertson, N., Dougherty, W. & Carrington, J. Distinct functions of capsid protein in assembly and movement of tobacco etch potyvirus in plants. EMBO J. 13, 1482 (1994).
Ivanov, K. I. & Mäkinen, K. Coat proteins, host factors and plant viral replication. Curr. Opin. Virol. 2, 712–718 (2012).
Soellick, T.-R., Uhrig, J., Bucher, G., Kellmann, J.-W. & Schreier, P. The movement protein NSm of tomato spotted wilt tospovirus (TSWV): RNA binding, interaction with the TSWV N protein, and identification of interacting plant proteins. Proc. Natl. Acad Sci. USA 97, 2373–2378 (2000).
Haupt, S. et al. Two plant–viral movement proteins traffic in the endocytic recycling pathway. Plant Cell 17, 164–181 (2005).
Mackenzie, J. Wrapping things up about virus RNA replication. Traffic 6, 967–977 (2005).
Salonen, A., Ahola, T. & Kaariainen, L. Viral RNA replication in association with cellular membranes. Curr. Top. Microbiol. Immunol. 285, 139–173 (2005).
Sanfacon, H. & Zhang, G. Analysis of interactions between viral replicase proteins and plant intracellular membranes. Methods Mol. Biol. 451, 361–375 (2008).
Schwartz, M., Chen, J., Lee, W. M., Janda, M. & Ahlquist, P. Alternate, virus-induced membrane rearrangements support positive-strand RNA virus genome replication. Proc. Natl. Acad Sci. USA 101, 11263–11268 (2004).
Kim, K. H. & Hemenway, C. The 5′ nontranslated region of potato virus X RNA affects both genomic and subgenomic RNA synthesis. J. Virol. 70, 5533–5540 (1996).
Kwon, S. J. & Kim, K. H. The SL1 stem-loop structure at the 5′-end of potato virus X RNA is required for efficient binding to host proteins and for viral infectivity. Mol. Cells 21, 63–75 (2006).
Miller, E. D., Kim, K. H. & Hemenway, C. Restoration of a stem-loop structure required for potato virus X RNA accumulation indicates selection for a mismatch and a GNRA tetraloop. Virology 260, 342–353 (1999).
Park, M. R., Kwon, S. J., Choi, H. S., Hemenway, C. L. & Kim, K. H. Mutations that alter a repeated ACCA element located at the 5′ end of the potato virus X genome affect RNA accumulation. Virology 378, 133–141 (2008).
Lough, T. J., Lee, R. H., Emerson, S. J., Forster, R. L. & Lucas, W. J. Functional analysis of the 5′ untranslated region of potexvirus RNA reveals a role in viral replication and cell-to-cell movement. Virology 351, 455–465 (2006).
Son, M., Choi, H. & Kim, K.-H. Specific binding of Fusarium graminearum Hex1 protein to untranslated regions of the genomic RNA of Fusarium graminearum virus 1 correlates with increased accumulation of both strands of viral RNA. Virology 489, 202–211 (2016).
Lian, S., Cho, W. K., Jo, Y., Kim, S.-M. & Kim, K.-H. Interaction study of rice stripe virus proteins reveals a region of the nucleocapsid protein (NP) required for NP self-interaction and nuclear localization. Virus Res. 183, 6–14 (2014).
Tzfira, T. et al. pSAT vectors: a modular series of plasmids for autofluorescent protein tagging and expression of multiple genes in plants. Plant Mol. Biol. 57, 503–516 (2005).
Kościańska, E., Kalantidis, K., Wypijewski, K., Sadowski, J. & Tabler, M. Analysis of RNA silencing in agroinfiltrated leaves of Nicotiana benthamiana and Nicotiana tabacum . Plant Mol. Biol. 59, 647–661 (2005).
Rao, A. Preparation and inoculation of mesophyll protoplasts from monocotyledenous jhand dicotyledenous hosts. Curr. Protoc. Microbiol. 16D, 12.11–16D. 12.18 (2007).
Takebe, I., OTSUKI, Y. & AOKI, S. Isolation of tobacco mesophyll cells in intact and active state. Plant Cell Physiol. 9, 115–124 (1968).
Chakrabarty, R. et al. PSITE vectors for stable integration or transient expression of autofluorescent protein fusions in plants: probing Nicotiana benthamiana-virus interactions. Mol. Plant Microbe Interact. 20, 740–750 (2007).
Tamura, K., Stecher, G., Peterson, D., Filipski, A. & Kumar, S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729 (2013).
Acknowledgements
This research was supported in part by grants from the Vegetable Breeding Research Center (No. 710001-07-5) through the Agriculture, Food and Rural Affairs Research Center Support Program from the Ministry of Agriculture, Food and Rural Affairs and the Next-Generation BioGreen 21 Program (No. PJ01101401), Rural Development Administration (RDA). HC and WKC were supported by research fellowships from the Brain Korea 21 Plus Project.
Author information
Authors and Affiliations
Contributions
H.C. and K.-H.K. designed the research; H.C. performed the research; H.C., W.K.C. and K.-H.K. analysed the data; and H.C. and W.K.C. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Choi, H., Cho, W. & Kim, KH. Two homologous host proteins interact with potato virus X RNAs and CPs and affect viral replication and movement. Sci Rep 6, 28743 (2016). https://doi.org/10.1038/srep28743
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep28743