Abstract
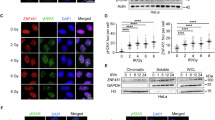
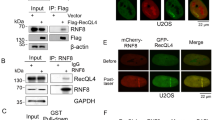
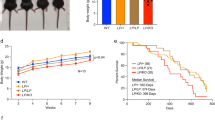
Unrepaired DNA double-strand breaks (DSBs) cause genetic instability that leads to malignant transformation or cell death. Cells respond to DSBs with the ordered recruitment of signaling and repair proteins to the sites of DNA lesions. Coordinated protein SUMOylation and ubiquitylation have crucial roles in regulating the dynamic assembly of protein complexes at these sites. However, how SUMOylation influences protein ubiquitylation at DSBs is poorly understood. We show herein that Rnf4, an E3 ubiquitin ligase that targets SUMO-modified proteins, accumulates in DSB repair foci and is required for both homologous recombination (HR) and non-homologous end joining repair. To establish a link between Rnf4 and the DNA damage response (DDR) in vivo, we generated an Rnf4 allelic series in mice. We show that Rnf4-deficiency causes persistent ionizing radiation-induced DNA damage and signaling, and that Rnf4-deficient cells and mice exhibit increased sensitivity to genotoxic stress. Mechanistically, we show that Rnf4 targets SUMOylated MDC1 and SUMOylated BRCA1, and is required for the loading of Rad51, an enzyme required for HR repair, onto sites of DNA damage. Similarly to inactivating mutations in other key regulators of HR repair, Rnf4 deficiency leads to age-dependent impairment in spermatogenesis. These findings identify Rnf4 as a critical component of the DDR in vivo and support the possibility that Rnf4 controls protein localization at DNA damage sites by integrating SUMOylation and ubiquitylation events.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
Abbreviations
- DSBs:
-
double-strand breaks
- HR:
-
homologous recombination
- NHEJ:
-
non-homologous end joining
- IR:
-
ionizing radiation
- UV:
-
ultraviolet
- IRIF:
-
ionizing radiation-induced foci
- DDR:
-
DNA damage response
- STUbl:
-
SUMO-targeted ubiquitin ligase
- SA:
-
splice acceptor
- PCR:
-
polymerase chain reaction
- MEFs:
-
mouse embryonic fibroblasts
- RT-qPCR:
-
reverse transcription quantitative PCR
- ROS:
-
reactive oxygen species
- PFGE:
-
pulsed-field gel electrophoresis
- IHC:
-
immunohistochemistry
- RING:
-
really interesting new gene
- H&E:
-
hematoxylin and eosin
- (E)GFP:
-
(enhanced) green fluorescent protein
- Hypo:
-
hypomorphic
- SIM:
-
SUMO interaction motif
References
Jackson SP, Bartek J . The DNA-damage response in human biology and disease. Nature 2009; 461: 1071–1078.
Ciccia A, Elledge SJ . The DNA damage response: making it safe to play with knives. Mol Cell 2010; 40: 179–204.
Wyman C, Kanaar R . DNA double-strand break repair: all's well that ends well. Annu Rev Genet 2006; 40: 363–383.
Rothkamm K, Kruger I, Thompson LH, Lobrich M . Pathways of DNA double-strand break repair during the mammalian cell cycle. Mol Cell Biol 2003; 23: 5706–5715.
Misteli T, Soutoglou E . The emerging role of nuclear architecture in DNA repair and genome maintenance. Nat Rev Mol Cell Biol 2009; 10: 243–254.
Polo SE, Jackson SP . Dynamics of DNA damage response proteins at DNA breaks: a focus on protein modifications. Genes Dev 2011; 25: 409–433.
Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM . DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem 1998; 273: 5858–5868.
Stucki M, Clapperton JA, Mohammad D, Yaffe MB, Smerdon SJ, Jackson SP et al. MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell 2005; 123: 1213–1226.
Panier S, Durocher D . Regulatory ubiquitylation in response to DNA double-strand breaks. DNA Repair (Amst) 2009; 8: 436–443.
Mailand N, Bekker-Jensen S, Faustrup H, Melander F, Bartek J, Lukas C et al. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell 2007; 131: 887–900.
Huen MS, Grant R, Manke I, Minn K, Yu X, Yaffe MB et al. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell 2007; 131: 901–914.
Bekker-Jensen S, Mailand N . The ubiquitin- and SUMO-dependent signaling response to DNA double-strand breaks. FEBS Lett 2011; 585: 2914–2919.
Bergink S, Jentsch S . Principles of ubiquitin and SUMO modifications in DNA repair. Nature 2009; 458: 461–467.
Galanty Y, Belotserkovskaya R, Coates J, Polo S, Miller KM, Jackson SP . Mammalian SUMO E3-ligases PIAS1 and PIAS4 promote responses to DNA double-strand breaks. Nature 2009; 462: 935–939.
Morris JR, Boutell C, Keppler M, Densham R, Weekes D, Alamshah A et al. The SUMO modification pathway is involved in the BRCA1 response to genotoxic stress. Nature 2009; 462: 886–890.
Sun H, Leverson JD, Hunter T . Conserved function of RNF4 family proteins in eukaryotes: targeting a ubiquitin ligase to SUMOylated proteins. EMBO J 2007; 26: 4102–4112.
Prudden J, Pebernard S, Raffa G, Slavin DA, Perry JJ, Tainer JA et al. SUMO-targeted ubiquitin ligases in genome stability. EMBO J 2007; 26: 4089–4101.
Tatham MH, Geoffroy MC, Shen L, Plechanovova A, Hattersley N, Jaffray EG et al. RNF4 is a poly-SUMO-specific E3 ubiquitin ligase required for arsenic-induced PML degradation. Nat Cell Biol 2008; 10: 538–546.
Kosoy A, Calonge TM, Outwin EA, O’Connell MJ . Fission yeast Rnf4 homologs are required for DNA repair. J Biol Chem 2007; 282: 20388–20394.
Barry KC, Abed M, Kenyagin D, Werwie TR, Boico O, Orian A et al. The Drosophila STUbL protein Degringolade limits HES functions during embryogenesis. Development 2011; 138: 1759–1769.
Hu XV, Rodrigues TM, Tao H, Baker RK, Miraglia L, Orth AP et al. Identification of RING finger protein 4 (RNF4) as a modulator of DNA demethylation through a functional genomics screen. Proc Natl Acad Sci USA 2011; 107: 15087–15092.
Migliorini D, Bogaerts S, Defever D, Vyas R, Denecker G, Radaelli E et al. Cop1 constitutively regulates c-Jun protein stability and functions as a tumor suppressor in mice. J Clin Invest 2011; 121: 1329–1343.
Hayashi S, Lewis P, Pevny L, McMahon AP . Efficient gene modulation in mouse epiblast using a Sox2Cre transgenic mouse strain. Mech Dev 2002; 119 (Suppl 1): S97–S101.
Celeste A, Petersen S, Romanienko PJ, Fernandez-Capetillo O, Chen HT, Sedelnikova OA et al. Genomic instability in mice lacking histone H2AX. Science 2002; 296: 922–927.
Lou Z, Minter-Dykhouse K, Franco S, Gostissa M, Rivera MA, Celeste A et al. MDC1 maintains genomic stability by participating in the amplification of ATM-dependent DNA damage signals. Mol Cell 2006; 21: 187–200.
Kolas NK, Chapman JR, Nakada S, Ylanko J, Chahwan R, Sweeney FD et al. Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase. Science 2007; 318: 1637–1640.
Ward IM, Minn K, van Deursen J, Chen J . p53 Binding protein 53BP1 is required for DNA damage responses and tumor suppression in mice. Mol Cell Biol 2003; 23: 2556–2563.
Bartek J, Bartkova J, Lukas J . DNA damage signalling guards against activated oncogenes and tumour progression. Oncogene 2007; 26: 7773–7779.
Lukas C, Falck J, Bartkova J, Bartek J, Lukas J . Distinct spatiotemporal dynamics of mammalian checkpoint regulators induced by DNA damage. Nat Cell Biol 2003; 5: 255–260.
Yin Y, Seifert A, Chua JS, Maure JF, Golebiowski F, Hay RT et al. SUMO-targeted ubiquitin E3 ligase RNF4 is required for the response of human cells to DNA damage. Genes Dev 2012; 26: 1196–1208.
Galanty Y, Belotserkovskaya R, Coates J, Jackson SP . RNF4, a SUMO-targeted ubiquitin E3 ligase, promotes DNA double-strand break repair. Genes Dev 2012; 26: 1179–1195.
Luo K, Zhang H, Wang L, Yuan J, Lou Z . Sumoylation of MDC1 is important for proper DNA damage response. EMBO J 2012; 31: 3008–3019.
Schimmel J, Larsen KM, Matic I, van Hagen M, Cox J, Mann M et al. The ubiquitin-proteasome system is a key component of the SUMO-2/3 cycle. Mol Cell Proteomics 2008; 7: 2107–2122.
Pierce AJ, Hu P, Han M, Ellis N, Jasin M . Ku DNA end-binding protein modulates homologous repair of double-strand breaks in mammalian cells. Genes Dev 2001; 15: 3237–3242.
Ogiwara H, Ui A, Otsuka A, Satoh H, Yokomi I, Nakajima S et al. Histone acetylation by CBP and p300 at double-strand break sites facilitates SWI/SNF chromatin remodeling and the recruitment of non-homologous end joining factors. Oncogene 2011; 30: 2135–2146.
San Filippo J, Sung P, Klein H . Mechanism of eukaryotic homologous recombination. Annu Rev Biochem 2008; 77: 229–257.
Longhese MP, Bonetti D, Guerini I, Manfrini N, Clerici M . DNA double-strand breaks in meiosis: checking their formation, processing and repair. DNA Repair (Amst) 2009; 8: 1127–1138.
Li L, Halaby MJ, Hakem A, Cardoso R, El Ghamrasni S, Harding S et al. Rnf8 deficiency impairs class switch recombination, spermatogenesis, and genomic integrity and predisposes for cancer. J Exp Med 2010; 207: 983–997.
Bohgaki T, Bohgaki M, Cardoso R, Panier S, Zeegers D, Li L et al. Genomic instability, defective spermatogenesis, immunodeficiency, and cancer in a mouse model of the RIDDLE syndrome. PLoS Genet 2011; 7: e1001381.
Fryrear KA, Guo X, Kerscher O, Semmes OJ . The Sumo-targeted ubiquitin ligase RNF4 regulates the localization and function of the HTLV-1 oncoprotein Tax. Blood 2011; 119: 1173–1181.
Eladad S, Ye TZ, Hu P, Leversha M, Beresten S, Matunis MJ et al. Intra-nuclear trafficking of the BLM helicase to DNA damage-induced foci is regulated by SUMO modification. Hum Mol Genet 2005; 14: 1351–1365.
Dou H, Huang C, Singh M, Carpenter PB, Yeh ET . Regulation of DNA repair through deSUMOylation and SUMOylation of replication protein A complex. Mol Cell 2010; 39: 333–345.
Arnaudeau C, Lundin C, Helleday T . DNA double-strand breaks associated with replication forks are predominantly repaired by homologous recombination involving an exchange mechanism in mammalian cells. J Mol Biol 2001; 307: 1235–1245.
Ouyang KJ, Woo LL, Zhu J, Huo D, Matunis MJ, Ellis NA . SUMO modification regulates BLM and RAD51 interaction at damaged replication forks. PLoS Biol 2009; 7: e1000252.
Acknowledgements
We thank Odessa Van Goethem for excellent technical assistance and Martijn Luijsterburg for help with the UV-A laser micro-irradiation experiments. This work was supported by the Belgian federation against cancer (BFK), the Association for International Cancer Research (AICR), the Flemish Organisation for Scientific Research (FWO ZKC2592-00_WO1), and by the Netherlands Organisation for Scientific Research (NWO).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by G Melino
Supplementary Information accompanies the paper on Cell Death and Differentiation website
Rights and permissions
About this article
Cite this article
Vyas, R., Kumar, R., Clermont, F. et al. RNF4 is required for DNA double-strand break repair in vivo. Cell Death Differ 20, 490–502 (2013). https://doi.org/10.1038/cdd.2012.145
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/cdd.2012.145
Keywords
This article is cited by
-
SENP5 promotes homologous recombination-mediated DNA damage repair in colorectal cancer cells through H2AZ deSUMOylation
Journal of Experimental & Clinical Cancer Research (2023)
-
RNF4 silencing induces cell growth arrest and DNA damage by promoting nuclear targeting of p62 in hepatocellular carcinoma
Oncogene (2022)
-
Crosstalk between SUMOylation and ubiquitylation controls DNA end resection by maintaining MRE11 homeostasis on chromatin
Nature Communications (2022)
-
RNF4~RGMb~BMP6 axis required for osteogenic differentiation and cancer cell survival
Cell Death & Disease (2022)
-
DNA damage-induced degradation of Sp1 promotes cellular senescence
GeroScience (2022)