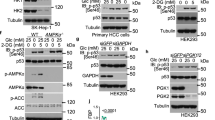
Abstract
Metabolic enzymes have been shown to function as transcriptional regulators. p53, a tumor-suppressive transcription factor, was recently found to regulate energy metabolism. These combined facts raise the possibility that metabolic enzymes may directly regulate p53 function. Here, we discover that nucleocytoplasmic malate dehydrogenase-1 (MDH1) physically associates with p53. Upon glucose deprivation, MDH1 stabilizes and transactivates p53 by binding to p53-responsive elements in the promoter of downstream genes. Knockdown of MDH1 significantly reduces binding of acetylated-p53 and transcription-active histone codes to the promoter upon glucose depletion. MDH1 regulates p53-dependent cell-cycle arrest and apoptosis in response to glucose deprivation, suggesting that MDH1 functions as a transcriptional regulator for a p53-dependent metabolic checkpoint. Our findings provide insight into how metabolism is directly linked to gene expression for controlling cellular events in response to metabolic stress.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
Abbreviations
- MDH1:
-
malate dehydrogenase-1
- GAPDH:
-
glyceraldehyde-3-phosphate dehydrogenase
- TCA:
-
tricarboxylic acid
- ChIP:
-
chromatin Immunoprecipitation
- GLUT:
-
glucose transporter
- HEK293:
-
human embryonic kidney 293
- p53-RE:
-
p53-responsive element
References
Shi Y, Shi Y . Metabolic enzymes and coenzymes in transcription-a direct link between metabolism and transcription? Trends Genet 2004; 20: 445–452.
Bhardwaj A, Wilkinson MF . A metabolic enzyme doing double duty as a transcription factor. Bioessays 2005; 27: 467–471.
Ladurner AG . Rheostat control of gene expression by metabolites. Mol Cell 2006; 24: 1–11.
Zheng L, Roeder RG, Luo YS . S phase activation of the histone H2B promoter by OCA-S, a coactivator complex that contains GAPDH as a key component. Cell 2003; 114: 255–266.
Sen N, Hara MR, Kornberg MD, Cascio MB, Bae BI, Shahani N et al. Nitric oxide-induced nuclear GAPDH activates p300/CBP and mediates apoptosis. Nat Cell Biol 2008; 10: 866–873.
Hall DA, Zhu H, Zhu X, Royce T, Gerstein M, Snyder M . Regulation of gene expression by a metabolic enzyme. Science 2004; 306: 482–484.
Cho YH, Yoo SD, Sheen J . Regulatory functions of nuclear hexokinase1 complex in glucose signaling. Cell 2006; 127: 579–589.
Warburg O . On respiratory impairment in cancer cells. Science 1956; 124: 269–270.
Gatenby RA, Gillies RJ . Why do cancers have high aerobic glycolysis? Nat Rev Cancer 2004; 4: 891–899.
Shaw RJ . Glucose metabolism and cancer. Curr Opin Cell Biol 2006; 18: 598–608.
Christofk HR, Vander Heiden MG, Harris MH, Ramanathan A, Gerszten RE, Wei R et al. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumor growth. Nature 2008; 452: 230–234.
Vogelstein B, Kinzler KW . Cancer genes and the pathways they control. Nat Med 2004; 10: 789–799.
Levine AJ, Hu W, Feng Z . The P53 pathway: what questions remain to be explored? Cell Death Differ 2006; 13: 1027–1036.
Kondoh H, Lleonart ME, Gil J, Wang J, Degan P, Peters G et al. Glycolytic enzymes can modulate cellular life span. Cancer Res 2005; 65: 177–185.
Bensaad K, Tsuruta A, Selak MA, Vidal MN, Nakano K, Bartrons R et al. TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell 2006; 126: 107–120.
Matoba S, Kang JG, Patino WD, Wragg A, Boehm M, Gavrilova O et al. p53 regulates mitochondrial respiration. Science 2006; 312: 1650–1653.
Kawauchi K, Araki K, Tobiume K, Tanaka N . p53 regulates glucose metabolism through an IKK-NF-κB pathway and inhibits cell transformation. Nat Cell Biol 2008; 10: 611–618.
Tyner SD, Venkatachalam S, Choi J, Jones S, Ghebranious N, Igelmann H et al. p53 mutant mice that display early ageing-associated phenotypes. Nature 2002; 415: 45–53.
Maier B, Gluba W, Bernier B, Turner T, Mohammad K, Guise T et al. Modulation of mammalian life span by the short isoform of p53. Genes Dev 2004; 18: 306–319.
Campisi J . Senescent cells, tumor suppression, and organismal aging: good citizens, bad neighbors. Cell 2005; 120: 513–522.
Schwartzenberg-Bar-Yoseph F, Armoni M, Karnieli E . The tumor suppressor p53 down-regulates glucose transporters GLUT1 and GLUT4 gene expression. Cancer Res 2004; 64: 2627–2633.
Katz EB, Stenbit AE, Hatton K, DePinho R, Charron MJ . Cardiac and adipose tissue abnormalities but not diabetes in mice deficient in GLUT4. Nature 1995; 377: 151–155.
Wadgaonkar R, Collins T . Murine double minute (MDM2) blocks p53-coactivator interaction, a new mechanism for inhibition of p53-dependent gene expression. J Biol Chem 1999; 274: 13760–13767.
Birktoft JJ, Banaszak LJ . The presence of a histidine-aspartic acid pair in the active site of 2-hydroxyacid dehydrogenases. X-ray refinement of cytoplasmic malate dehydrogenase. J Biol Chem 1983; 258: 472–482.
Assaily W, Benchimol S . Differential utilization of two ATP-generating pathways in regulated by p53. Cancer Cell 2006; 10: 4–6.
Bensaad K, Vousden KH . p53: new roles in metabolism. Trends Cell Biol 2006; 17: 286–291.
Green DR, Chipuk JE . p53 and metabolism: inside the TIGAR. Cell 2006; 126: 30–32.
Birktoft JJ, Fernley RT, Bradshaw RA, Banaszak LJ . Amino acid sequence homology among the 2-hydroxy acid dehydrogenases: mitochondrial and cytoplasmic malate dehydrogenases form a homologous system with lactate dehydrogenase. Proc Natl Acad Sci USA 1982; 79: 6166–6170.
Ronai Z . Glycolytic enzymes as DNA binding proteins. Int J Biochem 1993; 25: 1073–1076.
Shi Y, Sawada J, Sui G, Affar EB, Whetstine JR, Lan F et al. Coordinated histone modifications mediated by a CtBP co-repressor complex. Nature 2003; 422: 735–738.
Grooteclaes M, Deveraux Q, Hildebrand J, Zhang Q, Goodman RH, Frisch SM . C-terminal-binding protein corepresses epithelial and proapoptotic gene expression programs. Proc Natl Acad Sci USA 2003; 100: 4568–4573.
Kim JH, Cho EJ, Kim ST, Youn HD . CtBP represses p300-mediated transcriptional activation by direct association with its bromodomain. Nat Struct Mol Biol 2005; 12: 423–428.
Webb LE, Hill EJ, Banaszak LJ . Conformation of nicotinamide adenine dinucleotide bound to cytoplasmic malate dehydrogenase. Biochemistry 1973; 12: 5101–5109.
Grant PM, Roderick SL, Grant GA, Banaszak LJ, Strauss AW . Comparison of the precursor and mature forms of rat heart mitochondrial malate dehydrogenase. Biochemistry 1987; 26: 128–134.
Jones RG, Plas DR, Kubek S, Buzzai M, Mu J, Xu Y et al. AMP-activated protein kinase induces a p53-dependent metabolic checkpoint. Mol Cell 2005; 18: 283–293.
Thoreen CC, Sabatini DM . AMPK and p53 help cells through lean times. Cell Metab 2005; 1: 287–288.
Levine AJ, Feng Z, Mak TW, You H, Jin S . Coordination and communication between the p53 and IGF-1-AKT-TOR signal transduction pathways. Genes Dev 2006; 20: 267–275.
Kanai M, Hanashiro K, Kim SH, Hanai S, Boulares AH, Miwa M et al. Inhibition of Crm1-p53 interaction and nuclear export of p53 by poly(ADP-ribosyl)ation. Nat Cell Biol 2007; 9: 1175–1183.
Roe JS, Kim H, Lee SM, Kim ST, Cho EJ, Youn HD . p53 stabilization and transactivation by a von Hippel-Lindau protein. Mol Cell 2006; 22: 395–405.
He TC, Zhou S, da Costa LT, Yu J, Kinzler KW, Vogelstein B . A simplified system for generating recombinant adenoviruses. Proc Natl Acad Sci USA 1998; 95: 2509–2514.
Livak KJ, Schmittgen TD . Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 2001; 25: 402–408.
Acknowledgements
We thank Dr. H-W Lee and Dr. JH Chung for invaluable materials and JW Choi for initiating the cloning of metabolic enzymes. This work was supported by KOSEF grants from the National Research Laboratory (ROA-2007-000-20002-0), the Center for Aging and Apoptosis Research at Seoul National University (R11-2002-097-050050-0) and the Center for Functional Analysis for Human Genome (3344-20060070).
Author information
Authors and Affiliations
Corresponding author
Additional information
Edited KH Vousden
Supplementary Information accompanies the paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Supplementary information
Rights and permissions
About this article
Cite this article
Lee, S., Kim, J., Cho, E. et al. A nucleocytoplasmic malate dehydrogenase regulates p53 transcriptional activity in response to metabolic stress. Cell Death Differ 16, 738–748 (2009). https://doi.org/10.1038/cdd.2009.5
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/cdd.2009.5
Keywords
This article is cited by
-
Permission to pass: on the role of p53 as a gatekeeper for aneuploidy
Chromosome Research (2023)
-
Low glucose metabolite 3-phosphoglycerate switches PHGDH from serine synthesis to p53 activation to control cell fate
Cell Research (2023)
-
Cardio-onco-metabolism: metabolic remodelling in cardiovascular disease and cancer
Nature Reviews Cardiology (2022)
-
Dysregulated metabolic pathways in age-related macular degeneration
Scientific Reports (2020)
-
Maintaining homeostasis by controlled alternatives for energy distribution in plant cells under changing conditions of supply and demand
Photosynthesis Research (2019)