Abstract
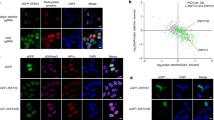
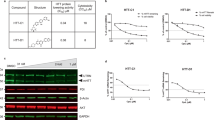
Aberrant chromatin remodeling is involved in the pathogenesis of Huntington's disease (HD) but the mechanism is not known. Herein, we report that mutant huntingtin (mtHtt) induces the transcription of alpha thalassemia/mental retardation X linked (ATRX), an ATPase/helicase and SWI/SNF-like chromatin remodeling protein via Cdx-2 activation. ATRX expression was elevated in both a cell line model and transgenic model of HD, and Cdx-2 occupancy of the ATRX promoter was increased in HD. Induction of ATRX expanded the size of promyelocytic leukemia nuclear body (PML-NB) and increased trimethylation of H3K9 (H3K9me3) and condensation of pericentromeric heterochromatin, while knockdown of ATRX decreased PML-NB and H3K9me3 levels. Knockdown of ATRX/dXNP improved the hatch rate of fly embryos expressing mtHtt (Q127). ATRX/dXNP overexpression exacerbated eye degeneration of eye-specific mtHtt (Q127) expressing flies. Our findings suggest that transcriptional alteration of ATRX by mtHtt is involved in pericentromeric heterochromatin condensation and contributes to the pathogenesis of HD.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
Abbreviations
- ATRX:
-
alpha thalassemia/mental retardation X linked
- CDX2:
-
caudal type homeobox 2
- ChIP:
-
chromatin immunoprecipitation
- CREB:
-
cAMP response element-binding
- Daxx:
-
death-domain associated protein
- Dnmt3:
-
DNA methyltransferase
- dXNP:
-
drosopila homolog of XNP/ATRX
- GATA2:
-
transcription factors GATA2
- GMR:
-
glass multimer repeat
- HD:
-
Huntington's disease
- H3K9me3:
-
trimethylation of H3K9
- Htt :
-
Huntingtin
- HP1α:
-
heterochromatin protein 1
- MeCP2:
-
methyl-CpG-binding protein 2
- mtHtt:
-
mutant huntingtin
- PML-NBs:
-
promyelocytic leukemia nuclear bodies
- SP1:
-
specificity protein1
- SWI/SNF:
-
switch/sucrose non-fermentable
References
Vonsattel JP, DiFiglia M . Huntington disease. J Neuropathol Exp Neurol 1998; 57: 369–384.
The Huntington’s Disease Collaborative Research Group. A nobel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. Cell 1993; 72: 971–983.
Steffan JS, Kazantsev A, Spasic-Boskovic O, Greenwald M, Zhu YZ, Gohler H et al. The Huntington’s disease protein interacts with p53 and CREB-binding protein and represses transcription. Proc Natl Acad Sci USA 2000; 97: 6763–6768.
Sugars KL, Rubinsztein DC . Transcriptional abnormalities in Huntington disease. Trends Genet 2003; 19: 233–238.
Ryu H, Lee J, Hargerty SW, Soh BY, Mcalpin SE, Cormier KA et al. ESET/SETDB1 gene expression and histone H3 (K9) trimethylation in Huntington’s disease. Proc Natl Acad Sci USA 2006; 103: 19176–19181.
Sadri-Vakili G, Cha JH . Mechanisms of disease: histone modifications in Huntington's disease. Nat Clin Pract Neurol 2006; 2: 330–338.
Varga-Weisz PD, Becker PB . Regulation of higher-order chromatin structures by nucleosome-remodelling factors. Curr Opin Genet Dev 2006; 16: 151–156.
Gibbons RJ, Picketts DJ, Villard L, Higgs DR . Mutations in a putative global transcriptional regulator cause X-linked mental retardation with alphathalassemia (ATR-X syndrome). Cell 1995; 80: 837–845.
Picketts DJ, Higgs DR, Bachoo S, Blake DJ, Quarrell OW, Gibbons RJ . ATRX encodes a novel member of the SNF2 family of proteins: mutations point to a common mechanism underlying the ATR-X syndrome. Hum Mol Genet 1996; 5: 1899–1907.
Gibbons RJ, McDowell TL, Raman S, O’Rourke DM, Garrick D, Ayyub H et al. Mutations in ATRX, encoding a SWI/SNF-like protein, cause diverse changes in the pattern of DNA methylation. Nat Genet 2000; 24: 368–371.
Baumann C, Viveiros MM, De La Fuente R . Loss of maternal ATRX results in centromere instability and aneuploidy in the mammalian oocyte and pre-implantation embryo. PLoS Genet 2010; 6: e1001137.
Berube NG, Jagla M, Smeenk C, De Repentigny Y, Kothary R, Picketts DJ . Neurodevelopmental defects resulting from ATRX overexpression in transgenic mice. Hum Mol Genet 2002; 11: 253–261.
Ishov AM, Vladimirova OV, Maul GG . Heterochromatin and ND10 are cell-cycle regulated and phosphorylation-dependent alternate nuclear sites of the transcription repressor Daxx and SWI/SNF protein ATRX. J Cell Sci 2004; 117: 3807–3820.
Tang J, Wu S, Liu H, Stratt R, Barak OG, Shiekhattar R et al. A novel transcription regulatory complex containing death domain-associated protein and the ATR-X syndrome protein. J Biol Chem 2004; 279: 20369–20377.
Nan X, Hou J, Maclean A, Nasir J, Lafuente MJ, Shu MJ et al. Interaction between chromatin proteins MECP2 and ATRX is disrupted by mutations that cause inherited mental retardation. Proc Natl Acad Sci USA 2007; 104: 2709–2714.
Berube NG, Mangelsdorf M, Jagla M, Vanderluit J, Garrick D, Gibbons RJ et al. The chromatin-remodeling protein ATRX is critical for neuronal survival during corticogenesis. J Clin Invest 2005; 115: 258–267.
Mangiarini L, Sathasivam K, Seller K, Cozens B, Harper A, Hetherington C et al. Exon1 of the HD gene with an expanded CAG repeat is sufficient to cause a progressive neurological phenotype in transgenic mice. Cell 1996; 87: 493–506.
Trettel F, Rigamonti D, Hilditch-Maguire P, Wheeler VC, Sharp AH, Persichetti F et al. Dominant phenotypes produced by the HD mutation in STHdh(Q111) striatal cells. Hum Mol Genet 2000; 9: 2799–2809.
McDowell TL, Gibbons RJ, Sutherland H, O’Rourke DM, Bickmore WA, Pombo A et al. Localization of a putative transcriptional regulator (ATRX) at pericentromeric heterochromatin and the short arms of acrocentric chromosomes. Proc Natl Acad Sci USA 1999; 96: 13983–13988.
Berube NG, Healy J, Medina CF, Wu S, Hodgson T, Jagla M et al. Patient mutations alter ATRX targeting to PML nuclear bodies. Eur J Hum Genet 2008; 16: 192–201.
Taylor JP, Taye AA, Campbell C, Kazemi-Esfarjani P, Fischbeck KH, Min KT . Aberrant histone acetylation, altered transcription, and retinal degeneration in a Drosophila model of polyglutamine disease are rescued by CREB-binding protein. Genes Dev 2003; 17: 1463–1468.
Lee NG, Hong YK, Yu SY, Han SY, Geum D, Cho D . dXNP, a Drosophila homolog of XNP/ATRX, induces apoptosis via Jun-N-terminal kinase activation. FEBS Lett 2007; 581: 2625–2632.
Hong YK, Lee NG, Lee MJ, Park MS, Choi MS, Suh YS et al. dXNP/DATRX increases apoptosis via the JNK and dFOXO pathway in Drosophila neurons. Biochem Biophys Res Commun 2009; 384: 160–166.
Borovecki F, Lovrecic L, Zhou J, Jeong H, Then H, Rosas HD et al. Genome-wide expression profiling of human blood reveals biomarkers for Huntington's disease. Proc Natl Acad Sci USA 2005; 102: 11023–11028.
Dhayalan A, Tamas R, Bock I, Tattermusch A, Dimitrova E, Kudithipudi S et al. The ATRX-ADD domain binds to H3 tail peptides and reads the combined methylation state of K4 and K9. Hum Mol Genet 2011; 20: 2195–2120.
Mallo GV, Soubeyran P, Lissitzky JC, André F, Farnarier C, Marvaldi J et al. Expression of the Cdx1 and Cdx2 homeotic genes leads to reduced malignancy in colon cancer-derived cells. J Biol Chem 1998; 273: 14030–14036.
Kazemi-Esfarjani P, Benzer S . Genetic suppression of polyglutamine toxicity in Drosophila. Science 2000; 287: 1837–1840.
Ryu H, Lee J, Impey S, Ratan RR, Ferrante RJ . Antioxidants modulate mitochondrial protein kinase A and increase CREB binding to D-Loop DNA of the mitochondrial genome in neurons. Proc Natl Acad Sci USA 2005; 102: 13915–13920.
Acknowledgements
We thank Dr. Marcy MacDonald for STHdhQ7/7 and STHdhQ111/111 cells and Soung Chan Kim for their technical assistance. This study was supported by NIH NS 067283-01A1 (HR), WCU Neurocytomics Program Grant (800-20080848) (HR), and SRC Grant (2010-0029-403) (HR) from NRF.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by N Bazan
Supplementary Information accompanies the paper on Cell Death and Differentiation website
Supplementary information
Rights and permissions
About this article
Cite this article
Lee, J., Hong, Y., Jeon, G. et al. ATRX induction by mutant huntingtin via Cdx2 modulates heterochromatin condensation and pathology in Huntington's disease. Cell Death Differ 19, 1109–1116 (2012). https://doi.org/10.1038/cdd.2011.196
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/cdd.2011.196
Keywords
This article is cited by
-
Decreased FAK activity and focal adhesion dynamics impair proper neurite formation of medium spiny neurons in Huntington's disease
Acta Neuropathologica (2022)
-
FUS(1-359) transgenic mice as a model of ALS: pathophysiological and molecular aspects of the proteinopathy
neurogenetics (2018)
-
Remodeling of heterochromatin structure slows neuropathological progression and prolongs survival in an animal model of Huntington’s disease
Acta Neuropathologica (2017)
-
Transcription, Epigenetics and Ameliorative Strategies in Huntington’s Disease: a Genome-Wide Perspective
Molecular Neurobiology (2015)