Abstract
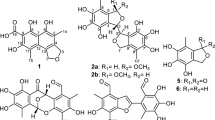
Two novel furaquinocin (FQ) analogues, I (1) and J (2), were isolated from Streptomyces reveromyceticus SN-593 strain NRM2. Their structures were elucidated by MS and NMR analyses. Similar to the previously described FQ D (3), both 1 and 2 possessed a dihydrofuran ring fused to a polyketide naphthoquinone skeleton. The main difference between 1, 2 and 3 was the type of residue attached to C-13; these were a carboxyl, a carboxamide and a methyl residue, respectively.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Bentley, S. D. et al. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417, 141–147 (2002).
Ikeda, H. et al. Complete genome sequence and comparative analysis of the industrial microorganism Streptomyces avermitilis. Nat. Biotech. 21, 526–531 (2003).
Ohnishi, Y. et al. Genome sequence of the streptomycin-producing microorganism Streptomyces griseus IFO 13350. J. Bacteriol. 190, 4050–4060 (2008).
Nogawa, T. et al. Verticilactam, a new macrolactam isolated from a microbial metabolite fraction library. Org. Lett. 12, 4564–4567 (2010).
Cheng, X.- C. et al. A new antibiotic, tautomycin. J. Antibiot. 40, 907–909 (1987).
Takahashi, S. et al. Biochemical characterization of a novel indole prenyltransferase from Streptomyces sp. SN-593. J. Bacteriol. 192, 2839–2851 (2010).
Osada, H., Koshino, H., Isono, K., Takahashi, H. & Kawanishi, G. Reveromycin A, a new antibiotic which inhibits the mitogenic activity of epidermal growth factor. J. Antibiot. 44, 259–261 (1991).
Chen, Y., Wendt-Pienkowski, E. & Shen, B. Identification and utility of FdmR1 as a Streptomyces antibiotic regulatory protein activator for fredericamycin production in Streptomyces griseus ATCC 49344 and heterologous hosts. J. Bacteriol. 190, 5587–5596 (2008).
Komatsu, M., Uchiyama, T., Omura, S., Cane, D. E. & Ikeda, H. Genome-minimized Streptomyces host for the heterologous expression of secondary metabolism. Proc. Natl. Acad. Sci. USA 107, 2646–2651 (2010).
Stutzman-Engwall, K. J., Otten, S. L. & Hutchinson, C. R. Regulation of secondary metabolism in Streptomyces spp. and overproduction of daunorubicin in Streptomyces peucetius. J. Bacteriol. 174, 144–154 (1992).
Komiyama, K. et al. Novel antibioitcs, furaquinocins A and B: taxonomy, fermentation, isolation and physico-chemical and biological characteristics. J. Antibiot. 43, 247–252 (1990).
Ishibashi, M., Funayama, S., Anraku, Y., Komiyama, K. & Omura, S. Novel antibiotics, furaquinocins C, D, E, F, G and H. J. Antibiot. 44, 390–395 (1991).
Funayama, S., Ishibashi, M., Anraku, Y., Komiyama, K. & Omura, S. Structures of novel antibiotics furaquinocins A and B. Tetrahedron Lett. 30, 7427–7430 (1989).
Kawasaki, T. et al. Biosynthesis of a natural polyketide-isoprenoid hybrid compound, furaquinocin A: identification and heterologous expression of the gene cluster. J. Bacteriol. 188, 1236–1244 (2006).
Kumano, T., Tomita, T., Nishiyama, M. & Kuzuyama, T. Functional characterization of the promiscuous prenyltransferase responsible for furaquinocin biosynthesis: identification of a physiological polyketide substrate and its prenylated reaction products. J. Biol. Chem. 285, 39663–39671 (2010).
Jiang, J. et al. Genome mining in Streptomyces avermitilis: a biochemical Baeyer-Villiger reaction and discovery of a new branch of the pentalenolactone family tree. Biochemistry 48, 6431–6440 (2009).
Sambrook, J. & Russell, D. W. Molecular Cloning: A Laboratory Manual. 3rd edn. (Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, 2001).
Kieser, T.,, Bibb, M. J., Buttner, M. J., Chater, K. F. & Hopwood, D. A. Practical Streptomyces Genetics (The John Innes Foundation: Norwich, United Kingdom, 2000).
Onaka, H., Taniguchi, S.-i., Ikeda, H., Igarashi, Y. & Furumai, T. pTOYAMAcos, pTYM18, and pTYM19, actinomycete-Escherichia coli integrating vectors for heterologous gene expression. J. Antibiot. 56, 950–956 (2003).
Takahashi, S. et al. Reveromycin A biosynthesis uses RevG and RevJ for stereospecific spiroacetal formation. Nat. Chem. Biol. (in press).
Muroi, M. et al. Application of proteomic profiling based on 2D-DIGE for classification of compounds according to the mechanism of action. Chem. Biol. 17, 460–470 (2010).
Acknowledgements
We are grateful to Dr Y Futamura for cytoxicity assays. This work was supported in part by a Grant-in-Aid for Creative Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on The Journal of Antibiotics website
Supplementary information
Rights and permissions
About this article
Cite this article
Panthee, S., Takahashi, S., Takagi, H. et al. Furaquinocins I and J: novel polyketide isoprenoid hybrid compounds from Streptomyces reveromyceticus SN-593. J Antibiot 64, 509–513 (2011). https://doi.org/10.1038/ja.2011.41
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ja.2011.41
Keywords
This article is cited by
-
Studies on Streptomyces sp. SN-593: reveromycin biosynthesis, β-carboline biomediator activating LuxR family regulator, and construction of terpenoid biosynthetic platform
The Journal of Antibiotics (2022)
-
β-carboline chemical signals induce reveromycin production through a LuxR family regulator in Streptomyces sp. SN-593
Scientific Reports (2020)
-
β-carboline biomediators induce reveromycin production in Streptomyces sp. SN-593
Scientific Reports (2019)
-
Kinanthraquinone, a new anthraquinone carboxamide isolated from Streptomyces reveromyceticus SN-593-44
The Journal of Antibiotics (2018)
-
Lysobacter species: a potential source of novel antibiotics
Archives of Microbiology (2016)