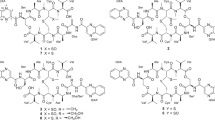
Abstract
Tunicamycins (TUN) are potent inhibitors of polyprenyl phosphate N-acetylhexosamine 1-phosphate transferases (PPHP), including essential eukaryotic GPT enzymes and bacterial HexNAc 1-P translocases. Hence, TUN blocks the formation of eukaryotic N-glycoproteins and the assembly of bacterial call wall polysaccharides. The genetic requirement for TUN production is well-established. Using two genes unique to the TUN pathway (tunB and tunD) as probes we identified four new prospective TUN-producing strains. Chemical analysis showed that one strain, Streptomyces niger NRRL B-3857, produces TUN plus new compounds, named quinovosamycins (QVMs). QVMs are structurally akin to TUN, but uniquely in the 1″,11′-HexNAc sugar head group, which is invariably d-GlcNAc for the known TUN, but is d-QuiNAc for the QVM. Surprisingly, this modification has only a minor effect on either the inhibitory or antimicrobial properties of QVM and TUN. These findings have unexpected consequences for TUN/QVM biosynthesis, and for the specificity of the PPHP enzyme family.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Walsh, C. T. & Wencewicz, T. A. Prospects for new antibiotics: a molecule-centered perspective. J. Antibiot. 67, 7–22 (2014).
Challis, G. L. Mining microbial genomes for new natural products and biosynthetic pathways. Microbiology 154, 1555–1569 (2008).
Doroghazi, J. R. et al. A roadmap for natural product discovery based on large-scale genomics and metabolomics. Nat. Chem. Biol. 10, 963–968 (2014).
Ju, K.-S., Doroghazi, J. R. & Metcalf, W. W. Genomics-enabled discovery of phosphonate natural products and their biosynthetic pathways. J. Ind. Microbiol. Biotechnol. 41, 345–356 (2014).
Wang, H., Fewer, D. P., Holm, L., Rouhiainen, L. & Sivonen, K. Atlas of nonribosomal peptide and polyketide biosynthetic pathways reveals common occurrence of non-modular enzymes. Proc. Natl Acad. Sci. USA 111, 9259–9264 (2014).
Zhang, Q., Doroghazi, J. R., Zhao, X., Walker, M. C. & van der Donk, W. A. Expanded natural product diversity revealed by analysis of lanthipeptide-like gene clusters in Actinobacteria. Appl. Environ. Microbiol. 81, 4339–4350 (2015).
Tamura, G. Tunicamycin, Japan Scientific Societies Press, Tokyo, Japan, (1982).
Eckardt, K. Tunicamycins, streptovirudins, and corynetoxins, a special subclass of nucleoside antibiotics. J. Nat. Prod. 46, 544–550 (1983).
Price, N. P. J. & Tsvetanova, B. C. Biosynthesis of the tunicamycins: a review. J. Antibiot. 60, 485–491 (2007).
Brandish, P. E. et al. Modes of action of tunicamycin, liposidomycin B, and mureidomycin A: inhibition of phospho-N-acetylmuramyl-pentapeptide translocase from Escherichia coli. Antimicrob. Agents Chemother. 40, 1640–1644 (1996).
Xu, L., Appell, M., Kennedy, S., Momany, F. A. & Price, N. P. Conformational analysis of chirally deuterated tunicamycin as an active site probe of UDP-N-acetylhexosamine:polyprenol-P N-acetylhexosamine-1-P translocases. Biochemistry 43, 13248–13255 (2004).
Tsvetanova, B. C., Keimle, D. J. & Price, N. P. J. Biosynthesis of tunicamycin and metabolic origin of the 11-carbon dialdose sugar, tunicamine. J. Biol. Chem. 277, 35289–35296 (2002).
Wyszynski, F. J., Hesketh, A. R., Bibb, M. J. & Davis, B. G. Dissecting tunicamycin biosynthesis by genome mining: cloning and heterologous expression of a minimal gene cluster. Chem. Sci. 1, 581–589 (2010).
Chen, W. et al. Characterization of the tunicamycin gene cluster unveiling unique steps involved in its biosynthesis. Protein Cell 1, 1093–1105 (2010).
Boyle, D. S. & Donachie, W. D. mraY is an essential gene for cell growth in Escherichia coli. J. Bacteriol. 180, 6429–6432 (1998).
Kimura, K.-I. & Bugg, T. D. H. Recent advances in antimicrobial nucleoside antibiotics targeting cell wall biosynthesis. Nat. Prod. Rep. 20, 252–273 (2003).
Dini, C. MraY inhibitors as novel antibacterial agents. Curr. Top. Med. Chem. 5, 1221–1236 (2005).
Doroghazi, J. R. et al. Genome sequences of three tunicamycin-producing streptomyces strains, S. chartreusis NRRL 12338, S. chartreusis NRRL 3882, and S. lysosuperificus ATCC 31396. J. Bacteriol. 193, 7021–7022 (2011).
Kapley, A. et al. Antimicrobial activity of Alcaligenes sp. HPC 1271 against multidrug resistant bacteria. Funct. Integr. Genomics 16, 57–65 (2015).
Wyszynski, F. J. et al. Biosynthesis of the tunicamycin antibiotics proceeds via unique exo-glycal intermediates. Nat. Chem. 4, 539–546 (2012).
Luo, Q., Hiessl, S., Poehlein, A. & Steinbüchel, A. Microbial gutta-percha degradation shares common steps with rubber degradation by Nocardia nova SH22a. Appl. Environ. Microbiol. 79, 1140–1149 (2013).
Tsvetanova, B. C. & Price, N. P. J. Liquid chromatography-electrospray mass spectrometry of tunicamycin-type antibiotics. Anal. Biochem. 289, 147–156 (2001).
Ponpipom, M. M. & Hanessian, S. A method for the selective bromination of primary alcohol groups. Carbohydr. Res. 18, 342–344 (1971).
Hanessian, S., Ponpipom, M. M. & Lavallee, P. Procedures for the direct replacement of primary hydroxyl groups in carbohydrates by halogen. Carbohydr. Res. 24, 45–56 (1972).
Eckardt, K., Ihn, W., Tresselt, D. & Krebs, D. The chemical structures of streptovirudins. J. Antibiot. 34, 1631–1632 (1981).
Li, T., Simonds, L., Kovrigin, E. L. & Noel., K. D. In vitro biosynthesis and chemical identification of UDP-N-acetyl-D-quinovosamine (UDP-D-QuiNAc). J. Biol. Chem. 289, 18110–18120 (2014).
Hwang, S., Aronov, A. & Bar-Peled, M. The biosynthesis of UDP-D-QuiAc in Bacillus cereus ATCC 14579. PLoS ONE 10, e133790 (2015).
Heifetz, A., Keenan, R. W. & Elbein, A. D. Mechanism of action of tunicamycin on the UDP-GlcNAc: dolichyl phosphate GlcNAc-1-phosphate transferase. Biochemistry 18, 2186–2192 (1979).
Price, N. P. & Momany, F. A. Modeling bacterial UDP-HexNAc: polyprenol-P HexNAc-1-P transferases. Glycobiology 15, 29–42 (2005).
Imperiali, B., O’Connor, S. E., Hendrickson, T. & Kellenberger, C. Chemistry and biology of asparagines-linked glycosylation. Pure Appl. Chem. 71, 777–787 (1999).
Lehrman, M. A. Biosynthesis of N-acetylglucosamine-P-P-dolichol, the committed step of asparagine-linked oligosaccharide assembly. Glycobiology 1, 553–623 (1991).
Lehrman, M. A. A family of UDP-GlcNAc/MurNAc: polyisoprenol-P GlcNAc/MurNAc-1-P transferases. Glycobiology 4, 768–771 (1994).
Bugg, T. D. H. & Brandish, P. E. From peptidoglycan to glycoproteins: common features of lipid-linked oligosaccharide biosynthesis. FEMS Microbiol. Lett. 119, 255–262 (1994).
Anderson, M. S., Eveland, S. & Price, N. P. J. Conserved cytoplasmic motifs that distinguish sub-groups of the polyprenol phosphate:N-acetylhexosamine-1-phosphate transferase family. FEMS Microbiol. Lett. 191, 169–175 (2000).
Amer, A. O. & Valvano, M. A. Conserved amino acid residues found in a predicted cytosolic domain of the lipopolysaccharide biosynthetic protein WecA are implicated in the recognition of UDP-N-acetylglucosamine. Microbiology 147, 3015–3025 (2001).
Swoboda, J. G., Campbell, J., Meredith, T. C. & Walker, S. Wall teichoic acid function, biosynthesis, and inhibition. ChemBioChem 11, 35–45 (2010).
Soldo, B., Lazarevic, V. & Karamata, D. tagO is involved in the synthesis of all anionic cell-wall polymers in Bacillus subtilis 168. Microbiology 148, 2079–2087 (2002).
Hancock, I. C., Wiseman, G. & Baddiley, J. Biosynthesis of the unit that links teichoic acid to the bacterial wall: inhibition by tunicamycin. FEBS Lett. 69, 75–80 (1976).
Belanger, M., Burrows, L. L. & Lam, J. S. Functional analysis of genes responsible for the synthesis of the B-band O antigen of Pseudomonas aeruginosa serotype O6 lipopolysaccharide. Microbiology 145, 3505–3521 (1999).
Raymond, C. K. et al. Genetic variation at the O-antigen biosynthetic locus in Pseudomonas aeruginosa. J. Bacteriol. 84, 3614–3622 (2002).
Naumann, T. A. & Price, N. P. J. Truncation of class IV chitinases from Arabidopsis by secreted fungal proteases. Mol. Plant Pathol. 13, 1135–1139 (2012).
Schwarz, F. & Aebi, M. Mechanisms and principles of N-linked protein glycosylation. Curr. Opin. Struct. Biol. 21, 576–582 (2011).
Larkin, A. & Imperiali, B. The expanding horizons of asparagine-linked glycosylation. Biochemistry 50, 4411–4426 (2011).
Chung, B. C. et al. Crystal structure of MraY, an essential membrane enzyme for bacterial cell wall synthesis. Science 341, 1012–1016 (2013).
DiGiandomenico, A. et al. Glycosylation of Pseudomonas aeruginosa 1244 pilin: glycan substrate specificity. Mol. Microbiol. 46, 519–530 (2002).
Dean, C. R. et al. Characterization of the serogroup O11 O-antigen locus of Pseudomonas aeruginosa PA103. J. Bacteriol. 181, 4275–4284 (1999).
Rocchetta, H. L., Burrows, L. L., Pacan, J. C. & Lam, J. S. Three rhamnosyltransferases responsible for assembly of the A-band D-rhamnan polysaccharide in Pseudomonas aeruginosa: a fourth transferase, WbpL, is required for the initiation of both A-band and B-band lipopolysaccharide synthesis. Mol. Microbiol. 28, 1103–1119 (1998).
Jolley, K. A. & Maiden, M. C. J. BIGSdb: Scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics 11, 595 (2010).
Price, N. P. J. Acylic sugar derivatives for GC/MS analysis of 13C-enrichment during carbohydrate metabolism. Anal. Chem. 76, 6566–6574 (2004).
Ciucanu, I. & Kerek, F. A simple and rapid method for the permethylation of carbohydrates. Carbohydr. Res. 131, 209–217 (1984).
Ciucanu, I. & Costello, C. E. Elimination of oxidative degradation during the per-O-methylation of carbohydrates. J. Am. Chem. Soc. 125, 16213–16219 (2003).
Sebban-Kreuzer, C., Deprez-Beauclair, P., Berton, A. & Crenon, I. High-level expression of nonglycosylated human pancreatic lipase-related protein 2 in Pichia pastoris. Protein Expr. Purif. 49, 284–291 (2006).
Acknowledgements
We thank Trina Hartman for technical assistance, Euan Price for help with drafting figures, and Dr Joseph Rich (USDA, NCAUR, Peoria, IL, USA) for pre-review of the manuscript. Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture. USDA is an equal opportunity provider and employer.
Author contributions
DPL, TAN and WWM designed, administered and performed the genomic database and searches. TAN also undertook the protein N-glycosylation assays. KEV provided analysis by NMR. MJB and MAB undertook permethylation and MS analysis. KMB designed and performed the antimicrobial assays. NPJP conceived the experiments, analyzed data and wrote the paper.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on The Journal of Antibiotics website
Supplementary information
Rights and permissions
About this article
Cite this article
Price, N., Labeda, D., Naumann, T. et al. Quinovosamycins: new tunicamycin-type antibiotics in which the α, β-1″,11′-linked N-acetylglucosamine residue is replaced by N-acetylquinovosamine. J Antibiot 69, 637–646 (2016). https://doi.org/10.1038/ja.2016.49
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ja.2016.49
This article is cited by
-
The tunicamycin derivative TunR2 exhibits potent antibiotic properties with low toxicity in an in vivo Mycobacterium marinum-zebrafish TB infection model
The Journal of Antibiotics (2024)
-
Synergistic enhancement of beta-lactam antibiotics by modified tunicamycin analogs TunR1 and TunR2
The Journal of Antibiotics (2019)
-
Tunicamycin: chemical synthesis and biosynthesis
The Journal of Antibiotics (2019)
-
Modified tunicamycins with reduced eukaryotic toxicity that enhance the antibacterial activity of β-lactams
The Journal of Antibiotics (2017)
-
Selective catalytic hydrogenation of the N-acyl and uridyl double bonds in the tunicamycin family of protein N-glycosylation inhibitors
The Journal of Antibiotics (2017)